Abstract
Extranodal natural killer/T-cell (ENK/T) lymphoma is a rare neoplasm, subcategorized into ENK/T-nasal (ENK/T-N) and ENK/T-nasal type (ENK/T-NT) lymphomas. ENK/T-NT lymphoma with initial presentation in the skin is known as primary cutaneous ENK/T-NT (PC-ENK/T-NT) lymphoma. The aim of this study was to investigate pathogenesis, genomic alterations, and prognosis of cutaneous ENK/T lymphomas to provide further insights into clinicopathologic features and genetic mechanism of lymphomagenesis. A retrospective case study of 5 white patients affected by ENK/T lymphoma (4 PC-ENK/T-NT and 1 ENK/T-N with cutaneous involvement) was performed. Most of the cases presented with multiple nodules and ulcerations localized on the extremities. A considerable percentage had disease in advanced stage with a 12-month survival rate of 40%. Genomic alterations were detected by array-based comparative genomic hybridization that showed gains of 1q, 7q and loss of 17p in the cases of PC-ENK/T-NT lymphomas and gain of 7q and loss of 9p, 12p, 12q in the case of ENK/T-N lymphoma. In conclusion, ENK/T lymphoma is a very aggressive entity, and, in our cases, the exclusively cutaneous presentation was not associated with a better prognosis. The results of our array comparative genomic hybridization analysis could be useful to better define the different ENK/T lymphoma subgroups with cutaneous involvement.
Introduction
According to the current World Health Organization (WHO) classification of hematolymphoid tumors, extranodal natural killer/T-cell lymphoma (ENK/T), nasal (ENK/T-N), and nasal type (ENK/T-NT) is a clinically aggressive entity, histologically characterized by an Epstein-Barr virus (EBV)–positive atypical lymphoid cytotoxic infiltrate, extensive vascular destruction, and prominent tissue necrosis.
ENK/T-N lymphoma usually involves the upper digestive tract, whereas ENK/T-NT frequently involves the skin, soft tissue, gastrointestinal tract, and testis also.1 ENK/T-NT lymphoma with initial presentation in the skin is known as primary cutaneous ENK/T-NT lymphoma (PC-ENK/T-NT).
ENK/T-NT lymphoma is a rare entity, accounting for less than 2% of non-Hodgkin lymphomas in Europe and North America,2 whereas it occurs with increased frequency in Asian and Central and South American populations (5%-10% of all non-Hodgkin lymphomas).3 PC-ENK/T-NT lymphoma is rarer; from a review of the literature, only approximately 57 cases are reported and less than 20 cases were in Western countries.4-10 However, its real frequency is difficult to establish correctly, because all required data are not always available.
ENK/T lymphomas show short survival time and poor response to therapy; the survival rate at 5 years ranges from 37.9% to 45.3%, even if it has improved in recent years with more intensive therapy.11,12
The common immunophenotype is CD2+, CD56+, surface CD3−, cytoplasmic CD3ϵ+, cytotoxic granules associated proteins (T-cell intracellular antigen-1 [TIA-1], granzyme, and perforin) and EBV-encoded RNA (EBER)–EBV+.13 T-cell receptor (TCR) and immunoglobulin genes are in germline configuration in most cases. In a small proportion of cases, the TCR genes show clonal rearrangement, presumably corresponding to the cases of cytotoxic T-lymphocyte derivation.1
In the current study, 4 cases of PC-ENK/T-NT lymphomas and 1 case of ENK/T-N lymphoma involving the skin were reported and retrospectively studied for the clinicopathologic features of the tumor and survival rate. Array-based comparative genomic hybridization (array CGH) was performed in 3 cases of PC-ENK/T-NT and in 1 case of ENK/T-N lymphoma to improve the knowledge in these particular entities. In fact, little is known about their genomic alterations, and these data could be relevant for a better understanding on genetic mechanism of lymphomagenesis.
Methods
Data from 4 white patients with PC-ENK/T-NT lymphomas and 1 patient with ENK/T-N lymphoma were collected from the cutaneous lymphomas center at the dermatology department of Milan between 2002 and 2009 (Table 1).
All cases started with a clinical presentation of lymphoma in the skin, but in one case the nasal and paranasal cavities were simultaneously involved (case 3). Histologic diagnosis of the tumor was based on the WHO–European Organization for Research and Treatment of Cancer classification of cutaneous lymphomas (2005)14 and the WHO classification for tumors of hematopoietic and lymphoid tissues (2008).1
The criteria for case inclusion were as follows: (1) the availability of all clinical data, (2) examination of histologic features, (3) complete immunophenotype analysis, (4) in situ hybridization for the presence of EBV, and (5) TCR gene rearrangement analysis.
The following clinical data were collected from the patient records: age, sex, clinical course, location and type of lesion; complete blood analysis; serum lactic dehydrogenase (LDH) level, Ann Arbor stage15 ; bone marrow biopsy and aspirate; computed tomographic scan of the thorax, abdomen, head, and neck; the presence of B symptoms; Eastern Cooperative Oncology Group performance score; International Prognostic Index score16 ; type of treatment; follow-up; and survival rate (measured from the date of diagnosis to the date of death). The 24-month survival rate was calculated according to the life table method.
Routine hematoxylin and eosin and Giemsa-stained sections were used to assess histologic features.
The following antibodies were used for immunohistochemistry analysis: cytoplasmic CD3ϵ (polyclonal; Dako), surface CD3 (UCHT1; Dako), CD4 (1F6; Novocastra), CD5 (4C7; Novocastra), CD8 (C8/144B; Dako), CD20 (L26; Dako), CD30 (Ber-H2; Dako), CD45RA(4KB5; Dako), CD45RO (UCHL1; Dako), CD56 (1B6; Novocastra), CD79a (JCB117; Dako), granzyme B (GrB-7; Monosan), TIA-1 (2G9; Immunotech), TdT (TdT-339; Novocastra), Mum-1 (MUM-1p; Dako), TCRβF1 (8A3; Thermo Scientific), TCRδ1 (TS8.24; Endogen), and ki-67 (MIB-1; Dako).
In situ hybridization was carried out with a fluorescein-labeled oligonucleotide probe complementary to 2 EBV-encoded small RNAs, EBER-1 and EBER-2 (Dako).
Array CGH was performed in 4 cases to compare their genomewide genomic alteration patterns. DNA samples were extracted from frozen lymphoma specimens with a standard phenol chloroform method.17 Array CGH protocol was elaborated by Agilent Technologies. The array consisted of 60 base pairs (Agilent 60-mer Whole Human Genome Microarrays) at a 75-kilobase resolution.
Results
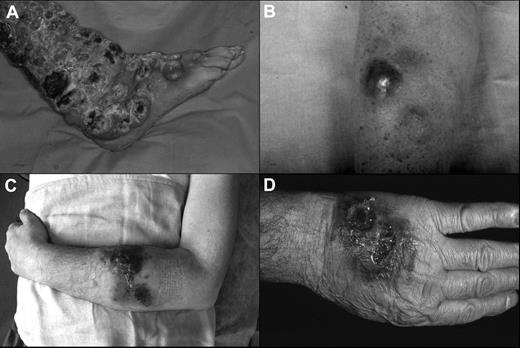
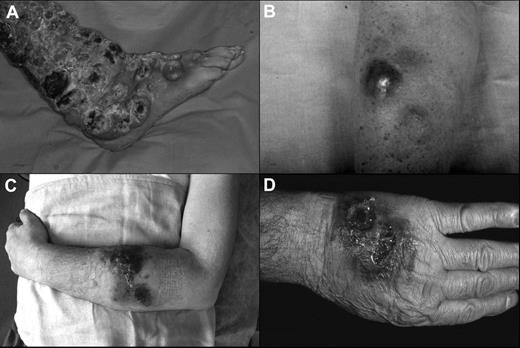
The clinical features of the patients are summarized in Tables 1 and 2. The median age was 60 years (range, 55-74 years) with a male-to-female ratio of 4:1. At the beginning, 4 patients showed exclusively cutaneous presentation (Figure 1A-C), whereas case 3 had both cutaneous (Figure 1D) and nasal-paranasal cavity involvement. The clinical presentation consisted of multiple nodules, confluent and diffused in 4 cases, and accompanied by ulceration in 2 cases. The distribution of the lesions was predominantly on the extremities (100%) and trunk (60%), especially the upper extremities (80%).
Clinical presentation of the patients. (A) Widespread ulcerated nodular lesions over the right leg (case 2); (B) erythematous-violaceous nodule on the arm at the onset (case 4); (C) plaques and nodules with ulceration and hemorrhagic aspects on the left arm (case 5); (D) nodular-ulcerative lesions localized on the left hand (case 3).
Clinical presentation of the patients. (A) Widespread ulcerated nodular lesions over the right leg (case 2); (B) erythematous-violaceous nodule on the arm at the onset (case 4); (C) plaques and nodules with ulceration and hemorrhagic aspects on the left arm (case 5); (D) nodular-ulcerative lesions localized on the left hand (case 3).
Extracutaneous lesions were observed in 2 cases, respectively 7 and 3 months after the onset of skin presentation (see Table 1): one had pharynx and pancreas involvement (case 1), the second had satellite lymph node localization, confirmed by histologic examination (case 5).
Most patients (60%) presented with a bad performance status (Eastern Cooperative Oncology Group performance score ≥ 2). Two patients (40%) were in stage III/IV, and systemic symptoms (B symptoms) such as fever, night sweats, and weight loss occurred in 40% of patients. Furthermore, a decreased white blood cell count and an elevated lactic dehydrogenase level were found in 40% of patients.
All patients received chemotherapy as primary treatment, only in one case was this followed by radiotherapy (case 2). One patient (case 4) was treated with antiviral therapy (butyrate plus ganciclovir for EBV high viral load) in association with chemotherapy without a favorable response. We observed progressive disease in all patients despite treatments. Follow-up data were available for all cases.
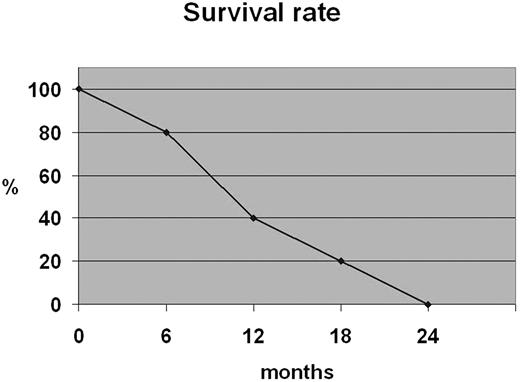
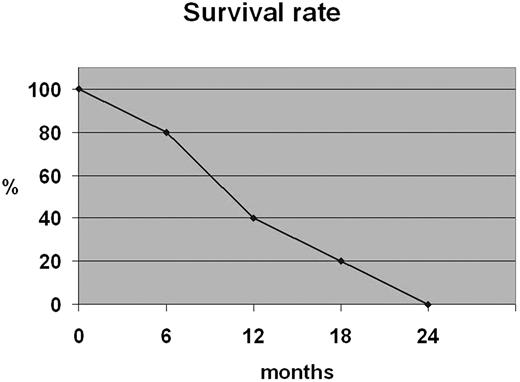
All patients died with a 12-month survival rate of 40% (Figure 2).
Histopathologically, all of the patients showed an atypical tumoral diffuse and angiocentric lymphoid infiltrate involving both the dermis and subcutaneous region, in association with coagulative necrosis in 2 cases. The neoplastic dermal infiltrate was consisted of medium-sized lymphocytes with scattered larger cells (Figure 3A), often accompanied by an inflammatory infiltrate containing histiocytes and neutrophils.
Morphologic and immunohistochemical features of case 5. (A) Histology showing a diffuse dermal infiltrate consisting of atypical medium-sized lymphocytes (hematoxylin and eosin stain; original magnification, ×400); Immunostaining for CD56 (B), granzyme B (C), and EBER signals by in situ hybridization (D; original magnifications ×400 [A,C] and ×200 [B,D]). Nikon Eclipse 50i microscope, objectives: Plan 20×/0.50 WD2.1; Plan 40×/0.75 WD0.72; Nikon DS-Fi1 camera, Nikon NIS-Elements F2.20 image acquisition software. Adobe Photoshop Elements 2.0 was used for image processing.
Morphologic and immunohistochemical features of case 5. (A) Histology showing a diffuse dermal infiltrate consisting of atypical medium-sized lymphocytes (hematoxylin and eosin stain; original magnification, ×400); Immunostaining for CD56 (B), granzyme B (C), and EBER signals by in situ hybridization (D; original magnifications ×400 [A,C] and ×200 [B,D]). Nikon Eclipse 50i microscope, objectives: Plan 20×/0.50 WD2.1; Plan 40×/0.75 WD0.72; Nikon DS-Fi1 camera, Nikon NIS-Elements F2.20 image acquisition software. Adobe Photoshop Elements 2.0 was used for image processing.
Immunohistochemical analysis showed the classical findings reported in literature with positivity of tumor cells of all patients for CD2, CD56 (Figure 3B), cytoplasmic CD3ϵ, granzyme B (Figure 3C), TIA-1, and negativity for surface CD3. CD45RO was expressed in 4 of 5 cases, whereas CD45RA was positive only in case 5. EBER signals were detected in all cases (Figure 3D), and clonal rearrangement of the TCR γ gene was always absent.
On the basis of clinical and histopathologic features, diagnosis of PC-ENK/T-NT lymphoma was made for cases 1, 2, 4, and 5 and of ENK/T-N lymphoma for case 3.
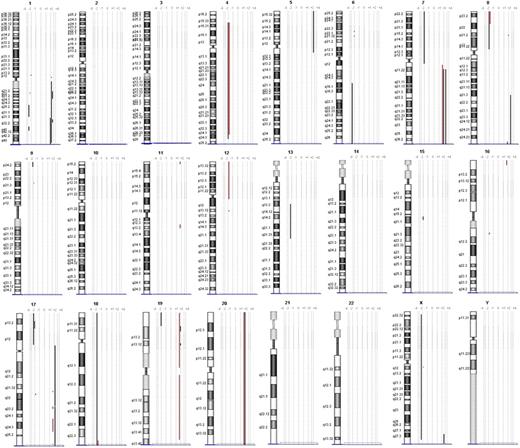
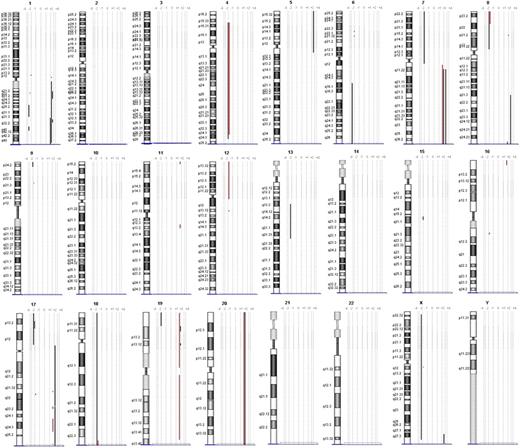
Array CGH was performed to analyze DNA samples of 3 PC-ENK/T-NT lymphoma cases (cases 1, 2, and 4) and 1 ENK/T-N lymphoma case (case 3). Chromosomal ideograms representing regions of gain or loss are summarized in Figure 4.
Chromosomal ideograms of genomic alterations detected by array CGH. Region of loss are indicated by bold lines on the left of each ideogram, and the lines on the right represent regions of gain. Genomic alterations of case 3 (ENK/T-N) are underlined with red.
Chromosomal ideograms of genomic alterations detected by array CGH. Region of loss are indicated by bold lines on the left of each ideogram, and the lines on the right represent regions of gain. Genomic alterations of case 3 (ENK/T-N) are underlined with red.
PC-ENK/T-NT lymphomas showed gain of 1q21.2-q23.3, 1q31.3-q32.1, 1q32.1-q44, 7q11.22-qter and loss of 17p13.2-p13.1 in 2 cases (66%); whereas gain of 1q32.1 was seen in 3 cases (100%). Deletion on chromosome 6 was found only in one case (6q16-qter). In the ENK/T-N case, region of gain was 7q11.22-qter and regions of loss were 9p21.3, 12pter-12p11.1, 12q13.12.
Discussion
ENK/T lymphoma is a rare neoplasm, especially uncommon in Western countries.
We collected data from 5 white patients born in Italy, in whom diagnosis of ENK/T lymphoma was thoroughly proven on histopathologic and immunophenotypical grounds. All cases showed an angiocentric growth pattern infiltrate of pleomorphic tumor cells with cytoplasmic CD3+ and CD56+ phenotype and showed the presence of EBV. Our cases showed similar clinicopathologic aspects to the cases reported in the literature8 : cutaneous onset was characterized by nodules preferentially localized on the extremities, and ulceration was observed in 2 cases.
Advanced stage of disease (III/IV) occurred in a considerable percentage of patients (40%). It was reported that localized cutaneous manifestations are related to a less-aggressive course and a better survival outcome.4,18 However, according to other investigators,1,8,9,19 in our cases the exclusively cutaneous presentation observed in 4 cases was not associated with a better prognosis; all patients died despite the therapy with a 12-month survival rate of 40%. Case 5 had an early lymph node involvement with a very short survival rate. Therefore, invasion of regional lymph nodes could induce an aggressive course as described in similar cases of nodal lymphomas.
From all the analyzed series in the literature, chemotherapy associated with radiotherapy seems to be the most efficacious treatment for ENK/T lymphomas. However, despite this, the prognosis is usually poor, with a median survival time of less than 15 months, confirming the highly aggressiveness of this particular lymphoma.20 Patients with poor prognostic features may be considered for an early autologous hematopoietic stem cell transplantation (HSCT). Allogenic HSCT seems to be an interesting alternative for the potential graft-versus-lymphoma effect. This is particularly relevant, because ENK/T lymphoma cells express EBV antigens that may be alloreactive targets; however, treatment is limited by a high transplantation-related mortality.21,22
Therefore, there are new experimental protocols ongoing that use monoclonal antibodies21,22 and L-asparaginase that appear to be promising and are now being tested in clinical trials.23 Prospective data on larger series of patients treated homogeneously also with innovative approaches are needed, to establish the best treatment for this very aggressive lymphoma.
Few reports in which array CGH was used have been published, although many of those reports did not distinguish between aggressive NK-cell leukemia and ENK/T lymphomas cases. They reported chromosome loss of 1p, 4q, 5g, 6q, 7q, 11q, 12q, 15q, 17q and gain of 2q, 10q, 13q. Several investigators have focused on loss of 6q, a region that includes many tumor suppressor genes.24-26
In our study we analyzed ENK/T genomic alterations with array CGH technique, to better characterize this distinct entity.
Gain of 1q was found in all the PC-ENK/T-NT lymphomas, whereas no gain was observed in the ENK/T-N case. Other interesting differences probably useful to distinguish between PC-ENK/T-NT and ENK/T-N lymphomas are the deletions on 9p, 12p, and 12q, which are present only in the ENK/T-N case. It is of interest that CDKN2A and CDKN2B are located on 9p21.3; these genes are involved in the cell cycle, and their loss is described in tumor formation in a wide range of tissues, particularly in cases of leukemia and lymphoma.27,28
Loss of 6q is a frequent reported genomic alteration in ENK/T lymphomas, but in our study it was present only in one case of PC-ENK/T-NT. However, as previously mentioned by Ko et al,25 the significance of 6q deletion on the pathogenesis remains unclear. Nakashima et al24 found loss of 17p13.1, on which region TP53 is located, in 40% of the aggressive NK-cell leukemia group, but not in the ENK/T lymphomas. Controversially, in other reports, abnormal expression of the p53 protein and mutation of the TP53 gene were detected in a wide range of cases.29 In our study, loss of 17q was found in 2 PC-ENK/T-NT cases (66%) but not in the ENK/T-N case. Thus, TP53 could play a relevant role in the PC-ENK/T-NT lymphomagenesis, even if its deletion is not always reported.
Our results could well provide further insights into the genomic mechanism of lymphomagenesis and a deeper understanding of the clinicopathologic features of ENK/T-N and -NT lymphomas.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank S. Gimelli, BSc, F. Novara, BSc, and Prof O. Zuffardi, MD (Biologia Generale e Genetica Medica, Università di Pavia, Pavia, Italy) for technical contribution and critical evaluation of array CGH results.
Authorship
Contribution: S.R. performed the research, collected and analyzed data, and wrote the paper; E.B. designed and performed research, collected and analyzed data, performed pathologic review, and wrote the paper; V.G. and P.V. collected and analyzed data and wrote the paper; and D.F. and L.V. performed immunohistochemical, molecular biology, and CGH analyses.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sebastiano Recalcati, via Pace 9, 20122 Milan, Italy; e-mail: immunopatologiacutanea@unimi.it.

![Figure 3. Morphologic and immunohistochemical features of case 5. (A) Histology showing a diffuse dermal infiltrate consisting of atypical medium-sized lymphocytes (hematoxylin and eosin stain; original magnification, ×400); Immunostaining for CD56 (B), granzyme B (C), and EBER signals by in situ hybridization (D; original magnifications ×400 [A,C] and ×200 [B,D]). Nikon Eclipse 50i microscope, objectives: Plan 20×/0.50 WD2.1; Plan 40×/0.75 WD0.72; Nikon DS-Fi1 camera, Nikon NIS-Elements F2.20 image acquisition software. Adobe Photoshop Elements 2.0 was used for image processing.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/2/10.1182_blood-2009-11-252957/4/m_zh89991054490003.jpeg?Expires=1767920578&Signature=cyAnykPxXrYvnApibSFAV3HggPJ8FWE4Wd9dEslbZbC~4Hg~LisLPMhR7u6~aGx4bu7G-kV7wuPd8Qu664hlPyggBCpjQxvzSu0gcuyqGMnEn9x27hxNwUiBszoM~c1gWU9ee0Pm9DthtAgxhCmVswxEQhf6WMiT2GfqW9xx~fY6MRruS4DXk7JsqPq8VpPzvH1Qd7N07j-rpZTq7jswVlWc--N1QAlX0BdoGtDZCGU~lzOQYs1MOvvLvmO-Usi1DSp~aI23wmheZ6MsKcQJIwez6ZSbM1RSK7E2V2t9ayP4ZK~BdwSfUPkV0A-IvlSQj-~Z4JMsNU5YR7bsaS1NBQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 3. Morphologic and immunohistochemical features of case 5. (A) Histology showing a diffuse dermal infiltrate consisting of atypical medium-sized lymphocytes (hematoxylin and eosin stain; original magnification, ×400); Immunostaining for CD56 (B), granzyme B (C), and EBER signals by in situ hybridization (D; original magnifications ×400 [A,C] and ×200 [B,D]). Nikon Eclipse 50i microscope, objectives: Plan 20×/0.50 WD2.1; Plan 40×/0.75 WD0.72; Nikon DS-Fi1 camera, Nikon NIS-Elements F2.20 image acquisition software. Adobe Photoshop Elements 2.0 was used for image processing.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/2/10.1182_blood-2009-11-252957/4/m_zh89991054490003.jpeg?Expires=1767920579&Signature=AFuQp6z0270lv6md5dIOo5bMj8kqIm2w2JQ2KWnUUQJhet7c6Hm8kKESEd8i8ijMsNl9sXnENkgdtCZ2OYqxu~sPEn-NGZhyuBgvSM4h4ioXGPBX~xP~eQnw2KX6g~UU8pHJW2yS-jOZ33pAhnPKEdVhqSYTNmx-Yh5VIvQcj8fj9AQ~g6pmyPxkToPSprI466ephqs~jHltL~mnsTC~sOu9gjveLhPE~bDqOM7qI8vAlFdqXSq9XzR-80gmAUYtU4WF8C0XvvZHExiSVmiGVoc3rmy2Ro-p0j~CM0fQ8Id24T6orZP~Pc3gQxENBHoDvOZBj7m-1EjuKGqktdd7XQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)