Abstract
Abstract 2505
Genetic alterations affecting the B cell transcription factor PAX5 were identified in over 30% of leukemic samples obtained from both pediatric and adult pro-B ALL patients in previous studies using high-resolution single-nucleotide polymorphism (SNP) microarrays. The clinical significance of PAX5 hemizygous deletion has not been fully characterized yet. In order to understand the role of PAX5 alteration in the context of pro-B ALL, we investigated the genetic changes as well as the relevant PAX5 transcript/protein expression in the leukemic cells.
Twenty-four children with pro-B ALL enrolled in Taiwan Pediatric Oncology Group (TPOG) were studied. RNA samples were prepared from the diagnostic bone marrow and PAX5 mRNA expression was analyzed by reverse transcriptase polymerase chain reaction (RT-PCR). The PCR products were subjected to cloning and nucleotide sequence analysis to detect mutations/micro-deletion in the PAX5 coding transcripts. PAX5 protein expression in leukemic cells was analyzed by Western blotting in 6 available samples. Mature B cells purified from PBMCs of healthy subjects were served as control in RT-PCR and Western blotting assays. Variation of copy number in PAX5 gene was analyzed in the above 6 patients. To assess the loss of heterozygosity (LOH) at/near PAX5 locus (PAX5/LOH) in the leukemic cells, length polymorphisms in 5 micro-satellite (STR) markers spanning chromosome 9p were compared between the leukemic cells and respective PBMCs obtained at the remission stage.
PAX5 transcripts were expressed at a reduced level (less than half of that in the normal mature B cells) in 13 of the 24 leukemic samples. While performing RT-PCR spanning exons 6 to 10 of PAX5, PCR products of different size (corresponding to full length, -E8, -E7/8, -E8/9 etc.) were frequently identified in the leukemic samples, compared to primarily full length- and little -E8-transcripts shown in the normal B cells. We did not identify any mutations or micro-deletion in the amplified PAX5 transcripts. In six leukemic samples available for both PAX5 protein Western blotting and DNA PAX5/LOH analysis, three showed a massive reduction in PAX5 protein while another 3 express PAX5 protein comparable to that of the normal mature B cells. The reduced PAX5 protein production correlated to the reduced level of full-length PAX5 transcripts. Three leukemic samples showed LOH at/near PAX5 locus. However, PAX5/LOH did not correlate with the reduced PAX5 mRNA and protein expression in the leukemic cells: 2 leukemic samples with PAX5/LOH showed PAX5 expression comparable to that of the normal mature B cells. Development of non-B-lineage differentiation was demonstrated in a mouse model with a conditionally knocked-out PAX5 in the committed B-cells. Although statistically non-significant, presence of non-B-lineage differentiation markers was more frequently present in the leukemic cells reduced PAX5 expression.
LOH at or near PAX5 locus is common, while point mutation or micro-deletion in PAX5 is rarely found in childhood pro-B ALL. Reduced PAX5 expression (in mRNA and protein) is common in leukemic cells. However, it does not correlate to the PAX5 hemizygous deletion status in the corresponding cells. Mechanisms other than haplo-insufficiency might determine PAX5 expression in the pro-B ALL cells. In leukemic samples, different variant forms at the C-terminal part of PAX5 transcript were frequently identified. The significance of the reduced PAX5 expression and the presence of the variant PAX5 transcripts, in leukemogenesis and the clinical settings in pro-B ALL remained to be studied.
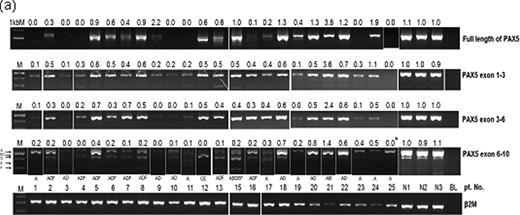
RT-PCR analysis of PAX5 mRNA. (a) The PAX5 expression level was normalized to that of the internal control beta-2 microglobulin. The PAX5 expression is displayed as a relative ratio to the average PAX5 expression in normal mature B cells.
RT-PCR analysis of PAX5 mRNA. (a) The PAX5 expression level was normalized to that of the internal control beta-2 microglobulin. The PAX5 expression is displayed as a relative ratio to the average PAX5 expression in normal mature B cells.
Comparison of DNA monoallelic deletion at PAX5 locus/ PAX5 mRNA expression/PAX5 protein expression in six B-ALL.
| Pt No. . | PAX5 DNA status* . | Relative PAX5 mRNA level . | Relative PAX5 protein level . |
|---|---|---|---|
| 23 | monoallelic deletion | 0.4 | 0.05 |
| 24 | monoallelic deletion | 0.6 | 0.82 |
| 20 | monoallelic deletion | 0.8 | 0.93 |
| 22 | unchanged | 0.6 | 0.21 |
| 25 | non-informative | 0 | 0.08 |
| 21 | non-informative | 1.4 | 0.96 |
| Pt No. . | PAX5 DNA status* . | Relative PAX5 mRNA level . | Relative PAX5 protein level . |
|---|---|---|---|
| 23 | monoallelic deletion | 0.4 | 0.05 |
| 24 | monoallelic deletion | 0.6 | 0.82 |
| 20 | monoallelic deletion | 0.8 | 0.93 |
| 22 | unchanged | 0.6 | 0.21 |
| 25 | non-informative | 0 | 0.08 |
| 21 | non-informative | 1.4 | 0.96 |
unchanged: both normal and leukemic cells showed heterozygous STR markers non-informative: both normal and leukemic cells showed mono-allele in STR markers
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.