Abstract 2503
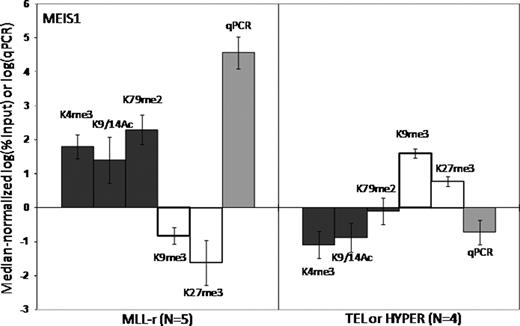
Epigenetic regulation of gene transcription is mediated both by methylation of DNA CpG islands and the local configuration of chromatin, which is dynamically regulated by post-translational modifications, or “marks”, of key lysines (K) of histones (especially H3). Some marks are associated with transcriptional repression [trimethylation (me3) of K9 and K27], and some with activation [me3 of K4, dimethylation (me2) of K79 and acetylation (Ac) of K9 and K14]. The MLL gene encodes a protein that functions as a master regulator of target gene expression by methylating H3K4 via its SET domain, and by interacting with other proteins with histone modifying properties. MLL is frequently rearranged (MLL-r) by translocations in acute leukemias, which exhibit a distinct global gene expression pattern. Many MLL-r partner genes form complexes that can methylate H3K79. Thus, histone modifications may be central to the function of both wild type (MLL-wt) and MLL-r. We hypothesized that aberrant histone coding of target genes contributes to MLL-r leukemogenesis. We characterized the histone code associated with the promoters of selected genes in n=5 MLL-r pre-B ALL samples (MLL-AF4 or MLL-ENL), n=4 MLL-wt pre-B ALL samples (TEL-AML1 or hyperdiploid) and normal control B-precursors (CD19+ cord blood cells). We selected 9 genes differentially overexpressed in MLL-r leukemia (HOXA7, HOXA9, MEIS1, FLT3, CCNA1, ZC3H12C, ATP8B4, C20orf103, and PROM1), and 3 control genes that are not MLL targets (HOXA1, HOXC8, LTF). We performed ChIP with antibodies specific for key H3 modifications (K4me3, K9me3, K9/14Ac, K27me3 and K79me2), followed by qPCR for the selected genes. Expression was measured by RT/qPCR. All 9 MLL target genes were significantly overexpressed in the MLL-r cohort, and this was associated with a specific “activating” histone code at the genes' promoters (fig 1 – MEIS1, e.g.). The opposite “repressive” code was found in the MLL-wt cohort, and in the MLL-r cohort at the promoters of the control genes. Compared to both sets of leukemias, normal B-precursors exhibited a paucity of histone modifications for all genes. For most genes, a specific developmental pattern of alterations in the histone code and corresponding relative change in expression could be traced from the normal B-precursors to the leukemia cells. This pattern was strikingly different in MLL-r leukemias than in MLL-wt leukemias, suggesting that the acquisition of MLL-r by normal B-precursors causes altered gene expression patterns via changes in the histone code. For most genes, normal B-precursors exhibit both the activating K4me3 mark and the repressive K27me3 mark, and express low but detectable levels of RNA. In MLL-r leukemias, upregulation of genes is associated with an increase in K4me3, loss of K27me3, and gain of K9/14Ac and/or K79me2. In MLL-wt leukemias, silencing of genes is associated with loss of K4me3 and gain of K9me3. To study the direct effects of MLL-wt and MLL-r on the histone code, we used 2 rounds of siRNA over 48 hours to knock down MLL-AF4 only, MLL-wt only or both in the RS4;11 cell line (MLL-AF4+ B-precursor ALL), then performed RT/qPCR and ChIP/qPCR. We achieved at least 60% knock down of MLL-AF4 and/or MLL-wt. Knock down of MLL-wt, with or without concomitant knock down of MLL-AF4, did not diminish the K4me3 mark for any genes, suggesting that MLL's SET domain is not required to maintain K4 methylation. While knock down of MLL-AF4 or MLL-wt alone did not diminish K79 methylation, knock down of both completely removed the K79me2 mark from all genes, suggesting that expression of either MLL-wt or MLL-AF4 is absolutely required for H3K79 methyltransferase activity. Two genes (HOXA7 and PROM1) demonstrated evidence of direct transcriptional regulation by MLL-AF4, since their expression decreased markedly after knock down of MLL-AF4 alone or with MLL-wt, but not with MLL-wt alone. In summary, primary MLL-r pre-B ALLs exhibit a distinct activating histone code at key overexpressed target genes when compared to MLL-wt pre-B ALLs and normal B-precursors. A causative role for MLL fusion proteins is suggested by the distinct pattern of histone code progression from normal B-precursors to MLL-r leukemias. Furthermore, knock down experiments provide direct evidence that some of the observed histone modifications in MLL-r leukemia, particularly H3K79 methylation, are directly downstream of wild type and mutant MLL.
Disclosures:
No relevant conflicts of interest to declare.
*
Asterisk with author names denotes non-ASH members.
© 2010 by The American Society of Hematology
2010