Abstract
A large number of alterations in genes encoding receptor tyrosine kinase (RTK), namely FLT3, c-KIT, platelet-derived growth factor (PDGF) receptors, fibroblast growth factor (FGF) receptors, and the anaplastic large cell lymphoma kinase (ALK), have been found in hematopoietic malignancies. They have drawn much attention after the development of tyrosine kinase inhibitors. RTK gene alterations include point mutations and gene fusions that result from chromosomal rearrangements. In both cases, they activate the kinase domain in the absence of ligand, producing a permanent signal for cell proliferation. Recently, this simple model has been refined. First, by contrast to wild-type RTK, many mutated RTK do not seem to signal from the plasma membrane, but from various locations inside the cell. Second, their signal transduction properties are altered: the pathways that are crucial for cell transformation, such as signal transducer and activator of transcription (STAT) factors, do not necessarily contribute to the physiologic functions of these receptors. Finally, different mechanisms prevent the termination of the signal, which normally occurs through receptor ubiquitination and degradation. Several mutations inactivating CBL, a key RTK E3 ubiquitin ligase, have been recently described. In this review, we discuss the possible links among RTK trafficking, signaling, and degradation in leukemic cells.
Introduction
Genetic alterations of receptor tyrosine kinase (RTK) in hematopoietic malignancies were first reported in 1994. Golub and colleagues1 found that platelet-derived growth factor receptor β (PDGFRβ) is fused to the TEL transcription factor (now called ETV6) in chronic myelomonocytic leukemia, as a result of a t(5;12) translocation. The same year, Morris et al2 identified a new RTK gene fused to nucleophosmin (NPM) in anaplastic large cell lymphoma. This new receptor was called anaplastic large cell lymphoma kinase (ALK) after the disease name. Later on, point mutations were described in c-KIT and FLT3, receptors that belong to the PDGFR family, which also includes the macrophage colony stimulating factor receptor c-FMS. Since then, more than 100 RTK alterations have been described in leukemia, lymphoma, myeloma, and other clonal hematologic disorders.
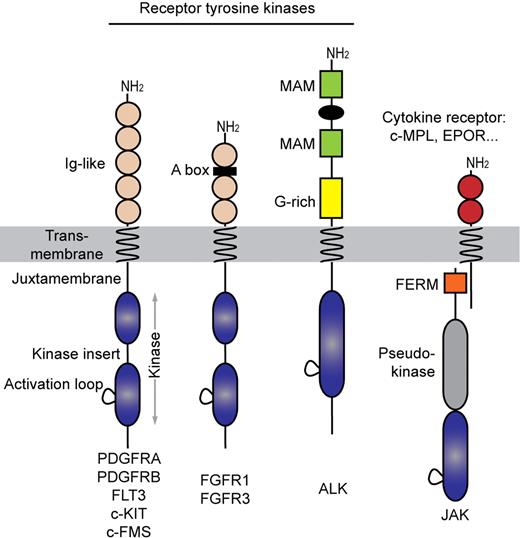
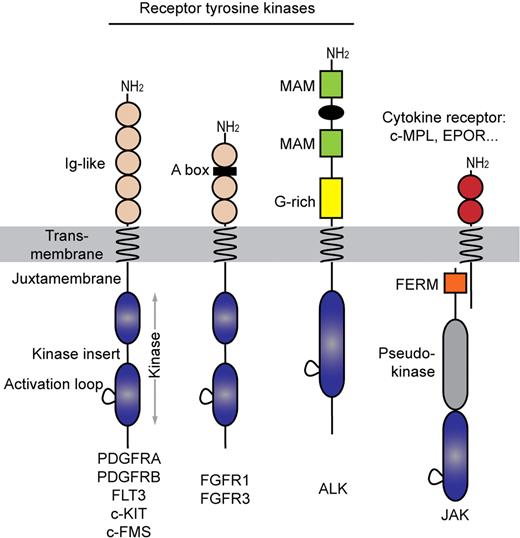
The PDGFR family of receptors, sometimes referred to as type III RTK, shares a common topology consisting of an extracellular ligand-binding domain, a single-spanning transmembrane domain, and an intracellular split kinase domain, which is divided in 2 parts connected by a flexible polypeptide linker, referred to as the kinase insert (Figure 1). These 2 parts will generate a complete functional kinase domain characterized by the typical bilobal folding consisting of the N-terminal and the C-terminal lobe.3 Three regions keep the kinase domain silent in the absence of ligand: the intracellular juxtamembrane domain, the activation loop of the kinase domain and, at least in PDGFRs, the C-terminal tail, which is the least conserved region. Fibroblast growth factor (FGF) receptors show a similar architecture with an additional heparin-binding motif in their extracellular part (Figure 1). By contrast, ALK has a unique extracellular domain among RTKs and an intracellular kinase domain that is reminiscent of the insulin receptor family. To our knowledge, mutations in other RTK families such as epidermal growth factor receptors have not been associated with hematopoietic malignancies. More recently, non-RTK of the Janus kinase family were found mutated in myeloproliferative disorders. Remarkably, the complexes formed by Janus kinases with cytokine receptors are structurally and functionally very similar to RTK (Figure 1).
Receptor tyrosine kinase families involved in hematopoietic malignancies. Acidic box (A box), meprin A5 and mu (MAM) proteins, and glycine-rich (G-rich) indicated.
Receptor tyrosine kinase families involved in hematopoietic malignancies. Acidic box (A box), meprin A5 and mu (MAM) proteins, and glycine-rich (G-rich) indicated.
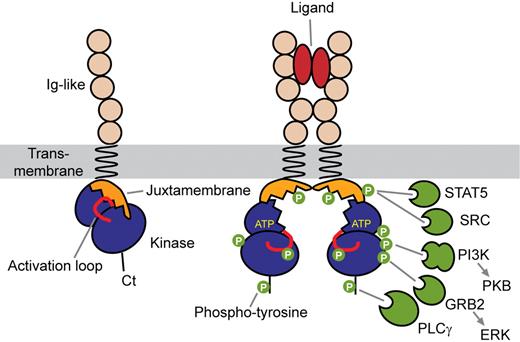
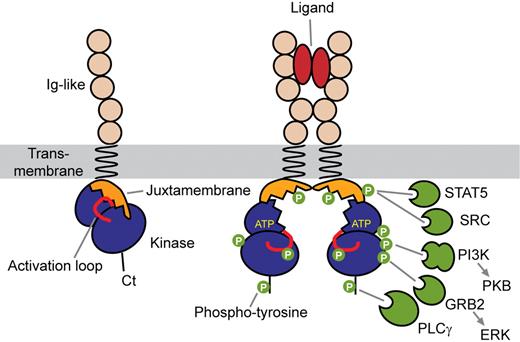
Receptor activation is initiated by ligand-induced receptor dimerization, which brings 2 kinase domains close to each other, allowing the transphosphorylation of critical regulatory tyrosine residues in the activation loop of the catalytic core and in the juxtamembrane domain (Figure 2).3,4 This is followed by transphosphorylation of multiple tyrosine residues of the receptor cytoplasmic part. These phosphotyrosines then act as docking sites for Src homology 2 (SH2) domains of a variety of signal transduction proteins, including phosphatidylinositol 3-kinase (PI3K), phospholipase Cγ (PLCγ), and SRC family kinases, which are shared by type III RTK. A plethora of adaptor molecules containing SH2 domains, such as Grb2, Grb7, Grb10, Shc, Crk, APS, and Nck, provide different connections between receptors and signaling pathways, such as mitogen-activated protein kinases.5-8 These receptors have drawn much attention as they can be efficiently targeted by tyrosine kinase inhibitors, the archetype of which is imatinib mesylate (Glivec, Gleevec; Novartis), which binds to the ATP-binding site of the inactive forms of PDGFRα, PDGFRβ, and c-KIT. The drug was initially designed as a BCR-ABL inhibitor, but PDGFR are approximately 100-fold more sensitive to imatinib than BCR-ABL in vitro.9 New inhibitors are being developed for other RTKs. This topic, which has been extensively discussed (see, eg, Tefferi10 ), will not be the subject of the present review, which will focus on the mechanisms of RTK activation.
Mechanism of type III receptor activation. Ligand binding causes receptor dimerization and phosphorylation of regulatory tyrosine redsidues. Phosphotyrosines then act as docking sites for SH2 domains of a variety of signal transduction proteins.
Mechanism of type III receptor activation. Ligand binding causes receptor dimerization and phosphorylation of regulatory tyrosine redsidues. Phosphotyrosines then act as docking sites for SH2 domains of a variety of signal transduction proteins.
Physiologic functions
FLT3, c-KIT, and c-FMS have been shown to play critical roles in hematopoiesis. FLT3 is predominantly expressed in hematopoietic progenitors and regulates their survival and expansion in synergy with c-KIT. This was demonstrated in FLT3-deficient mice, which have a reduced ability in reconstituting the lymphoid and myeloid lineages after irradiation,11 whereas double-knockout mice for FLT3 and c-KIT display a more severe pancytopenia, indicating that the 2 receptors have partially overlapping roles in hematopoietic stem cell maintenance and differentiation.12 c-KIT expression is down-regulated during cell differentiation with the exception of mast cells that retain a high level of expression.13 c-KIT is also expressed in melanocytes, testis, brain, vascular endothelial cells, interstitial cells of Cajal, breast glandular epithelial cells, and sweat glands. Remarkably, mutated forms of c-KIT were identified in tumors arising from most of these cell types.6 C-FMS can stimulate differentiation and proliferation of macrophages and monocytes and therefore contributes to innate immunity and inflammatory diseases.14,15
Although PDGFRs may be expressed at low levels on some hematopoietic cells,16,17 the receptors do not seem to play an important role in normal hematopoiesis. Knockout mice for PDGF-B or PDGFRβ have hematopoietic defects but these seem to be secondary to the impaired vascularization.18 Similarly, normal hematopoiesis does not require the presence of ALK or FGF receptors.19,20 FGF receptors and PDGFRs are crucial for the proper development of many other organs in the embryo.19,21
Molecular alterations of RTK in hematopoietic neoplasms
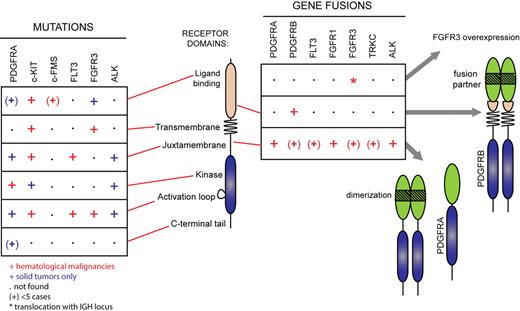
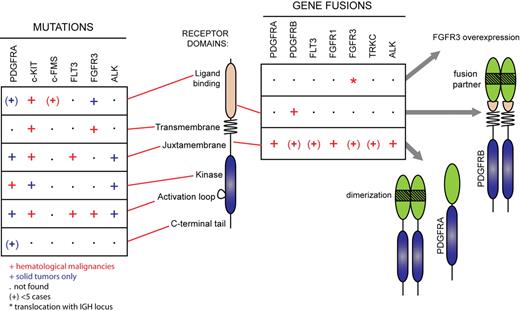
Alterations in the genes encoding c-KIT and FLT3 usually affect only a few amino acids in the sequence, keeping the overall architecture of the receptor intact (Figure 3; supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). These activating mutations fall mainly in 2 hot spots: the juxtamembrane domain and the activation loop. Mutations have also been identified in the other domains at a lower frequency. Although PDGFR, FGF receptor, and ALK mutations similar to those found in c-KIT and FLT3 have been identified in solid tumors, they have not been reported in hematologic disorders so far, most likely because these receptors are poorly expressed in hematopoietic cells. Instead, these receptors are subjected to chromosomal rearrangements that create chimeric oncoproteins. Their expression is always controlled by the promoter of the 5′ partner gene. The importance of the level of expression of RTK for transformation is also illustrated by the t(4;14) translocation in multiple myeloma, leading to the overexpression of wild-type FGF receptor 3 (FGFR3).19
RTK gene alterations in hematopoietic malignancies and other cancer types. Mutations found in hematopoietic malignancies (red symbols) and solid tumors (blue symbols) are shown. The symbol (+) indicates less than 5 reported cases. Asterisk (*) indicates translocation with the IGH locus not affecting the open reading frame.
RTK gene alterations in hematopoietic malignancies and other cancer types. Mutations found in hematopoietic malignancies (red symbols) and solid tumors (blue symbols) are shown. The symbol (+) indicates less than 5 reported cases. Asterisk (*) indicates translocation with the IGH locus not affecting the open reading frame.
Mutations in the extracellular domain
The RTK extracellular domain contains sequences devoted to ligand binding and to receptor dimerization. Insights into the mechanism of ligand-induced activation of RTKs have been provided by the description of the crystal structure of the extracellular domain of c-KIT before and after stimulation by its ligand, stem cell factor (SCF), also called steel factor.4 SCF binds to a concave surface formed by the immunoglobulin (Ig)–like domains 1, 2, and 3 and causes the dimerization of the receptor. SCF-induced dimerization is followed by a large conformational change of the Ig-like domains 4 and 5, which is crucial for receptor activation. The rearrangement of these domains creates interfaces of interaction between 2 adjacent receptor molecules. Point mutations of key amino acids within these interfaces impair the activation of the receptor in cells. Because of the presence of conserved residues in this region, a similar mechanism of activation is predicted for the other members of the family, including PDGFRβ.22
In cancer patients, several gain-of-function mutations have been described in this region of the extracellular domain (Figure 3). These alterations target the Ig-like domains 4 and 5 of c-KIT in gastrointestinal stromal tumors (GIST)23 and in the core binding factor type of acute myeloid leukemia (AML), which features inv(16) or t(8;21). Although this subtype of AML is generally associated with a favorable prognosis, KIT mutations in the extracellular domain confer a greater probability of relapse after remission.24-28 The speculation that these relatively rare mutations lock the receptors in the active conformation awaits experimental confirmation.
Mutations in the transmembrane domain
A few germline mutations within the transmembrane region of c-KIT have been reported in familial mastocytosis (supplemental Table 1).29-31 Some of these mutations only weakly activate c-KIT. For instance, cell lines expressing the M541L receptor showed higher growth rates in the presence of SCF and higher sensitivity to imatinib mesylate but were not transformed. A somatic I530 V mutation was found in a patient with AML associated with inv(16).28 A Y373C mutation in the transmembrane domain of FGFR3 was also found in a myeloma cell line that overexpresses FGFR3 as a result of t(4;14).32 Altogether, these mutations suggest that the transmembrane domain is more than a hydrophobic linker between extracellular and intracellular parts, and can play a regulatory role in receptor activation.
Mutations in the juxtamembrane domain
In the type III RTK family, the juxtamembrane region plays an inhibitory role on the kinase domain.3,33,34 The crystal structure of the autoinhibited form of FLT3 shows that the juxtamembrane domain stabilizes the inactive form of the receptor by contacting the C-terminal lobe of the kinase domain. The conformational changes that are induced by ligand-binding release this inhibition and allow the activation of the kinase activity. FLT3 mutations in the juxtamembrane domain were first identified during a screening for FLT3 expression in mRNA samples from AML patients.35 Elongated fragments of the juxtamembrane domain were detected in the polymerase chain reaction products and sequencing revealed the presence of sequences duplicated in tandem (internal tandem duplication [ITD]) sometimes with the insertion of a few nucleotides. Such mutants are active in a ligand-independent manner. They are found in approximately 30% of all the cases of AML and confer a poor prognosis. Point mutations in FLT3 juxtamembrane domain have also been described.36 A single case involving ITD in c-KIT has been reported for a patient with AML,27 and point mutations were found in systemic mastocytosis,37,38 sinonasal natural killer/T-cell lymphoma,39 in GIST, in seminomas and melanoma (supplemental Table 1). Several activating mutations in the juxtamembrane domain of PDGFRα have also been identified in patients with GIST who responded well to imatinib therapy.
Mutations in the kinase domain, including the activation loop
The activation loop of the kinase domain is located within the C-terminal lobe and contains 1 to 3 tyrosine residues, which can be phosphorylated. In the inactive state, the activation loop assumes a typical closed conformation by folding into the cleft between the N- and C-terminal lobe, thereby blocking access to the substrate pocket and ATP binding site. This conformation is disrupted by phosphorylation of the activation loop, which steps aside and no longer restricts the peptide substrate and ATP from binding.40 This mechanism was validated for most RTK but remains controversial for PDGFRβ.41 The first report of an activating mutation in the activation loop was D816V in c-KIT.37 The alteration was initially identified in a human cell line derived from mast cell leukemia, and was then described in several malignancies like AML, mastocytosis, sinonasal natural killer/T-cell lymphoma, GIST, and seminoma.39,42-48 Different mutations in that loop have now been reported (supplemental Table 1). A study, designed to analyze the analogous position in FLT3, uncovered another group of mutations of this receptor found in approximately 7% of AML cases.49 Similar mutations in the activation loop of PDGFRα, such as D842V, are found in patients with GIST that are negative for c-KIT mutations.50 It is generally believed that activation of these mutated receptors does not require dimerization because the kinase domain never adopts the inactive conformation. For the same reason, these mutations in c-KIT and PDGFRα are often resistant to imatinib mesylate treatment because they force the kinase domain into an active conformation that can no longer bind imatinib.
A few studies described mutations of residues in the first part of the split kinase domain. A report in GIST identified a K642E mutation in c-KIT and demonstrated the ligand-independent activation of the receptor.51 Substitution of asparagine 659 in PDGFRα with lysine or tyrosine was found in less than 1% of GIST. These tumors were considered to have low malignant potential and cells expressing the mutant were shown to be sensitive to imatinib.52,53 A few cases of mutations of the FGFR3 activation loop in myeloma with t(4;14) were also described.19
Mutations in the C-terminal tail
The region located after the kinase domain is highly variable. An oncogenic variant of c-FMS, v-FMS, was identified in the McDonough strain of feline sarcoma virus.54 Compared with wild-type receptor, v-FMS has lost 50 C-terminal residues, which are replaced by 14 unrelated amino acids.55 The deletion causes the loss of tyrosine 969, which negatively regulates the activity of the kinase domain. Deletion of this residue is enough to activate the receptor but additional mutations in the extracellular domain, which are believed to mimic the effects of ligand binding, are required to induce full cell transformation.56 The existence of c-FMS mutations at codons 969 in human cancer has been debated (supplemental Table 1). Several other mutations that have been described in human myeloid malignancies seem to represent rare polymorphisms or nonpathogenic mutations.57 To our knowledge, the existence of acquired transforming mutations of c-FMS has not been confirmed in human neoplasms.
A truncated form of PDGFRα, which has lost 40 C-terminal amino acids, has been found in a case of glioblastoma.58 Although a negative regulatory role for the C-terminal tail of PDGFRα and PDGFRβ has already been suggested,59,60 we did not find evidence that the truncation found in glioblastoma activates the receptor (F.T. and J.-B.D., unpublished observations).
Chromosomal rearrangements
PDGFRA, PDGFRB, and FGFR1 genes are subjected to chromosomal translocations and other rearrangements, producing fusion genes in several related myeloid neoplasms associated with eosinophilia (Figure 3 and supplemental Table 2). ALK fusion, such as NPM-ALK, is observed in a significant proportion of non-Hodgkin lymphoma, particularly anaplastic large cell lymphoma in children and young adults.20
The general structure of a fusion receptor consists of an N-terminal domain derived from a partner protein, the most common being ETV6, and the C-terminal part of the receptor, which includes an intact kinase domain (Figure 3). The expression of these chimeric oncogenes is always controlled by the promoter of the partner gene, allowing the activation of receptors that are poorly expressed in normal hematopoietic cells. These receptors are no longer responsive to the ligand and are constitutively phosphorylated. The paradigm for constitutive activation of fusion receptors is enforced dimerization mediated by specific domains present in the N-terminal portion of the partner protein.19,20,61,62 Most of the fusion partners identified contain self-association domains such as a pointed domain or coiled-coil repeats.
ETV6-PDGFRβ is one of the best characterized fusion RTK.1,61,63-65 Its activation is mediated by the pointed domain (PNT, also known as helix-loop-helix domain, or sterile alpha motif domain) of the transcription repressor TEL (now called ETV6).61 Expression of the hybrid in Ba/F3 cells allows them to grow in the absence of growth factors.61 Furthermore, ETV6-PDGFRβ expression in mice induces a fatal myeloproliferative disorder that resembles the human malignancy except that eosinophilia was absent.66
The most recurrent PDGFR rearrangement is FIP1L1-PDGFRα, which was described in 2003 by Cools et al.9,67 It is found in 10% to 20% of patients with idiopathic hypereosinophilia and in a few cases of systemic mastocytosis.68 The fusion protein is created by an internal deletion of 800 kb on chromosome 4, which is undetectable by classical cytogenetic methods. The FIP1L1 coding sequence, which does not contain any oligomerization motif, is dispensable for Ba/F3 cell transformation.69 However, a study performed in human progenitor cells suggested that it may play a role in signaling.70 The PDGFRα breakpoint is always located within exon 12, which encodes the juxtamembrane domain. The disruption of this region alone is enough to activate the receptor in a ligand-independent manner.69,71 This domain is disrupted in all known PDGFRα fusion proteins, whereas this region is intact in most PDGFRβ hybrid proteins. In addition, PDGFRβ hybrids retain the transmembrane domain (Figure 3), which is required for cell transformation, even though ETV6-PDGFRβ is not a membrane protein.64
Patients harboring PDGFR rearrangements have been diagnosed with chronic myelomonocytic leukemia, juvenile chronic myelomonocytic leukemia, chronic eosinophilic leukemia, atypical chronic myelogenous leukemia. In one case the rearrangement appeared during the relapse phase of a patient with AML. We also reported one case of thrombocythemia.72 Remarkably, most of these patients were males, showed significant eosinophilia and were highly responsive to imatinib therapy.9,73 Relapses are not common but have been reported in few cases because of the acquisition of extra-mutations conferring resistance to the drug, such as the T674I in FIP1L1-PDGFRα.9 Patients with eosinophilia in the absence of PDGFR rearrangement do not usually respond to imatinib therapy, with a few exceptions that might present cryptic PDGFR activation.74 In the new World Health Organization classification, patients harboring rearrangements in PDGFR have been grouped in a single clinical entity referred to as myeloid neoplasms associated with eosinophilia and PDGFR rearrangement.74 A related disorder, the 8p11 myeloproliferative syndrome, which is also associated with eosinophilia, is caused by FGFR1 fusion, such as ZNF198-FGFR1.19
Passenger mutations
Recent studies based on cancer genome sequencing suggested that cancer cells harbor many passenger mutations (also known as bystander mutations), which do not confer any advantage to cancer cells. In these studies, several additional mutations have been described in the PDGFR family.57,75,76 Some mutations were located in mutation hotspots and are likely or proven transforming mutations. However, other somatic mutations were silent at the protein level, raising the possibility that even well-characterized proto-oncogenes such as RTK can contain somatic passenger mutations. Therefore, the impact of newly identified mutations should be systematically tested in cell transformation assays before being considered as therapeutic targets.
Molecular mechanisms of transformation by mutated RTK
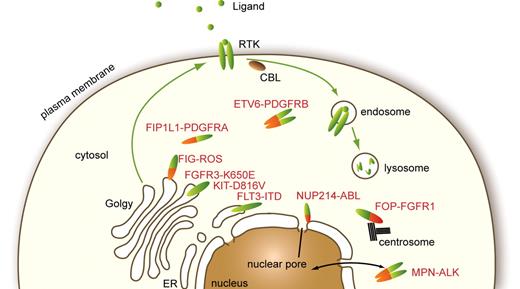
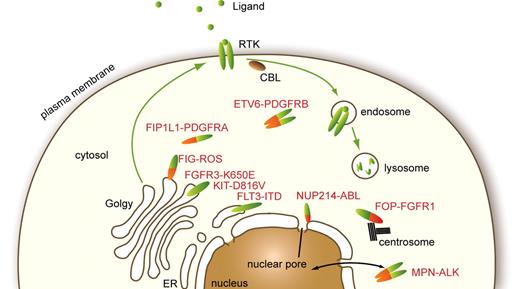
Receptors that are activated by mutations can start signaling as soon as they are synthesized in the endoplasmic reticulum (ER). Studies have shown that premature signaling hinders normal maturation and routing of receptors from the ER through the Golgi to the plasma membrane (Figure 4). Hybrid receptors that have lost the signal peptide and, except in the case of PDGFRβ, the transmembrane domain are excluded from the ER and Golgi. Mislocalization affects the nature of the signaling cascades activated by mutants compared with wild-type receptors and also perturbs their degradation.
Cellular localization of normal and mutated RTK. Physiologic RTK traffic within the cell and reported subcellular localizations of a few oncogenes found in hematopoietic malignancies are shown.
Cellular localization of normal and mutated RTK. Physiologic RTK traffic within the cell and reported subcellular localizations of a few oncogenes found in hematopoietic malignancies are shown.
Altered subcellular localization
First experimental evidence of the importance of the localization of oncogenes on cell transformation has been provided by the work of Xiang et al77 on the D816V c-KIT mutant. They showed that the receptor is mainly localized in the Golgi and only a minor fraction reaches the cell surface. In these experiments, only signaling from the Golgi was essential for cell transformation in vitro and development of myeloproliferative disorder in mice after bone marrow transplantation. Perinuclear localization of c-KIT was also observed in GIST samples from patients harboring c-KIT mutations but could not be detected in patients without KIT mutations. Furthermore, phosphorylation of the immature form of the receptor, which reside in the ER/Golgi compartment, was detected in GIST with c-KIT mutation but was absent in GIST with wild-type c-KIT.78
Similarly, hyperphosphorylated FLT3 ITD mutants undergo aberrant intracellular trafficking and are not properly expressed at cell surface.79,80 The protein is mainly located in the ER in a hypoglycosylated state. Inhibition of its phosphorylation can restore full maturation and surface localization. Furthermore, FLT3 ITD could activate STAT5 when targeted to the ER via an ER retention sequence, indicating that this mutant can transform cells from an intracellular compartment.81 One PDGFRα mutant found in glioblastoma was also shown to localize in the cytoplasm.82
The localization of hybrid RTK is imposed by the partner protein (Figure 4). Many of them, including ALK and PDGFR hybrids, were detected in the cytosol.20,83 The fusion of FOP, a centrosomal protein, to FGFR1 was shown to be located at the centrosome, which was required for activation of PI3K and PLCγ.84 This may apply to a significant number of hybrid RTK as several fusion partners of FGFR1 and PDGFRβ are also centrosomal proteins, including CEP110, NIN, PDE4DIP, PCM1, and TRIP11. NPM-ALK is present both in the cytosol and in the nucleus, but the latter localization is not essential for cell transformation.20 ALK fusion proteins with clathrin and moesin are located close to the plasma membrane, in structures that may play a role in ALK constitutive activation.20,83 FIG-ROS, a hybrid RTK identified in glioblastoma, is targeted to the Golgi apparatus.85 Another striking example of mislocalized tyrosine kinase is the fusion of the nonreceptor kinase ABL1 with the nucleoporin NUP214, identified in patients with T-lineage acute lymphoblastic leukemia. NUP214-ABL1 oligomerizes as a consequence of incorporation into the nuclear pore complex, which is essential for kinase activation, signaling, and transformation.86 It was suggested that this particular localization may account for the differences in signaling and oncogenic activity between NUP214-ABL1 and BCR-ABL1.86 Altogether these reports indicate that aberrant localization is a frequent feature of activated kinase mutants, which was shown in many cases to be critical for cell transformation.
Altered signal transduction
The role of the STAT pathway in leukemia development and in normal hematopoiesis has been well documented during the last few years. STATs are strongly activated by mutant forms of ALK, FLT3, c-KIT, PDGFR, and FGFR.9,63,79,87-94 Activation of STAT3 by c-KIT mutants and ALK hybrids is required to stimulate cell growth.20,95 STAT5 is essential for the proliferation of Ba/F3 cells expressing ZNF198-FGFR191 and human CD34+ cells expressing FIP1L1-PDGFRα.70 In vivo, the onset of a myeloproliferative disease by ETV6-PDGFRβ was much delayed in STAT5A- and STAT5B-deficient mice.63 Mice transplanted with bone marrow transduced with FLT3-ITD mutated at tyrosines 589 and 591, which act as STAT5 docking sites, do not develop a myeloproliferative disorder.96 Thus, transformation of hematopoietic cells by RTK mutants generally requires the activation of STAT3 or STAT5. However, kinase mutations in FLT3 seem to create weaker oncogenes, which produce a reduced STAT5 phosphorylation that is not dependent on tyrosines 589 and 591 in mice.79,97 In patients with AML, these mutations have also been associated with a less aggressive disease compared with FLT3-ITD, as reviewed in detail.98
Surprisingly, although wild-type RTK can activate STAT to some extent, it is usually rather weak and has not been ascribed to any key physiologic function, as STAT-deficient mice phenotypes have not suggested any defect in RTK signaling so far. Several studies have shown that stimulation of wild-type FLT3 does not lead to STAT5 phosphorylation by contrast to FLT3-ITD.79,89,99 Unlike c-KIT-D816 mutants, activation of wild-type c-KIT does not induce STAT3 phosphorylation.95 Similarly, wild-type PDGFRβ only weakly activates STAT5 in hematopoietic cells by contrast to PDGFR hybrids (Wilbanks et al87 and F.T. and J.-B.D., unpublished observations). Altogether, these studies indicate that RTK mutations produce a major switch in signaling regarding the activation of STATs. Choudhary et al100 demonstrated that FLT3-ITD and KIT-ITD aberrantly activates STAT5 from the ER, whereas membrane-targeted FLT3 did not. This effect was enough to support cell survival and growth.81,100 In line with this recent report, Kermorgant and Parker101 have shown that the c-MET receptor is unable to activate STAT3 from the plasma membrane, but can induce nuclear accumulation of this transcription factor only if it is activated from a perinuclear endosomal compartment. The study suggested that the shorter travel distance between the place of activation and the nucleus reduces the probability of STAT3 dephosphorylation by cytosolic phosphatases, allowing the weak STAT3 signal emanating from c-MET to be delivered to the nucleus. Future work will be needed to validate this hypothesis in hematopoietic cells. One of the key target genes of STAT3 and STAT5 in the context of hematopoietic transformation is the Pim kinase, which phosphorylates a set of proteins that overlaps partially with AKT.100
Three major signaling cascades for RTK are PI3K, PLCγ, and mitogen-activated protein kinase. It was shown that FLT3-ITD and KIT-ITD activate these pathways preferentially from the plasma membrane, which is consistent with the localization of several components of these pathways, such as phosphatidylinositols, diacylglycerol, and farnesylated RAS. Activation of the PI3K/AKT pathway is found in many cases of AML included those associated with FLT3-ITD.102 Treatment with PI3K inhibitors, in combination with FLT3 inhibitors, seems to have a synergist role in the treatment of AML.103 Although activation of PI3K may not be essential in ETV6-PDGFRβ–mediated cell transformation, mice reconstituted with tyrosine to phenylalanine mutants of ETV6-PDGFRβ at specific sites responsible for the activation of the PI3K pathway developed a delayed myeloproliferative disease.66 This correlates with the decreased proliferation of ETV6-PDGFRβ–transformed 32D cells upon pharmacologic inhibition of the PI3K pathway.104 The SH2-containing inositol 5-phosphatase-1, which is thought to be a negative regulator of the PI3K, has been found physically associated with the Huntingtin interacting protein 1 (HIP1)-PDGFRβ fusion protein in Ba/F3 cells. Ross and colleagues105 proposed that the interaction of HIP1-PDGFRβ with SH2-containing inositol 5-phosphatase-1 might sequester the latter from its substrates, enhancing phosphatidylinositol-dependent signaling. Downstream PDGFR, PI3K activates AKT (protein kinase B), which modulates several transcription factors such as FOXO, a key cell cycle and apoptosis regulator.94,106
PI3K was shown to be activated in primary GIST cell cultures associated with c-KIT mutations.107 However, PI3K activation was crucial only for the survival of the imatinib-resistant forms of GIST.108 Constitutive association of the regulatory subunit p85 with the D816V c-KIT mutant was demonstrated in immortalized murine progenitor cells.109 In that case, activation of PI3K strongly supported growth factor–independent proliferation of the cells. Mutation of the PI3K binding site in c-KIT-D816V (Y720F) abolished the tumor development in syngenic mice injected with cells expressing the c-KIT-D816V-Y720F mutant, confirming the important role of PI3K activation. As discussed, FOP-FGFR1 is also signaling via PLCγ and PI3K from centrosomes.84 How these enzymes access their lipid substrates from the centrosome is not clear.
This spatial regulation of signaling pathways within cells might also be linked to differences in the pattern of tyrosine phosphorylation of the receptors themselves.110 For instance, FLT3-ITD was shown to be phosphorylated on tyrosine 842 only at the plasma membrane.100 However, the importance of this specific phosphorylation has not been established. Fusion proteins such as of ETV6-PDGFRβ are phosphorylated on additional tyrosine residues present in the fusion partner.111
Another example of altered signaling was recently reported by the Rönnstrand group.112 RTK activates kinases of the SRC family. By contrast to wild-type receptor, c-KIT-D816V does not require SRC activity to sustain proliferation. The study suggested that the D816V mutation alters the kinase substrate specificity, which resembles the SRC members and thus circumvents the need of SRC activation to induce proliferation of cell. This could apply to other kinase domain mutations that are located in the vicinity of the substrate pocket and may affect substrate specificity.
Evasion from receptor down-regulation
Inappropriate RTK activation by loss of negative regulators is increasingly recognized as an alternative mechanism leading to aberrant activity in cells. A major down-regulation pathway involves ligand-induced receptor endocytosis and degradation in lysosomes. This process is initiated upon receptor ubiquination by the E3 ubiquitin ligase CBL, whose substrates include PDGFR, c-FMS, c-KIT, and FLT3.113-118 Inactivating mutations in CBL have been described in several myeloid neoplasms, including atypical CML, chronic myelomonocytic leukemia, myelodysplastic syndrome, and AML.118-121 Most mutations inactivate the CBL RING domain, which is essential for its ubiquitin ligase function. It was shown that expression of mutant CBL together with wild-type FLT3 can transform cells.119,120 It is likely that other RTK expressed in hematopoietic cells, such as c-KIT and c-FMS, also contribute to the effect of mutated CBL. These mutants seem to have a dominant negative effect over wild-type CBL. In addition, the wild-type allele is frequently lost as a result of uniparental disomy. CBL also plays a positive role in RTK signaling as an adaptor protein. This role is retained in CBL mutants.120 As an alternative mechanism to escape CBL-mediated down-regulation, mutations in RTK such as c-MET and c-FMS were shown to prevent CBL recruitment.116,122 Such modifications have not been identified yet in hematologic disorders.
A slightly different case is represented by cytosolic hybrid RTK. We have shown that several fusion proteins, namely FIP1L1-PDGFRα, ETV6-PDGFRβ, and ZNF198-FGFR1, escape ubiquitination and degradation in comparison to the corresponding wild-type receptors, both in cell lines and in leukemia samples.65 In this case, the CBL pathway seems intact but is unable to ubiquitinate the hybrid oncoproteins, perhaps because of their altered cytosolic localization. Restoration of ETV6-PDGFRβ degradation dramatically reduced its ability to stimulate proliferation.65 A few proteins that are components of the endocytic machinery are found as oncogenic fusion proteins together with PDGFRβ (ie, HIP1, Rab5). The resulting receptors are constitutively active because of forced dimerization mediated by the coiled-coil domains in the HIP1 or Rab5 protein. In addition, it is possible that the fusion proteins also interfere with the normal activity of the endocytic machinery and, by altering down regulation of signaling, contribute to cell transformation.
One would expect that the abnormal traffic of RTK harboring activating point mutations may hinder the normal degradation of activated receptors in lysosomes. However, this effect has not been much studied so far.
Conclusions
In this review, we discuss studies showing that mutant RTK acts from unusual subcellular localizations, which may explain at least in part the alterations in signal transduction that have been reported, concerning in particular the activation of STAT transcription factors. Altered localization may also be responsible for the evasion from ubiquitination and degradation, in addition to mutations in key components of the RTK degradation pathway, such as CBL. Altogether, the study of oncogenic RTK variants shed new light on the importance of receptor traffic on signal transduction.
Although the mechanisms by which mutated RTK stimulate proliferation have been largely elucidated, it is still unclear how they affect cell differentiation. Each mutant RTK is indeed associated with a particular type of hematopoietic neoplasm. Although the expression pattern of each receptor influences this process, it is also likely that each RTK can produce a specific signal for differentiation. Further work in more physiologic models, such as purified transduced human stem cells and progenitors will have to tackle this issue.
The discovery and characterization of mutated RTK in leukemia has already led to major advances and new hopes for the treatment of chronic myeloid neoplasms. However, resistance does occur and kinase inhibitors are much less efficient against acute diseases. A better understanding of the mechanism of RTK activation and signaling may lead to the development of new drug families with synergistic effects.
The online version of this article contains a data supplement.
Acknowledgments
We thank Dr Hélène Schoemans for critical reading of the manuscript and the members of the Experimental Medicine Unit for constant support. We apologize to the authors whose excellent work could not be cited due to space limitations.
This work was supported by grants from the Salus Sanguinis foundation and Action de Recherches Concertées (Communauté Française de Belgique).
Authorship
Contribution: F.T. and J.-B.D. wrote the paper together.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jean-Baptiste Demoulin, de Duve Institute, Université Catholique de Louvain, UCL 74.30, ave Hippocrate 75, BE-1200 Brussels, Belgium; e-mail: jb.demoulin@uclouvain.be.