Abstract
Clinical application of retinoic acids (RAs) and demethylation agents has proven to be effective in treating certain myeloid leukemia patients. However, the target genes that mediate these antileukemia activities are still poorly understood. In this study, we identified olfactomedin 4 (OLFM4), a myeloid-lineage–specific gene from the olfactomedin family, as a novel target gene for RAs and the demethylation agent, 5-aza-2′-deoxycytidine. We demonstrated that the retinoic acid receptor alpha/retinoic X receptor alpha heterodimer binds to a retinoic acid response-element (DR5) site in the OLFM4 promoter and mediates all-trans-retinoic acid (ATRA)–induced transactivation of the OLFM4 gene. OLFM4 overexpression in HL-60 cells led to growth inhibition, differentiation, and apoptosis, and potentiated ATRA induction of these effects. Conversely, down-regulation of endogenous OLFM4 in acute myeloid leukemia-193 cells compromised ATRA-induced growth inhibition, differentiation, and apoptosis. Overexpression of OLFM4 in HL-60 cells inhibited constitutive and ATRA-induced phosphorylation of the eukaryote initiation factor 4E-binding protein 1 (4E-BP1), whereas down-regulation of OLFM4 protein in acute myeloid leukemia-193 cells increased 4E-BP1 phosphorylation, suggesting that OLFM4 is a potent upstream inhibitor of 4E-BP1 phosphorylation/deactivation. Thus, our study demonstrates that OLFM4 plays an important role in myeloid leukemia cellular functions and induction of OLFM4-mediated effects may contribute to the therapeutic value of ATRA.
Introduction
Acute myeloid leukemia (AML) is a hematologic malignancy characterized by a block of hematopoietic progenitor cell differentiation at the early stages of myelopoiesis, proliferation of immature blasts, and invasion of the bone marrow. Despite our advanced understanding of the molecular mechanisms of myeloid leukemias, current therapeutic approaches to these diseases have not significantly improved, except for acute promyelocytic leukemia (APL). Retinoic acids (RAs) are natural, synthetic derivatives of vitamin A that have been shown to induce cellular differentiation of APL cells in vitro and in vivo.1,2 Treatment of APL with all-trans-retinoic acid (ATRA) has proven to be extremely successful in inducing clinical remission in most patients.2 The demethylation agent, 5-aza-2′-deoxycytidine, has been shown to improve hematopoiesis in leukemia patients.3 In addition, the combination of 5-aza-2′-deoxycytidine, valproic acid (a histone deacetylase inhibitor), and ATRA has been found to have synergistic antileukemia activity.4 However, the molecular mechanisms by which ATRA or other agents regulate terminal myeloid-cell differentiation and apoptosis still remain incompletely understood. Identification of the genes regulated by RA may open up new prospects for understanding the mechanisms of ATRA therapy for myeloid leukemia.
Olfactomedin 4 (OLFM4, also called GW112 and hGC-1) was first identified in human myeloid cells induced by granulocyte colony-stimulating factor (G-CSF).5 OLFM4 is a member of the olfactomedin gene family, whose members share a common C-terminal olfactomedin domain.5 At least 13 olfactomedin domain–containing proteins exist in mammals.6 Recent studies have revealed that olfactomedin-containing proteins play important roles in a variety of cellular functions, including neurogenesis, cell adhesion, cell-cycle regulation, and tumorigenesis, and may serve as modulators of critical signaling pathways.6 The OLFM4 gene encodes a 510-amino-acid N-linked glycoprotein with a multimer structure.7 OLFM4 is constitutively expressed in the bone marrow, gastrointestinal tract, and prostate,5 and its expression is regulated by the myeloid ets transcription factor, PU1,8 and nuclear factor (NF)–κB.9 It has been shown that OLFM4 interacts with GRIM-19 (gene associated with retinoid-interferon–induced mortality-19), which is a component of respiratory complex I of mitochondria,10 and has an antiapoptotic role in prostate cancer cells.11 Recently, OLFM4 overexpression in stomach12-14 and colon cancers15-17 has been reported and associated with clinical outcome. Reduction of OLFM4 protein level has been correlated with poor prognosis for colon cancer patients.16 However, the expression, regulation, and potential biologic functions of OLFM4 in myeloid leukemia still remain unknown.
Methods
Human samples
Genomic DNA and cDNA from peripheral blood leukocytes of AML patients with detailed clinical data and normal individuals were purchased from Capital Biosciences. Peripheral blood samples were collected from patients or normal individuals with informed consent in accordance with the Declaration of Helsinki. The mononuclear cell fraction was separated by sedimentation through Percoll gradients. Red blood cells were lysed in 0.84% ammonium chloride, and the leukocytes were used for the preparation of genomic DNA and cDNA. Peripheral blood mononuclear cells from AML patients and normal individuals, and neutrophils from normal individuals, were obtained commercially from AllCells. The neutrophils were isolated with the EasySep Human Neutrophil Enrichment kit (StemCell Technologies) after removing mononuclear cells with Ficoll-Paque PLUS and red blood cells with HetaSep (StemCell Technologies). The purity of isolated neutrophils was confirmed to be more than 95% by flow cytometry.
Cell transfection
HL-60 cells were transfected using the Cell Line Nucleofector Kit V from Amaxa, according to the manufacturer's instructions, and selected with G418 (500 μg/mL). COS-7 cells were transfected with Lipofectamine 2000 from Invitrogen.
qRT-PCR
Total RNAs were extracted with Trizol (Invitrogen). Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was performed in an Mx 3000P detector from Stratagene, using the following parameters: 50°C (2 minutes), 95°C (2 minutes), followed by 40 cycles of 95°C (15 seconds), and 60°C (1 minute). The primers and probe to amplify OLFM4 were as follows: sense: 5′-AGATCAAAACACCCCTGTC-3′, anti-sense: 5′-CACACCACCATGACCACA-3′, probe (6-carboxy-fluorescein–labeled): 5′-CCACCCTCCTCCCACTCCA-3′. β-actin primers and probe were purchased from Applied Biosystems.
Quantitative methylation analysis
The methylation status of the OLFM4 promoter and exon 1 was analyzed by pyrosequence using Pyro Q-CpG software (EpigenDx) as previously described.16
Luciferase assay
The DNA fragment from −969 to +63 (0 indicates the transcription start site of the OLFM4 gene) was amplified from genomic DNA of HL-60 cells and was cloned into pGL3-Basic Vector (Promega) to create the OLFM4-Luc reporter plasmid. Deletion of RARE sites in the promoter of the OLFM4 gene was performed using the QuikChange Lightning Site-Directed Mutagenesis kit from Stratagene. The luciferase activities were measured with the Dual-Luciferase Reporter Assay system (Promega), according to the manufacturer's instructions.
Electrophoretic mobility-shift assay
Retinoic acid receptor (RAR)–α, retinoic X receptor (RXR)–α, and RXR-β were in vitro–translated using the TNT Quick Coupled Transcription/Translation Systems (Promega) from 1 μg of the corresponding expression vector in a 50-μL final volume. For the electrophoretic mobility-shift assays, annealed oligonucleotides containing the DR5 sequence (5′-GGCTGGCTCACAAGAAGCTCAGGGG-3′; 5′-CCCCTGAGCTTCTTGTGAGCCAGCC-3′) and DR1 sequence (5′-GAAGAAGGCAGAGGTCACAAAGGTG-3′; 5′-CACCTTTGTGACCTCTGCCTTCTTC-3′) were radiolabeled with γ-32P ATP (adenosine triphosphate) using T4 polynucleotide kinase (Promega). Approximately 0.5 ng of the 32P-labeled oligonucleotide probe was added to the reaction mixture containing 2 μL of in vitro–translated protein and incubated at room temperature for 30 minutes. The subsequent gel-shift assays were performed as previously described.9
Western blotting analysis and immunocytochemistry
RNA interference
Lentiviral shRNA against human OLFM4 and control shRNA were purchased from Santa Cruz Biotechnology and transduced into cells according to the manufacturer's instructions. The transduced cells were treated with puromycin (5 μg/mL) for 5 days, then the puromycin-resistant cells were selected.
Statistical analysis
Statistical differences were analyzed with a 2-tail Student t test. A P value < .05 was considered statistically significant.
Sources of reagents and antibodies, protocols for cell culture, and methods for the luciferase reporter methylation assay and the cell proliferation, differentiation, and apoptosis assays are provided in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Results
OLFM4 expression is up-regulated in the leukocytes of a subset of AML patients
Recent studies have shown that OLFM4 is up-regulated in stomach and colon tumor tissues of gastric cancer12-14 and colon cancer15-17 patients, respectively. We wanted to investigate whether OLFM4 gene expression is altered in AML patients. We first examined OLFM4 mRNA and protein expression in neutrophils from the peripheral blood and bone marrow of normal individuals. The expression of OLFM4 mRNA was very low in peripheral blood neutrophils, but high in bone marrow neutrophils, as determined by qRT-PCR (Figure 1A). Similarly, OLFM4 protein expression in the neutrophils from the peripheral blood was remarkably lower than in neutrophils from bone marrow (Figure 1B). OLFM4 protein was found to be present in multiple subcellular compartments of neutrophils, including the cytoplasm, plasma membrane, and mitochondria (Figure 1C).
Up-regulation of OLFM4 in a subset of AML patients. (A) OLFM4 mRNA expression in human bone marrow (BM) and peripheral blood (PB) neutrophils from normal individuals, as determined by qRT-PCR. OLFM4 expression is shown relative to β-actin mRNA expression. *P < .01. (B) OLFM4 protein expression in BM and PB neutrophils from normal individuals, as determined by Western blot analysis. β-actin expression was used as a loading control. (C) OLFM4 protein expression in subcellular fractions of human BM neutrophils from normal individuals, as determined by Western blot analysis. Expressions of Apaf-1 and MnSOD were used to confirm the identity of the cytosolic and mitochondria fractions, respectively. (D) OLFM4 expression in peripheral blood leukocytes from M1 (n = 7), M2 (n = 10), M3 (n = 12), M4 (n = 10), or M5 (n = 3) subtypes of acute myeloid leukemia (AML) patients and normal individuals (N, n = 10) was analyzed by qRT-PCR. The bar represents the mean level of OLFM4 expression relative to β-actin expression. The values in the M4 group is significantly different from each of the other groups using 1-way ANOVA analysis, followed by Bonferroni multiple comparison test; P < .05. (E) Peripheral blood mononuclear cells from a normal individual (N), and 3 AML patients (P1 [M4 subtype], P2 [M3 subtype], and P3 [M1 subtype]) were immunostained with OLFM4 antibody (1:100) and counterstained with Giemsa. Brown color (arrows) indicates the OLFM4-positive cells.
Up-regulation of OLFM4 in a subset of AML patients. (A) OLFM4 mRNA expression in human bone marrow (BM) and peripheral blood (PB) neutrophils from normal individuals, as determined by qRT-PCR. OLFM4 expression is shown relative to β-actin mRNA expression. *P < .01. (B) OLFM4 protein expression in BM and PB neutrophils from normal individuals, as determined by Western blot analysis. β-actin expression was used as a loading control. (C) OLFM4 protein expression in subcellular fractions of human BM neutrophils from normal individuals, as determined by Western blot analysis. Expressions of Apaf-1 and MnSOD were used to confirm the identity of the cytosolic and mitochondria fractions, respectively. (D) OLFM4 expression in peripheral blood leukocytes from M1 (n = 7), M2 (n = 10), M3 (n = 12), M4 (n = 10), or M5 (n = 3) subtypes of acute myeloid leukemia (AML) patients and normal individuals (N, n = 10) was analyzed by qRT-PCR. The bar represents the mean level of OLFM4 expression relative to β-actin expression. The values in the M4 group is significantly different from each of the other groups using 1-way ANOVA analysis, followed by Bonferroni multiple comparison test; P < .05. (E) Peripheral blood mononuclear cells from a normal individual (N), and 3 AML patients (P1 [M4 subtype], P2 [M3 subtype], and P3 [M1 subtype]) were immunostained with OLFM4 antibody (1:100) and counterstained with Giemsa. Brown color (arrows) indicates the OLFM4-positive cells.
Next, we investigated OLFM4 mRNA expression in the peripheral blood leukocytes of 42 AML patients and 10 normal individuals by qRT-PCR and determined the correlation of OLFM4 mRNA expression with AML subtype (Figure 1D). OLFM4 mRNA expression was substantially elevated in 9 cases of the 42 patients (21.4%). Among the 9 patients with elevated OLFM4, 7 were M4 subtype (77.8%) and 2 were M3 subtpye (22.2%; Figure 1D). The OLFM4 expression values in the M4-subtype patients were found to be significantly different from each of the other subtype patients using 1-way analysis of variance (ANOVA) analysis, followed by Bonferroni's multiple comparison tests (P < .05). No significant increase in OLFM4 mRNA expression, compared with that of normal individuals, was observed in peripheral blood leukocytes from M1, M2, or M5 patients (Figure 1D).
Finally, we examined OLFM4 protein expression in peripheral blood mononuclear cells from AML patients and normal individuals. Immunohistochemistry staining demonstrated a stronger, more prevalent OLFM4 expression in an M4-subtype patient (80% cells were positive) and a lack of expression in an M1-subtype patient as well as a normal individual (Figure 1E). Approximately 40% of cells were positively stained in an M3-subtype patient (Figure 1E). Thus, OLFM4 up-regulations appeared to occur preferentially in the M4-subtype of AML patients, suggesting that OLFM4 mRNA expression is correlated with subtypes in AML patients.
OLFM4 expression is negatively regulated by methylation in AML patients
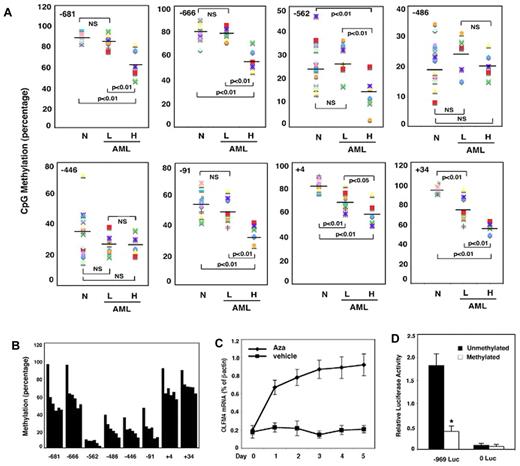
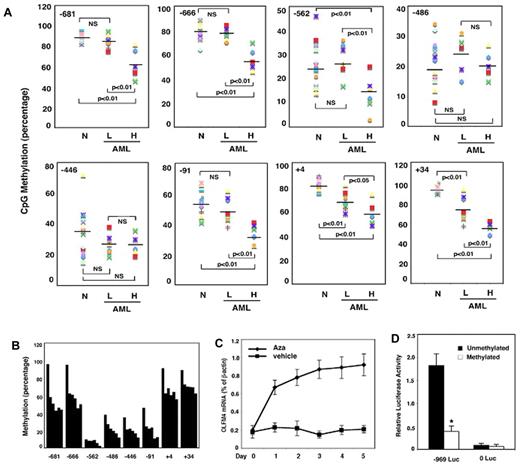
To understand the mechanism of up-regulation of OLFM4 expression in AML patients, we investigated the OLFM4 promoter, CpG, methylation status in AML patients. The methylation analysis was performed on DNA samples obtained from 12 AML patients whose cDNA samples were used for qRT-PCR analysis (Figure 1D). In 6 of the 8 CpG sites we studied (−681, −666, −562, −91, +4, and +34 relative to the OLFM4 transcription start site), the methylation level was significantly lower in the 5 AML patients with higher OLFM4 mRNA expression than in the 7 AML patients with absent or lower OLFM4 mRNA expression and normal individuals (Figure 2A). These results suggest that hypomethylation of some CpG sites in the promoter of the OLFM4 gene could contribute to the enhanced OLFM4 transcription in AML patients.
OLFM4 promoter CpG methylation status in AML patients and effect of 5-aza-2′-deoxycytidine on OLFM4 transcription in HL-60 cells. (A) Methylation status (percentage) of 8 CpG sites (−681, −666, −562, −486, −446, −91, +4, and +34, relative to the OLFM4 transcription start site) in the OLFM4 promoter in the DNA isolated from the peripheral blood leukocytes of AML patients. Methylation levels in AML patients with low OLFM4 mRNA expression (L, n = 7), AML patients with high OLFM4 mRNA expression (H, n = 5), and normal individuals (N, n = 10) were measured with pyrosequence. (B) Methylation (percentage) of the 8 CpG sites in the OLFM4 promoter of HL-60 cells was determined with pyrosequence. Each bar (from left to right) in every CpG site group represents 5-aza-2′-deoxycytidine (5μM) treatment for days 0, 1, 2, 3, 4, and 5. Medium was replaced with new medium with freshly added 5-aza-2′-deoxycytidine every 48 hours. (C) HL-60 cells were treated with 5-aza-2′-deoxycytidine (Aza, 5μM) or vehicle for 5 days. Medium was replaced with new medium with freshly added 5-aza-2′-deoxycytidine or vehicle every 48 hours. OLFM4 expression was determined by qRT-PCR. Values relative to β-actin represent the mean ± SD of 3 experiments performed in triplicate. (D) Promoter activity of the 5′-flanking region of the OLFM4 gene (−969 Luc) in HL-60 cells transfected with OLFM4 promoter-reporter constructs that were treated with or without Sss I CpG methylase. 0 Luc represents parental luciferase reporter construct without the OLFM4 promoter. Data represent the relative activities to the Renilla luciferase activities of phRL-TK, which was transfected together with each OLFM4 plasmid construct. Values represent the mean ± SD of 3 experiments performed in triplicate. *P < .05, compared with unmethylated promoter.
OLFM4 promoter CpG methylation status in AML patients and effect of 5-aza-2′-deoxycytidine on OLFM4 transcription in HL-60 cells. (A) Methylation status (percentage) of 8 CpG sites (−681, −666, −562, −486, −446, −91, +4, and +34, relative to the OLFM4 transcription start site) in the OLFM4 promoter in the DNA isolated from the peripheral blood leukocytes of AML patients. Methylation levels in AML patients with low OLFM4 mRNA expression (L, n = 7), AML patients with high OLFM4 mRNA expression (H, n = 5), and normal individuals (N, n = 10) were measured with pyrosequence. (B) Methylation (percentage) of the 8 CpG sites in the OLFM4 promoter of HL-60 cells was determined with pyrosequence. Each bar (from left to right) in every CpG site group represents 5-aza-2′-deoxycytidine (5μM) treatment for days 0, 1, 2, 3, 4, and 5. Medium was replaced with new medium with freshly added 5-aza-2′-deoxycytidine every 48 hours. (C) HL-60 cells were treated with 5-aza-2′-deoxycytidine (Aza, 5μM) or vehicle for 5 days. Medium was replaced with new medium with freshly added 5-aza-2′-deoxycytidine or vehicle every 48 hours. OLFM4 expression was determined by qRT-PCR. Values relative to β-actin represent the mean ± SD of 3 experiments performed in triplicate. (D) Promoter activity of the 5′-flanking region of the OLFM4 gene (−969 Luc) in HL-60 cells transfected with OLFM4 promoter-reporter constructs that were treated with or without Sss I CpG methylase. 0 Luc represents parental luciferase reporter construct without the OLFM4 promoter. Data represent the relative activities to the Renilla luciferase activities of phRL-TK, which was transfected together with each OLFM4 plasmid construct. Values represent the mean ± SD of 3 experiments performed in triplicate. *P < .05, compared with unmethylated promoter.
To further elucidate the involvement of the CpG methylation status of the OLFM4 promoter in the regulation of OLFM4 expression, we determined OLFM4 mRNA expression in HL-60 cells (derived from an AML patient) treated with 5-aza-2′-deoxycytidine, a demethylating agent, using qRT-PCR. The 5-aza-2′-deoxycytidine effectively demethylated all the 8 CpG sites in the OLFM4 promoter (Figure 2B) in HL-60 cells, which exhibit strong methylation in the CpG sites of the OLFM4 promoter and lack constitutive OLFM4 expression. OLFM4 mRNA expression was observed after treatment of HL-60 cells with 5-aza-2′-deoxycytidine for 1 day and reached a peak at day 5 (Figure 2C).
To analyze the effects of DNA methylation on promoter activity, we used an OLFM4 promoter luciferase construct (−969, relative to the transcription start site) treated with or without Sss I methylase to methylate the promoter in vitro. In vitro–methylated or –unmethylated OLFM4 promoter luciferase reporter constructs were transfected into HL-60 cells, along with a Renilla luciferase expression-control plasmid. OLFM4 promoter activity was strongly repressed by methylation (Figure 2D). Taken together, these data suggest that CpG methylation status in the OLFM4 promoter plays an important role in gene transcription and expression.
A positive (DR5) and a negative (DR1) RARE are located in the OLFM4 promoter
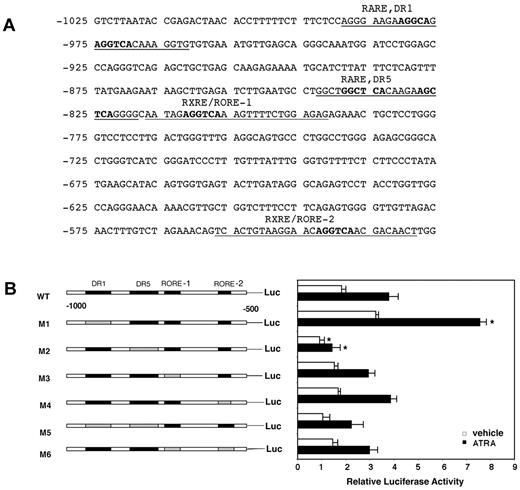
To determine other factors that may regulate OLFM4 gene expression, we examined the promoter region of OLFM4 for putative binding sites using Genomatix v2.0 software (Genomatix). One RA responsive element (RARE)–DR1 site (−989 to −962), 1 RARE-DR5 site (−843 to −819), and 2 retinoid X responsive elements (RXREs) or retinoid-acid receptor-related orphan receptor elements (ROREs; −817 to −797 and −557 to −529) were identified (Figure 3A). We first investigated the potential contribution of these elements to ATRA- mediated OLFM4 transcription in HL-60 cells. We transfected the cells with plasmids containing a deletion of each element (alone or in combination) in a parental reporter construct of the OLFM4 promoter, followed by the luciferase reporter gene, −969 OLFM4-Luc, then exposed the cells to ATRA or vehicle (Figure 3B). OLFM4 transcription activity in HL-60 cells was induced 2.1-fold by treatment with ATRA. In the absence of RARE-DR5, both the basal and ATRA-induced luciferase activities were significantly reduced, compared with the levels observed in cells transfected with the parental OLFM4 promoter construct (Figure 3B). When the RARE-DR1 site was deleted, an increase in basal luciferase activity level was observed, and the OLFM4 transcription-activity response to ATRA was conserved (Figure 3B). Deletion of the 2 RXRE or RORE sites alone or in combination did not change the basal and ATRA-induced OLFM4 transcription levels significantly. Thus, we identified RARE-DR5 as a positive element and RARE-DR1 as a negative element for the regulation of ATRA-induced OLFM4 transcription activity in HL-60 cells.
Identification of a positive and a negative RARE in the OLFM4 promoter. (A) The potential retinoic acid responsive elements (RAREs) and retinoid X responsive elements (RXREs) binding sites identified by Genomatix software in the proximal promoter of the OLFM4 gene are underlined. The bold characters represent nucleotides that were deleted. (B) OLFM4 promoter activity in HL-60 cells transfected with various promoter constructs and then treated with either vehicle or ATRA. Luciferase reporter activities of OLFM4 promoter with serial RARE or RXRE/RORE deletions were analyzed with a dual-reporter system. The black boxes represent RARE or RXRE/RORE binding sites. The gray boxes represent corresponding deleted sites. Values were normalized to Renilla luciferase internal controls. Data represent the mean ± SD of 3 independent experiments performed in triplicates. *P < .05, compared with corresponding wild-type (WT)–Luc control.
Identification of a positive and a negative RARE in the OLFM4 promoter. (A) The potential retinoic acid responsive elements (RAREs) and retinoid X responsive elements (RXREs) binding sites identified by Genomatix software in the proximal promoter of the OLFM4 gene are underlined. The bold characters represent nucleotides that were deleted. (B) OLFM4 promoter activity in HL-60 cells transfected with various promoter constructs and then treated with either vehicle or ATRA. Luciferase reporter activities of OLFM4 promoter with serial RARE or RXRE/RORE deletions were analyzed with a dual-reporter system. The black boxes represent RARE or RXRE/RORE binding sites. The gray boxes represent corresponding deleted sites. Values were normalized to Renilla luciferase internal controls. Data represent the mean ± SD of 3 independent experiments performed in triplicates. *P < .05, compared with corresponding wild-type (WT)–Luc control.
RAs induced OLFM4 expression in HL-60 cells
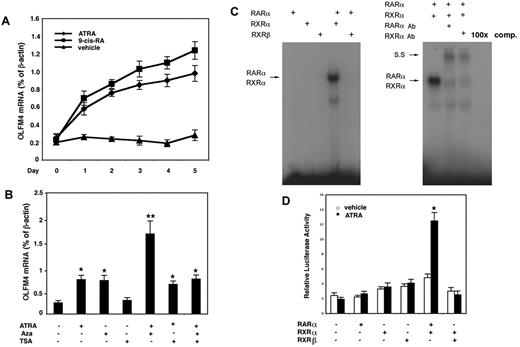
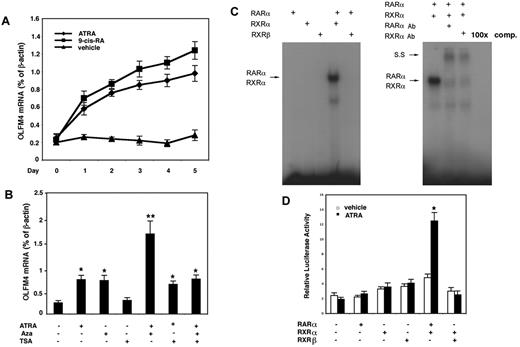
Next, we determined whether RAs could induce OLFM4 mRNA expression in HL-60 cells, which lack constitutive OLFM4 expression.5 Both ATRA and 9-cis-RA induced OLFM4 expression, with 9-cis-RA having a slightly higher induction effect than ATRA (Figure 4A). Because our data to date indicated that OLFM4 could be up-regulated by 2 factors, DNA demethylation and RAs, we next further studied the effect of a combination of the RA, ATRA, with the demethylation agent, 5-aza-2′-deoxycytidine, or trichostatin A (a histone deacetylase inhibitor) on the expression of OLFM4. All 3 of these reagents have been reported to have antileukemia activities,4 and a recent clinical study has shown that the combination of DNA demethylator, histone deacetylase inhibitor, and ATRA was effective in treating myeloid leukemia.4 When we treated HL-60 cells with ATRA and 5-aza-2′-deoxycytidine, we observed a synergistic effect on the induction of OLFM4 gene expression (Figure 4B). No synergistic effect on OLFM4 expression was observed with a combination of ATRA and trichostatin A, and trichostatin A alone did not induce OLFM4 expression (Figure 4B).
ATRA induces OLFM4 expression and transactivation of the OLFM4 promoter by RARα/RXRα heterodimer. (A) HL-60 cells were treated with ATRA (1μM), 9-cis-RA (1μM), or vehicle control for 5 days. Medium was replaced with new medium with freshly added RAs or vehicle every 48 hours. OLFM4 mRNA expression relative to β-actin expression was determined by qRT-PCR. (B) HL-60 cells were treated with ATRA (1μM), 5-aza-2′-deoxycytidine (Aza, 5μM), or trichostatin A (TSA, 1μM) alone or in combination for 2 days. OLFM4 mRNA expression relative to β-actin expression was determined by qRT-PCR. *P < .05, **P < .01, when compared with vehicle treatment. (C) Left panel: in vitro–transcribed and –translated RAR-α, RXR-α, or RXR-β protein alone or in combination was incubated with γ-32P-labeled RARE-DR5 probe of the OLFM4 promoter, then analyzed by electrophoretic mobility-shift assay. Right panel: the in vitro–transcribed and –translated RAR-α and RXR-α protein mixture was incubated with RARE-DR5 probe and subjected to electrophoretic mobility-shift assay. SS indicates supershift band. 100× comp, 100× cold-probe competitions. (D) COS-7 cells were cotransfected for 48 hours with the OLFM4 promoter, luciferase-reporter plasmid (OLFM4-Luc, −959), phRL-TK vector, and expression vectors expressing no cDNA (leftmost set of bars), RAR-α, RXR-α, RXR-β, or combinations of cDNA, as indicated. For the last 24 hours, the cells were treated with 1μM ATRA or vehicle control, then luciferase activities in cell extracts were determined. Values represent the OLFM4 promoter activity relative to TK activity (Renilla luciferase as internal control). Data represent the mean ± SD of 3 independent experiments performed in triplicate. *P < .05, compared with empty vector (no cDNA: leftmost set of bars).
ATRA induces OLFM4 expression and transactivation of the OLFM4 promoter by RARα/RXRα heterodimer. (A) HL-60 cells were treated with ATRA (1μM), 9-cis-RA (1μM), or vehicle control for 5 days. Medium was replaced with new medium with freshly added RAs or vehicle every 48 hours. OLFM4 mRNA expression relative to β-actin expression was determined by qRT-PCR. (B) HL-60 cells were treated with ATRA (1μM), 5-aza-2′-deoxycytidine (Aza, 5μM), or trichostatin A (TSA, 1μM) alone or in combination for 2 days. OLFM4 mRNA expression relative to β-actin expression was determined by qRT-PCR. *P < .05, **P < .01, when compared with vehicle treatment. (C) Left panel: in vitro–transcribed and –translated RAR-α, RXR-α, or RXR-β protein alone or in combination was incubated with γ-32P-labeled RARE-DR5 probe of the OLFM4 promoter, then analyzed by electrophoretic mobility-shift assay. Right panel: the in vitro–transcribed and –translated RAR-α and RXR-α protein mixture was incubated with RARE-DR5 probe and subjected to electrophoretic mobility-shift assay. SS indicates supershift band. 100× comp, 100× cold-probe competitions. (D) COS-7 cells were cotransfected for 48 hours with the OLFM4 promoter, luciferase-reporter plasmid (OLFM4-Luc, −959), phRL-TK vector, and expression vectors expressing no cDNA (leftmost set of bars), RAR-α, RXR-α, RXR-β, or combinations of cDNA, as indicated. For the last 24 hours, the cells were treated with 1μM ATRA or vehicle control, then luciferase activities in cell extracts were determined. Values represent the OLFM4 promoter activity relative to TK activity (Renilla luciferase as internal control). Data represent the mean ± SD of 3 independent experiments performed in triplicate. *P < .05, compared with empty vector (no cDNA: leftmost set of bars).
RAR-α/RXR-α heterodimer transactivates the OLFM4 promoter
RAs exert their effects through their binding and activation of specific nuclear receptors, RARs and RXRs, to promote the formation of heterodimeric RAR/RXR and homodimeric RXR/RXR complexes. Because RAR-α, RXR-α, and RXR-β are major forms of RA receptors in HL-60 cells,18 we tested the ability of RAR-α, RXR-α, and RXR-β to bind the RARE-DR5 in electrophoretic mobility-shift assays using in vitro–transcribed and –translated RAR-α, RXR-α, and RXR-β proteins. In gel-shift assays, the formation of a retardation complex was detected only when the RAR-α and RXR-α combination was incubated with the RARE-DR5 probe (Figure 4C left). The complex's mobility was shifted by anti–RAR-α– or –RXR-α–specific antibodies, and complex formation was eliminated by incubation with 100× cold probe (Figure 4C right). These results demonstrated that the RAR-α/RXR-α heterodimer recognizes the RARE-DR5 motif within the OLFM4 promoter.
To further confirm that ATRA effects on the transcription of the OLFM4 were mediated through the RAR-α/RXR-α heterodimer, we cotransfected in COS-7 cells with the OLFM4 promoter-reporter plasmid (−969-Luc) and expression vectors for RAR-α, RXR-α, or RXR-β alone or in combination. Overexpression of RAR-α and RXR-α in combination resulted in a 6-fold induction of luciferase activity in cells treated with ATRA, whereas this combination weakly stimulated (2-fold) OLFM4 promoter activity in the absence of ATRA treatment (Figure 4D). These results suggest that ATRA induced OLFM4 transcription through a mechanism mediated by the RAR-α/RXR-α heterodimer.
OLFM4 expression induced by ATRA is also mediated by the NF-κB pathway
In addition to binding to RA nuclear receptors, ATRA also can activate other signaling pathways. Our previous study demonstrated that the NF-κB pathway mediated OLFM4 gene expression in HL-60 cells.9 Therefore, we wanted to determine whether NF-κB activation also contributed to OLFM4 expression during ATRA-induced HL-60 cell differentiation. Using specific NF-κB pathway inhibitors, Bay 11-7082 (an inhibitor of IκBα phosphorylation) and sulfasalazine (an inhibitor of IκB kinase), and dominant negative plasmids for IκBα, IKK1, and IKK2, we found that inhibition of the NF-κB pathway significantly suppressed ATRA-induced OLFM4 expression in HL-60 cells (supplemental Figure 1), suggesting that the IKK–NF-κB pathway also contributes to ATRA-induced OLFM4 expression in HL-60 cells.
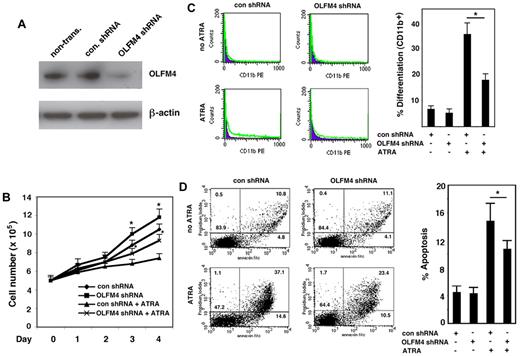
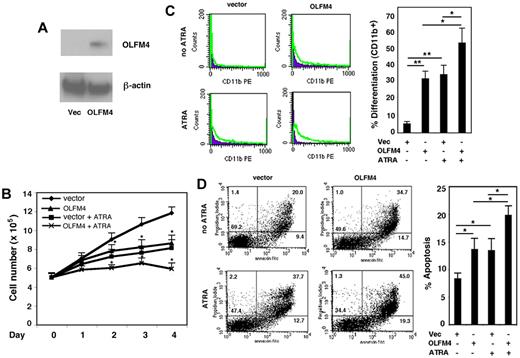
Expression of OLFM4 inhibits cell growth and induces cell differentiation and apoptosis in HL-60 cells
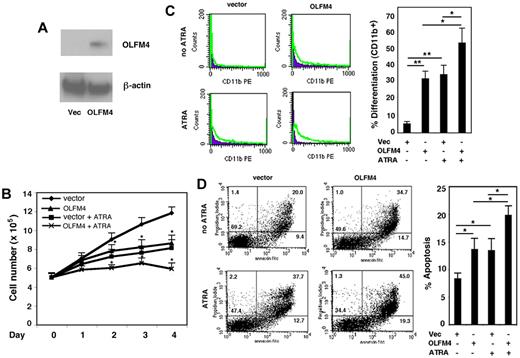
Having established that OLFM4 is a target gene of RAs, we wanted to determine whether OLFM4 is involved in myeloid leukemia cell growth, differentiation, and apoptosis. HL-60 cells were transfected with OLFM4 expression vector or an empty control vector and selected with G418. The expression of OLFM4 protein in these cells was confirmed by Western blotting (Figure 5A). Proliferation of the OLFM4-transfected HL-60 cells was determined on days 1-4. The cell growth of HL-60 cells transfected with OLFM4 was significantly inhibited, compared with that of cells transfected with empty vector (Figure 5B). As expected, treatment of HL-60 cells transfected with empty vector with ATRA inhibited their growth (Figure 5B). When cells transfected with OLFM4 were treated with ATRA, inhibition of cell growth was more significant than that of cells transfected with empty vector and treated with ATRA (Figure 5B).
Effect of OLFM4 overexpression on HL-60 cell growth, differentiation, and apoptosis. (A) The cell lysates of HL-60 cells transfected with either OLFM4 expression vector or empty vector (Vec) were subjected to Western blotting analysis with OLFM4 antibody, then stripped and reprobed with β-actin antibody. (B) HL-60 cells were transfected with either OLFM4 expression plasmid or empty vector, then stably transfected cells were treated with ATRA (1μM) or vehicle for 4 days. Cell numbers were counted with trypan blue exclusion. Data represent the mean ± SD of 3 independent experiments *P < .05, when OLFM4 was compared with vector, and when OLFM4 + ATRA was compared with vector + ATRA. (C) Stably transfected HL-60 cells were treated with ATRA (1μM) or vehicle for 4 days. The expression of CD11b was analyzed by flow cytometry. A representative experiment is shown in the left panel, and the percentage of CD11b+ cells is presented in the right panel. Data represent mean ± SD of 3 independent experiments. *P < .05, **P < .01. (D) Stably transfected HL-60 cells were treated with ATRA (1μM) or vehicle for 4 days. Cell apoptosis was analyzed with annexin V–propidium iodide staining using flow cytometry. A representative analysis data are shown in the left panel, and the percentage of apoptotic cells is shown in the right panel. Data represent the mean ± SD of 3 independent experiments. *P < .05.
Effect of OLFM4 overexpression on HL-60 cell growth, differentiation, and apoptosis. (A) The cell lysates of HL-60 cells transfected with either OLFM4 expression vector or empty vector (Vec) were subjected to Western blotting analysis with OLFM4 antibody, then stripped and reprobed with β-actin antibody. (B) HL-60 cells were transfected with either OLFM4 expression plasmid or empty vector, then stably transfected cells were treated with ATRA (1μM) or vehicle for 4 days. Cell numbers were counted with trypan blue exclusion. Data represent the mean ± SD of 3 independent experiments *P < .05, when OLFM4 was compared with vector, and when OLFM4 + ATRA was compared with vector + ATRA. (C) Stably transfected HL-60 cells were treated with ATRA (1μM) or vehicle for 4 days. The expression of CD11b was analyzed by flow cytometry. A representative experiment is shown in the left panel, and the percentage of CD11b+ cells is presented in the right panel. Data represent mean ± SD of 3 independent experiments. *P < .05, **P < .01. (D) Stably transfected HL-60 cells were treated with ATRA (1μM) or vehicle for 4 days. Cell apoptosis was analyzed with annexin V–propidium iodide staining using flow cytometry. A representative analysis data are shown in the left panel, and the percentage of apoptotic cells is shown in the right panel. Data represent the mean ± SD of 3 independent experiments. *P < .05.
We further investigated whether OLFM4-mediated cell growth inhibition was associated with cell differentiation and apoptosis. The expression of CD11b, a granulocytic differentiation marker, was used to determine cell differentiation in HL60 cells. Expression of OLFM4 in HL-60 cells induced the cell expression of CD11b, similar to ATRA treatment of HL-60 cells (Figure 5C). Expression of CD11b was significantly increased in HL-60 cells transfected with OLFM4 and treated with ATRA, compared with either HL-60 cells transfected with OLFM4 and not treated with ATRA or HL-60 cells transfected with empty vector and treated with ATRA (Figure 5C). The effect of overexpression of OLFM4 on HL-60 cell differentiation was further confirmed by morphologic observation using May-Grunwald-Giemsa staining (supplemental Figure 2). Finally, we analyzed cell apoptosis using annexin V and propidium iodide staining. Expression of OLFM4 induced significantly more cell apoptosis, compared with that seen in control vector-transfected cells (Figure 5D). Treatment of the cells transfected with OLFM4 with ATRA significantly increased apoptosis, compared with OLFM4-transfected cells without ATRA treatment or cells transfected with empty vector and treated with ATRA (Figure 5D). These results suggest that OLFM4 overexpression affects multiple functions of HL-60 cells and could potentiate ATRA-induced cell growth inhibition, differentiation, and apoptosis.
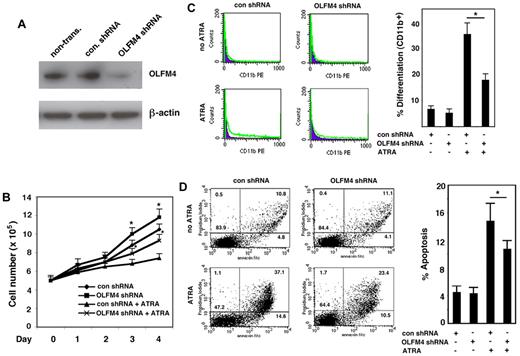
Down-regulation of OLFM4 compromises ATRA-induced cell growth inhibition, differentiation, and apoptosis in AML-193 cells
Next, we wanted to confirm whether OLFM4 mediated ATRA-induced myeloid leukemia cell growth inhibition, differentiation, and apoptosis. We used a lentivirus shRNA approach to knock down OLFM4 expression in AML-193 cells, an AML cell line that has endogenous OLFM4 expression. If OLFM4 were involved in mediating ATRA effects, then suppression of OLFM4 expression in AML-193 cells would be expected to attenuate these effects. For these studies, AML-193 cells transduced with either lentiviral shRNA against OLFM4 or control shRNA were selected with puromycin and then treated with ATRA. Suppression of ATRA-induced OLFM4 expression in OLFM4 shRNA-transduced cells was verified by Western blotting analysis (Figure 6A). Cell growth assays showed that AML-193 cells transduced with OLFM4 shRNA demonstrated significantly enhanced growth compared with cells transduced with control shRNA (Figure 6B). ATRA inhibited the growth of control shRNA-transduced cells, but these inhibitions were compromised in the OLFM4 shRNA-transduced cells treated with ATRA (Figure 6B). When AML-193 cells were treated with ATRA, expression of CD11b in cells transduced with OLFM4 shRNA was significantly decreased compared with cells transduced with control shRNA (Figure 6C). The effect of OLFM4 shRNA on ATRA-induced AML-193 cell differentiation was further confirmed by morphologic observation using May-Grunwald-Giemsa staining (supplemental Figure 3A). In addition, suppression of ATRA-induced OLFM4 expression in HL-60 (supplemental Figure 3B) and CD34+ cells (supplemental Figure 4C) using OLFM4 shRNA also compromised cell differentiation. Finally, when we evaluated ATRA-induced apoptosis in AML-193 cells transduced with OLFM4 shRNA, compared with cells transduced with control shRNA, we found that ATRA-treated cells transduced with OLFM4 shRNA demonstrated significantly decreased apoptosis, compared with ATRA-treated cells transduced with control shRNA (Figure 6D). Our results demonstrated that suppression of OLFM4 expression by OLFM4 shRNA attenuated ATRA-induced AML-193 cell death, including early (lower right quadrant) and late (upper right quadrant) apoptosis (Figure 6D left). These results suggest that OLFM4 has an inhibitory role for AML-193 cell growth, and that OLFM4 mediates ATRA-induced cell growth inhibition, differentiation, and apoptosis in AML-193 cells.
Effect of OLFM4 shRNA on ATRA-mediated AML-193 cell growth, differentiation, and apoptosis. (A) AML-193 cells were transduced with lentiviral OLFM4 shRNA or control shRNA, and puromycin-resistant cell populations were selected. Blots of cell lysates were subjected to Western blotting with OLFM4 antibody, then stripped and reprobed with β-actin antibody. (B) Puromycin-enriched AML-193 cells transduced with OLFM4 shRNA or control shRNA were treated with ATRA (1μM) or vehicle for 4 days. Cell numbers were counted with trypan blue exclusion. Data represent the mean ± SD of 3 independent experiments. *P < .05, when OLFM4 shRNA was compared with control shRNA, and when OLFM4 shRNA + ATRA was compared with control shRNA + ATRA. (C) Puromycin-resistant AML-193 cells transduced with OLFM4 shRNA or control shRNA were treated with ATRA (1μM) or vehicle for 4 days. The expression of CD11b was analyzed by flow cytometry. A representative experiment is shown in the left panel, and the percentage of CD11b+ cells is presented in the right panel. Data represent the mean ± SD of 3 independent experiments. *P < .05. (D) Puromycin-resistant AML-193 cells transduced with OLFM4 shRNA or control shRNA were treated with ATRA (1μM) or vehicle for 4 days. Cell apoptosis was analyzed with annexin V–propidium iodide staining using flow cytometry. A representative analysis is shown in the left panel, and the percentage of apoptotic cells is shown in the right panel. Data represent the mean ± SD of 3 independent experiments. *P < .05.
Effect of OLFM4 shRNA on ATRA-mediated AML-193 cell growth, differentiation, and apoptosis. (A) AML-193 cells were transduced with lentiviral OLFM4 shRNA or control shRNA, and puromycin-resistant cell populations were selected. Blots of cell lysates were subjected to Western blotting with OLFM4 antibody, then stripped and reprobed with β-actin antibody. (B) Puromycin-enriched AML-193 cells transduced with OLFM4 shRNA or control shRNA were treated with ATRA (1μM) or vehicle for 4 days. Cell numbers were counted with trypan blue exclusion. Data represent the mean ± SD of 3 independent experiments. *P < .05, when OLFM4 shRNA was compared with control shRNA, and when OLFM4 shRNA + ATRA was compared with control shRNA + ATRA. (C) Puromycin-resistant AML-193 cells transduced with OLFM4 shRNA or control shRNA were treated with ATRA (1μM) or vehicle for 4 days. The expression of CD11b was analyzed by flow cytometry. A representative experiment is shown in the left panel, and the percentage of CD11b+ cells is presented in the right panel. Data represent the mean ± SD of 3 independent experiments. *P < .05. (D) Puromycin-resistant AML-193 cells transduced with OLFM4 shRNA or control shRNA were treated with ATRA (1μM) or vehicle for 4 days. Cell apoptosis was analyzed with annexin V–propidium iodide staining using flow cytometry. A representative analysis is shown in the left panel, and the percentage of apoptotic cells is shown in the right panel. Data represent the mean ± SD of 3 independent experiments. *P < .05.
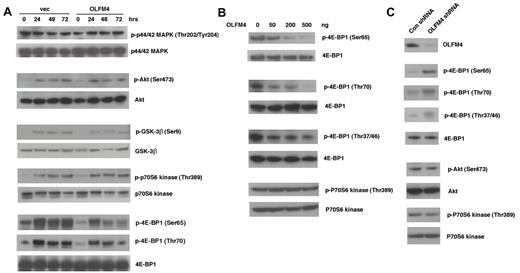
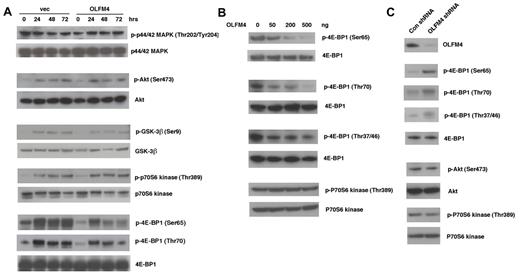
OLFM4 is associated with phosphorylation of the translation repressor 4E-BP1
To further investigate the molecular mechanism that mediates the effect of OLFM4 on leukemia cells, we examined whether OLFM4 could affect several important signal transduction pathways for cell proliferation, differentiation, and apoptosis. Overexpression of OLFM4 in HL-60 cells did not change constitutive or ATRA-induced activation of p44/42 mitogen-activated protein kinase (MAPK), Akt, glycogen synthase kinase 3-beta (GSK-3-β), or p70S6 kinase, but markedly reduced the phosphorylation of 4E-BP1 (Ser65 and Thr70), a translation repressor downstream of the mammalian target of rapamycin (mTOR; Figure 7A). To further confirm the effect of OLFM4 on 4E-BP1 phosphorylation, we cotransfected human embryonic kidney (HEK)–293T cells with a 4E-BP1 expression plasmid and varying amounts of OLFM4 expression plasmid. OLFM4 inhibited phosphorylation of 4E-BP1 (Ser65 and Thr70) in a dose-dependent manner (Figure 7B). OLFM4 also inhibited phosphorylation of 4E-BP1 (Thr37/46), but not in a dose-dependent manner (Figure 7B). Cotransfection of OLFM4 and p70S6 kinase expression plasmids into HEK-293T cells did not affect the phosphorlylation of p70S6 kinase (Figure 7B). Transduction of lentiviral shRNA against OLFM4 into AML-193 cells enhanced the phosphorylation of 4E-BP1 at all phosphorylation sites (Ser65, Thr70, and Thr37/46), compared with phosphorylation levels after transduction with control shRNA (Figure 7C). However, no difference was observed in the phosphorylation levels of Akt and p70S6 kinase in cells transduced with OLFM4 shRNA, compared with levels after transduction with control shRNA. To determine whether OLFM4 directly associates with 4E-BP1, we contransfected HEK-293T cells with OLFM4 and 4E-BP1 expression plasmids and performed coimmunoprecipitation assays. No coimmunoprecipitation between OLFM4 and 4E-BP1 was detected (data not shown). Collectively, these results indicate that OLFM4 acts as an upstream inhibitor of phosphorylation of 4E-BP1, but it does not affect the protein level of 4E-BP1. OLFM4 overexpression or knockdown did not affect phosphorylation of Akt and p70S6 kinase (Figure 7A-C), suggesting that OLFM4 inhibition of 4E-BP1 phosphorylation occurs downstream of Akt and mTOR.
OLFM4 inhibits 4E-BP1 phosphorylation. (A) HL-60 cells were transfected with OLFM4 expression plasmid (OLFM4) or empty vector (vec) and selected by G418. G418-resistant HL-60 cells were treated with ATRA (1μM) for 3 days. Total cell lysates were subjected to Western blotting analysis with antibodies for phospho-p44/42 MAPK (Thr202/Tyr204), phospho-Akt (Ser473), phospho-GSK3β (Ser9), phopho-p70S6 kinase (Thr389), and phospho-4E-BP1 (Ser65 or Thr70). The blots were stripped and reprobed with corresponding antibodies for p44/42 MAPK, Akt, GSK-3β, p70S6 kinase, and 4E-BP1. (B) HEK-293T cells were cotransfected with 4E-BP1 or p70S6 kinase expression plasmid together with OLFM4 expression plasmid in different amounts, as indicated. After 24 hours, total cell lysates were subjected to Western blotting analysis with antibodies for phospho-4E-BP1 (Ser65, Thr70, or Thr37/46) or phospho-p70S6 kinase (Thr389). The blots were stripped and reprobed with total anti–4E-BP1 or anti-p70S6 kinase. (C) AML-193 cells were transduced with lentiviral shRNA against OLFM4 or control shRNA. After 48 hours, total cell lysates were subjected to Western blotting analysis with antibodies for OLFM4, phospho-4E-BP1 (Ser65, Thr70, or Thr37/46), phospho-Akt (Ser473), or phospho-p70S6 kinase (Thr389). The blots were stripped and reprobed with corresponding 4E-BP1, Akt, and p70S6 kinase antibodies.
OLFM4 inhibits 4E-BP1 phosphorylation. (A) HL-60 cells were transfected with OLFM4 expression plasmid (OLFM4) or empty vector (vec) and selected by G418. G418-resistant HL-60 cells were treated with ATRA (1μM) for 3 days. Total cell lysates were subjected to Western blotting analysis with antibodies for phospho-p44/42 MAPK (Thr202/Tyr204), phospho-Akt (Ser473), phospho-GSK3β (Ser9), phopho-p70S6 kinase (Thr389), and phospho-4E-BP1 (Ser65 or Thr70). The blots were stripped and reprobed with corresponding antibodies for p44/42 MAPK, Akt, GSK-3β, p70S6 kinase, and 4E-BP1. (B) HEK-293T cells were cotransfected with 4E-BP1 or p70S6 kinase expression plasmid together with OLFM4 expression plasmid in different amounts, as indicated. After 24 hours, total cell lysates were subjected to Western blotting analysis with antibodies for phospho-4E-BP1 (Ser65, Thr70, or Thr37/46) or phospho-p70S6 kinase (Thr389). The blots were stripped and reprobed with total anti–4E-BP1 or anti-p70S6 kinase. (C) AML-193 cells were transduced with lentiviral shRNA against OLFM4 or control shRNA. After 48 hours, total cell lysates were subjected to Western blotting analysis with antibodies for OLFM4, phospho-4E-BP1 (Ser65, Thr70, or Thr37/46), phospho-Akt (Ser473), or phospho-p70S6 kinase (Thr389). The blots were stripped and reprobed with corresponding 4E-BP1, Akt, and p70S6 kinase antibodies.
Discussion
AML is a heterogeneous disease with an overall poor prognosis. We have shown that OLFM4 mRNA expression was up-regulated in the leukocytes of peripheral blood in a subset of AML patients. The preferential up-regulation of OLFM4 mRNA in the M4 subtype of AML suggests that OLFM4 expression is limited to specific myeloid cell-differentiation stages. Therefore, OLFM4 could prove to be a useful differentiation marker to define the subtype of AML.
The biologic significance of DNA methylation in the regulation of gene expression, and its role in cancer, has been recognized.19 Although a general genomic hypomethylation has been considered to be a potential precursor to leukemogenesis,20 only a few reports have demonstrated that hypomethylation mediates the activation of genes in cancer cells.21,22 In this study, our methylation analysis of leukocytes from normal individuals demonstrated a hypermethylation status in most of the CpG sites in the promoter of the OLFM4 gene. The significant hypomethylation of these CpG sites in AML patients was inversely correlated with enhanced OLFM4 gene expression. This result suggested that the hypomethylation status of the OLFM4 promoter region might be a necessary condition for constitutive OLFM4 gene overexpression in AML patients. This possibility was further supported by 2 lines of evidence. First, 5-aza-2′-deoxycytidine, an inhibitor of DNA methyltransferase, induced OLFM4 gene expression in HL-60 cells that demonstrated a hypermethylation status in the OLFM4 promoter and lacked constitutive expression of OLFM4. Second, methylation of the OLFM4 promoter by Sss I methylase treatment strongly repressed OLFM4 promoter transcription activity.
From a mechanistic point of view, 2 aspects have been shown to be involved in the transcriptional regulation of OLFM4. First, the methylation status of CpG in the promoter has an inverse correlation with OLFM4 gene expression. Second, our experiments indicated that trans-activation of the OLFM4 gene is associated with exogenous stimulation, including stimulation by RAs. This activation appears to be mediated through nuclear trans-acting factors. Our results suggest that ATRA-mediated up-regulation of the OLFM4 gene is due to the binding of RAR-α/RXR-α to a positive RARE. A functional RARE has been identified in the OLFM4 promoter and is required to confer ATRA induction through RAR-α/RXR-α heterodimers. In this study, we demonstrated that OLFM4 is a common target of ATRA and 5-aza-2′-deoxycytidine. We also showed that the combined application of ATRA and 5-aza-2′-deoxycytidine increased OLFM4 expression synergistically. The coregulation of the OLFM4 gene by ATRA and 5-aza-2′-deoxycytidine provides another example of a direct mechanism by which these 2 agents can synergize at the transcriptional level and, potentially, contribute to higher cellular growth inhibition.
In this study, we tested the potential involvement of OLFM4 in cell growth, differentiation, and apoptosis of myeloid leukemia cells. Several lines of evidence in this study point to a critical role of OLFM4 in the modulation of myeloid cell activities. First, induction of OLFM4 expression by ATRA correlates with the growth-inhibitory effect and differentiation response of ATRA. Second, overexpression of OLFM4 in HL-60 cells inhibits cell growth and induces cell differentiation and apoptosis. Its effect to induce cell differentiation is similar to that of ATRA application alone. Third, down-regulation of OLFM4 by shRNA in AML-193 cells compromised ATRA-mediated cell proliferation, differentiation, and apoptosis. Collectively, these results suggest that OLFM4 plays important roles in cell growth, differentiation, and apoptosis in myeloid leukemia cells.
In contrast to a previous report that suggested OLFM4 has an antiapoptotic effect in prostate cancer cell lines,11 our result showed that OLFM4 has a proapoptotic role in myeloid leukemia cells. The previous study was based on overexpression of OLFM4 together with GRIM19, a proapoptosis gene in prostate cell lines. In this study, in addition to the overexpression model, we also used shRNA to down-regulate OLFM4 expression in myeloid leukemia cells. In addition to using different technical approaches, the different results in the 2 studies may also suggest that OLFM4 may function as either a pro- or antiapoptosis factor, depending on the cellular context and gene-expression level. This phenomenon has been demonstrated for some other growth-related genes.23-25 For example, p21 can inhibit cell growth and survival, but under some circumstances, it can also promote cell survival.23,26
The regulation of protein translation plays an important role in controlling cell proliferation,27 differentiation,28 and apoptosis.29 4E-BP1 plays important roles in the regulation of the cap-dependent initiation of translation30 and in the generation of ATRA-mediated biologic responses.31 The activity of 4E-BP1 is crucially regulated by protein phosphorylation: For example, hypophosphorylated 4E-BP1 binds with high affinity to the eukaryotic translation initiation factor 4E and inhibits protein translation, and, conversely, the hyperphosphorylation of 4E-BP1 prevents this interaction.32 The protein kinase mTOR directly phosphorylates Thr37/46 of 4E-BP1, but the identities of the kinases acting at Ser65 and Thr70 of 4E-BP1 are not known.33 The precise mechanisms and kinases responsible for the direct phosphorylation and deactivation of 4E-BP1 downstream of mTOR remain to be determined.
In this study, we identified OLFM4 as a novel inhibitory signal upstream of 4E-BP1 phosphorylation/deactivation, as evidenced by the results that overexpression or down-regulation of OLFM4 influences the status of 4E-BP1 phosphorylation at multiple sites. This effect may happen downstream of mTOR, as OLFM4 did not affect the phosphorylation/activation of p70S6 kinase. This result suggests that OLFM4 is a factor that contributes to the inhibition of cap-dependent mRNA protein initiation and adds a new dimension to mTOR–4E-BP1 pathway regulation. Inhibition of translation is a common mechanism in terminal myeloid differentiation. Induction of granulocytic differentiation of HL-60 cells has been shown to be associated with significant inhibition of protein synthesis by 85%-90%.34 Therefore, it is possible that up-regulation of the expression of OLFM4, a novel cap-dependent translation inhibitor, by ATRA may be one of the mechanisms responsible for the ATRA-induced terminal granulocytic differentiation of APL cells. It has been reported that other olfactomedin-domain proteins may be scaffolds for other enzymes and substrates.35 Given this evidence, OLFM4 might interact with the kinases responsible for the phosphorylation of 4E-BP1 and inhibit their effects. Whatever the mechanism of OLFM4-induced 4E-BP1 hypophosphorylation may be, the fact that OLFM4 negatively acts on 4E-BP1 phosphorylation identifies 4E-BP1 as a likely target of the intracellular activity of OLFM4.
In summary, we show that OLFM4 is a novel target gene of RAs and DNA demethylation modification in myeloid leukemia cells. Both the overexpression and down-regulation experiments demonstrate that OLFM4 plays a role in the cellular functions of human myeloid leukemia cells. These effects of OLFM4 in myeloid leukemia cells appear to be associated with its inhibition of 4E-BP1 phosphorylation/deactivation. OLFM4 could represent a therapeutic target in myeloid leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Fellow Editorial Board of the National Institutes of Health for their helpful review and comment on the manuscript.
This work is supported by the intramural research fund of the National Heart, Lung, and Blood Institute, National Institutes of Health.
National Institutes of Health
Authorship
Contribution: W.L. designed experiments, performed research, analyzed data, and wrote the manuscript; H.W.L. and Y.L. performed experiments; R.W. contributed analytical tools and analyzed data; and G.P.R. designed experiments and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Griffin P. Rodgers, Molecular and Clinical Hematology Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Bldg 10, Rm 9N111, 9000 Rockville Pike, Bethesda, MD 20892; e-mail: gr5n@nih.gov.

![Figure 1. Up-regulation of OLFM4 in a subset of AML patients. (A) OLFM4 mRNA expression in human bone marrow (BM) and peripheral blood (PB) neutrophils from normal individuals, as determined by qRT-PCR. OLFM4 expression is shown relative to β-actin mRNA expression. *P < .01. (B) OLFM4 protein expression in BM and PB neutrophils from normal individuals, as determined by Western blot analysis. β-actin expression was used as a loading control. (C) OLFM4 protein expression in subcellular fractions of human BM neutrophils from normal individuals, as determined by Western blot analysis. Expressions of Apaf-1 and MnSOD were used to confirm the identity of the cytosolic and mitochondria fractions, respectively. (D) OLFM4 expression in peripheral blood leukocytes from M1 (n = 7), M2 (n = 10), M3 (n = 12), M4 (n = 10), or M5 (n = 3) subtypes of acute myeloid leukemia (AML) patients and normal individuals (N, n = 10) was analyzed by qRT-PCR. The bar represents the mean level of OLFM4 expression relative to β-actin expression. The values in the M4 group is significantly different from each of the other groups using 1-way ANOVA analysis, followed by Bonferroni multiple comparison test; P < .05. (E) Peripheral blood mononuclear cells from a normal individual (N), and 3 AML patients (P1 [M4 subtype], P2 [M3 subtype], and P3 [M1 subtype]) were immunostained with OLFM4 antibody (1:100) and counterstained with Giemsa. Brown color (arrows) indicates the OLFM4-positive cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/23/10.1182_blood-2009-10-246439/4/m_zh89991060950001.jpeg?Expires=1765898672&Signature=KYFtWcjsWLy9Dbn4P7aMZMtxe6VoQCpKUQjCFVkVum7CPI2IvvkzfHm2G3ZYFgDmU3RDWV0~M4UFnU8oKTQfuf1-zN45zAbiY6Hl8JlziOATAoOEfAAxuI77qsrj6~hjvGSqRxblyDANQi1XjTbv-TdxoxN9qBeymmG824FFtHaO6xjK1faJyqQgeipLnE5yqgMXqEbT7oYbenOokeNTcf2IdF5GqhcdAxb3zPqqx4-CQERJX6AS9p021tVIkrGM72a-FkhtsLlUIFTCqiu57TaZvPzt0~vLwv~X-MUDDGtajFxl3g9v-fDwW0uDbp3xR30P7N3eezp-W0XRqyH8KQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 1. Up-regulation of OLFM4 in a subset of AML patients. (A) OLFM4 mRNA expression in human bone marrow (BM) and peripheral blood (PB) neutrophils from normal individuals, as determined by qRT-PCR. OLFM4 expression is shown relative to β-actin mRNA expression. *P < .01. (B) OLFM4 protein expression in BM and PB neutrophils from normal individuals, as determined by Western blot analysis. β-actin expression was used as a loading control. (C) OLFM4 protein expression in subcellular fractions of human BM neutrophils from normal individuals, as determined by Western blot analysis. Expressions of Apaf-1 and MnSOD were used to confirm the identity of the cytosolic and mitochondria fractions, respectively. (D) OLFM4 expression in peripheral blood leukocytes from M1 (n = 7), M2 (n = 10), M3 (n = 12), M4 (n = 10), or M5 (n = 3) subtypes of acute myeloid leukemia (AML) patients and normal individuals (N, n = 10) was analyzed by qRT-PCR. The bar represents the mean level of OLFM4 expression relative to β-actin expression. The values in the M4 group is significantly different from each of the other groups using 1-way ANOVA analysis, followed by Bonferroni multiple comparison test; P < .05. (E) Peripheral blood mononuclear cells from a normal individual (N), and 3 AML patients (P1 [M4 subtype], P2 [M3 subtype], and P3 [M1 subtype]) were immunostained with OLFM4 antibody (1:100) and counterstained with Giemsa. Brown color (arrows) indicates the OLFM4-positive cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/23/10.1182_blood-2009-10-246439/4/m_zh89991060950001.jpeg?Expires=1766078968&Signature=0TJ6~O2OOYb2LZLe8QKVIphtacjkem6LYYWIhmkPVk104nTv6AVwY0fkrP3Lk5DW0icj4QxJPA9~lA-yUy4wgj-razS4L0Os0PJTkicpX0ncvnV9x18ZIoyvT7iYE2ujj~AQCg9BnUlFHyOIY~xvduSrXt6zhWTE--NYUJ2utD5BTNVfS85XBFM3apMszdKC6OVnnn6L7pkX~c36YMwpkrVzbkYS2VXbRhDkaABLjLuCx0LWFakgTA0YVMRRtrX5cYY~MHR9pSxhWh~ddw9FGnka~Pl--w~Sv9QuYd6aUbNsImY9j5beacEqSDAF9UDUNeRGqeGsyW2~3w5Z26htvg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)