In this issue of Blood, 2 articles define a new class of somatic mutations that allow transcription of oncogenic Notch1 from cryptic internal promoters.
Notch1 signaling is essential for T-lineage commitment as well as for survival and proliferation of committed T-cell progenitors in the thymus. Not surprisingly, Notch1-activating mutations are found in more than 50% of human T-cell acute lymphoblastic leukemias (T-ALLs).1 Rodent T-ALLs induced by a variety of genetic manipulations also exhibit frequent Notch1 mutations that render them Notch1-dependent for survival and proliferation in vitro.1 Thus, aberrant Notch1 activation represents a key event in the multistep pathway of T-cell leukemogenesis in rodents and humans. Two articles in this issue report the unexpected finding that spontaneous or Cre-LoxP–targeted deletion of the 5′ Notch1 promoter allows generation of leukemogenic Notch1 from “cryptic” internal promoters.2,3 Interestingly, loss-of-function mutations in Ikaros, a transcriptional repressor and T-cell tumor suppressor, potently enhance transcription from these cryptic internal Notch1 promoters.3
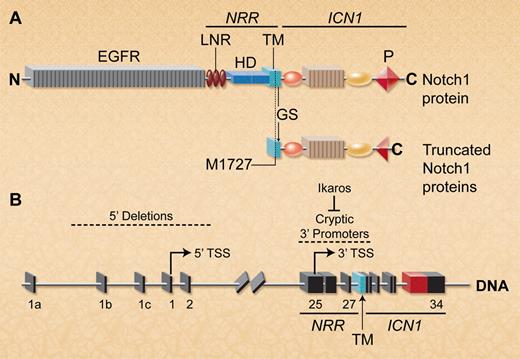
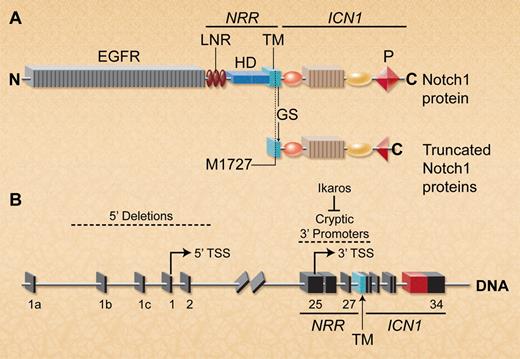
Deletion of the 5′ Notch1 promoter activates cryptic 3′ internal promoters to allow expression of truncated Notch1 proteins lacking the ectodomain. (A) Schematic depiction of full-length wild-type and mutant Notch1 proteins. The ectodomain includes the EGF repeats and the NRR, which contains 3 Lin/Notch repeats (LNR) and the heterodimerization domain (HD). The Notch1 intracellular region (ICN1) contains a C-terminal PEST domain (P). Notch1 transcripts lacking most of the ectodomain are translated using a conserved internal initiator methionine (M1727) that is just upstream of the GS cleavage site in the TM region. In T-ALLs characterized by both groups, these truncated Notch1 proteins also frequently exhibited PEST domain truncations within ICN1. (B) Spontaneous or targeted Notch1 deletions that include the 5′ promoter and transcriptional start site (TSS) in exon 1 allow transcription of truncated Notch1 mRNAs that initiate in exons 25-27 (indicated by arrow). Ikaros loss facilitates epigenetic remodeling to permit transcription from 3′ cryptic promoters in this region. (Professional illustration by A. Y. Chen.)
Deletion of the 5′ Notch1 promoter activates cryptic 3′ internal promoters to allow expression of truncated Notch1 proteins lacking the ectodomain. (A) Schematic depiction of full-length wild-type and mutant Notch1 proteins. The ectodomain includes the EGF repeats and the NRR, which contains 3 Lin/Notch repeats (LNR) and the heterodimerization domain (HD). The Notch1 intracellular region (ICN1) contains a C-terminal PEST domain (P). Notch1 transcripts lacking most of the ectodomain are translated using a conserved internal initiator methionine (M1727) that is just upstream of the GS cleavage site in the TM region. In T-ALLs characterized by both groups, these truncated Notch1 proteins also frequently exhibited PEST domain truncations within ICN1. (B) Spontaneous or targeted Notch1 deletions that include the 5′ promoter and transcriptional start site (TSS) in exon 1 allow transcription of truncated Notch1 mRNAs that initiate in exons 25-27 (indicated by arrow). Ikaros loss facilitates epigenetic remodeling to permit transcription from 3′ cryptic promoters in this region. (Professional illustration by A. Y. Chen.)
Notch receptors have large extracellular regions consisting of many epidermal growth factor repeats (EGFR) and a negative regulatory region (NRR) that autoinhibits Notch activation in the absence of Notch ligands (see figure panel A). Notch binding to its ligands triggers a conformational change in the NRR, allowing gamma secretase (GS)–mediated cleavage within the transmembrane (TM) region, releasing the Notch1 intracellular (ICN1) domain to travel to the nucleus and regulate transcription of Notch target genes.1 ICN1 does not bind DNA directly, but interacts with DNA-bound RPBJk to induce expression of Notch target genes. Many T-ALL cases have NRR mutations that allow ligand-independent Notch1 activation.1 A subset has truncating mutations within the ICN1 C-terminal PEST domain, increasing ICN1 stability. However, PEST mutations only weakly activate Notch1 signaling in reporter assays and are poorly leukemogenic in mice,4 most likely because Notch1 activation remains ligand-dependent.
The strong selection for ligand-independent Notch1 mutant alleles during human T-cell leukemogenesis makes sense because intrathymic Notch ligands required for Notch1 activation are functionally limiting for normal T-cell development.5 Surprisingly, however, PEST truncations are common but NRR mutations are rare in murine T-ALLs.1 Therefore, Ashworth and colleagues examined a panel of murine T-ALL cell lines for other abnormalities that could render Notch1 activation ligand-independent. They identified 2 types of truncated Notch1 transcripts that lacked most of the extracellular domain but retained the TM and ICN1 regions. These transcripts arose from mutant Notch1 alleles that had large 5′ genomic deletions (see figure, panel B). The most common deletion removed the 5′ Notch1 promoter as well as the initiator ATG codon in exon 1. Interestingly, deletion breakpoints bore features suggesting that they arose by illegitimate V(D)J recombination, similar to those characterized by Tsuji and colleagues in radiation-induced and Atm−/− thymic lymphomas.6 A few cell lines had other types of large intragenic Notch1 deletions that lacked hallmarks of illegitimate V(D)J recombination. In transient assays, transcripts derived from both types of 5′-deleted Notch1 alleles strongly activated transcription of a Notch reporter construct, results comparable to those induced by a strong human NRR mutant allele. Thus, this study describes a novel mechanism in which 5′ Notch1 genomic deletions generate highly active Notch1 proteins lacking most of the Notch1 ectodomain, explaining the paucity of NRR Notch1 mutations in murine T-ALLs.
The study by Jeannet et al supports these conclusions but also demonstrates that loss of Ikaros strongly potentiates transcription of Notch1 alleles harboring 5′ Notch1 deletions. Loss-of-function Ikaros mutations significantly cooperate with activating Notch1 mutations to promote murine T-ALL.7,8 Ikaros can bind RBPJk sites and repress certain Notch-regulated genes, suggesting that Ikaros loss may promote T-cell leukemogenesis by enhancing expression of Notch target genes. In support of this notion, the Jeannet study demonstrated that conditional CD4-Cre–mediated deletion of RBPJk markedly delayed T-ALL onset in IkL/L mice expressing hypomorphic Ikaros. Thus, T-cell leukemogenesis induced by Ikaros loss requires canonical Notch signaling. Surprisingly, however, T-ALL onset in IkL/L mice was greatly accelerated, rather than delayed, by CD4-Cre–mediated deletion of the 5′ end of a floxed Notch1 allele. This targeted deletion is smaller than the spontaneous ones found by Ashworth et al, but still removes the 5′ Notch1 promoter and leader peptide encoded by exon 1. Nonetheless, T-ALLs from IkL/L Notch1f/f CD4-Cre+ mice expressed truncated Notch1 transcripts lacking most of the Notch1 ectodomain and high levels of active and frequently PEST-mutated ICN1 protein. Furthermore, T-ALLs arising in IkL/L Notch1+/+ mice had spontaneously deleted the 5′ end of Notch1, similar to the T-ALL cell lines described by Ashworth.
How is Notch1 transcribed from alleles bearing targeted or spontaneous deletions of the Notch1 promoter? The truncated transcripts described in both articles typically initiated in exons 25-27, suggesting the existence of cryptic promoters in this region (figure panel B). Using ChIP-Seq analysis, Jeannet et al identified high levels of acetylated histone 3, an epigenetic mark associated with active chromatin near promoters, centered over the entire 3′ end of Notch1 beginning abruptly around exon 25. This region lacked acetylated histone 3 in IkL/L T-ALLs with intact 5′ Notch1. Thus, deletion of the 5′ Notch1 promoter facilitates epigenetic remodeling at the 3′ end of the locus, allowing transcription from cryptic internal promoters. Although this remodeling was also seen in nontransformed Notch1-deleted thymocytes expressing wild-type Ikaros, it was greatly increased in the absence of Ikaros, suggesting that Ikaros represses transcription from 3′ cryptic promoters (figure panel B). However, some murine T-ALLs in the Ashworth study expressed wild-type Ikaros, indicating that other factors likely regulate transcription from the cryptic promoters.
Ashworth et al demonstrated that mutant Notch1 transcripts lacking exon 1 could be translated using an internal initiator methionine (M1727). This residue lies within the Notch TM domain just upstream of the GS cleavage site (figure panel A), and thus generates a Notch1 protein lacking the entire ectodomain but which should still require cleavage by GS. Indeed in both studies, activation of 5′ truncated Notch1 proteins in T-ALLs was blocked by GS inhibitors. Interestingly, this internal methionine is conserved in the Notch TM across many vertebrate species.2 It will thus be important to determine whether generation of ligand-independent Notch1 proteins has a role in normal T-cell development.
The absence of the Notch1 ectodomain likely allows ligand-independent GS cleavage of these mutant Notch proteins. Nonetheless, PEST mutations were also strongly selected during leukemogenesis in both studies. Some human T-ALLs exhibit NRR and PEST Notch1 mutations in cis, and these synergistically increased Notch-dependent transcription as well as leukemogenesis in mice.4 Therefore, even when Notch1 activation is rendered ligand-independent by deletion of the ectodomain, increasing ICN1 stability by PEST domain truncation further increases Notch transcriptional activity and leukemogenic potential in mice and humans.
Despite the high frequency of 5′ Notch1 deletions in murine T-ALL, such lesions were not detected in a large panel of human T-ALLs,2 although small deletions could have been missed. This observation most likely reflects poor conservation of the “cryptic” recombination signal sequences that mediate illegitimate V(D)J recombination at the 5′ end of murine Notch1. Nonetheless, in rare cases of human T-ALL with (7,9) chromosomal translocations, Notch1 transcription appears to initiate from internal promoters around exon 25, and the conserved TM methionine can be used to generate human Notch1 protein lacking its ectodomain.2 It is therefore important to identify other mechanisms by which 5′-truncated Notch proteins can be generated in human T-ALL, as this would allow cells to evade therapies currently in development that target Notch ectodomains.9,10
Conflict-of-interest disclosure: The author declares no competing financial interests. ■