Abstract
Epidermal growth factor-like domain 7 (Egfl7) is important for regulating tubulogenesis in zebrafish, but its role in mammals remains unresolved. We show here that endothelial overexpression of Egfl7 in transgenic mice leads to partial lethality, hemorrhaging, and altered cardiac morphogenesis. These defects are accompanied by abnormal vascular patterning and remodeling in both the embryonic and postnatal vasculature. Egfl7 overexpression in the neonatal retina results in a hyperangiogenic response, and EGFL7 knockdown in human primary endothelial cells suppresses endothelial cell proliferation, sprouting, and migration. These phenotypes are reminiscent of Notch inhibition. In addition, our results show that EGFL7 and endothelial-specific NOTCH physically interact in vivo and strongly suggest that Egfl7 antagonizes Notch in both the postnatal retina and in primary endothelial cells. Specifically, Egfl7 inhibits Notch reporter activity and down-regulates the level of Notch target genes when overexpressed. In conclusion, we have uncovered a critical role for Egfl7 in vascular development and have shown that some of these functions are mediated through modulation of Notch signaling.
Introduction
Blood vessel development is regulated by several known signaling pathways, including Notch.1 In mammals, the Notch pathway is composed of 4 membrane-bound Notch receptors (1-4) and 5 ligands. Of these, Notch1 and Notch4 and their ligands Jagged1, Jagged2, Delta-like 1, and Delta-like 4 (Dll4) are expressed in the vasculature. Binding of a ligand to the extracellular domain of the Notch receptor triggers a series of proteolytic cleavages that leads to the release of the Notch intracellular domain. The Notch intracellular domain is then translocated to the nucleus where it interacts with the transcription factor CSL (CBF1/Su(H)/LAG1), displaces transcriptional repressor complexes, and recruits transcriptional activation machinery. This results in the expression of Notch target genes, including transcription factors Hairy/Enhancer of Split (HES) and the HES-related genes, Hey1 and Hey2.1
Epidermal growth factor-like domain 7 (Egfl7), a 41-kDa secreted factor that is highly conserved in vertebrates,2-4 is a potential novel Notch ligand. In support, we have previously reported that EGFL7 contains a putative Delta/Serrate/LAG-2 (DSL) domain,2 which is required for Notch receptor-ligand interactions.1 Egfl7 has also been shown to regulate Notch signaling in vitro in adult neural stem cells.5
Evidence suggests a role for Egfl7 in vascular development. Egfl7 expression is largely restricted to endothelial cells and their progenitors.2,3,6 Expression is highest when the endothelium is in an actively proliferating state, as is the case during embryonic vascular development, physiologic angiogenesis, and in response to vascular injury.2,3,6 EGFL7 acts a chemoattractant for endothelial cells6 and binds to components of the extracellular matrix.7 Knockdown of Egfl7 in zebrafish results in disruption of vascular tube formation.3,8 The role of Egfl7 in mammalian vasculature, however, is less clear and is complicated by the presence of the microRNA, miR126, within the Egfl7 gene.9-11 miR126 expression is highly enriched in endothelial cells,9,11 and mice that lack this microRNA but express Egfl7 display partial embryonic lethality caused by a loss of vascular integrity.10,11 Two distinct Egfl7 knockout mouse models show vascular defects.7 However, it is unclear whether the phenotype is the result of the lack of Egfl7 or a reduction in miR126 expression.7 A third Egfl7 knockout model does not exhibit any obvious phenotype.10
We determined the role of Egfl7 by generating transgenic mice and modulating the level of EGFL7 in human umbilical vein endothelial cells (HUVECs). Using both in vivo and in vitro analysis, our study elucidates a clear function for Egfl7 in the mammalian vasculature and defines its ability to regulate Notch target gene expression in vivo.
Methods
Generation of Tie2-Egfl7 transgenic mice
A Myc/His-tagged murine Egfl7 cDNA was cloned into the pT2HLacZpA1 vector backbone.12 Two independent transgenic lines were generated (Scripps Research Institute Mouse Genetic Core Facility). All animal protocols included in the research were approved by the Animal Care and Use Committee at Weill Cornell Medical College.
Quantitative RT-PCR
RNA was isolated using either Trizol reagent (Invitrogen), RNeasy (QIAGEN) or RNaqueous kit (Ambion). First-strand synthesis was performed using Superscript Reverse Transcriptase III (Invitrogen), and gene expression was measured using specific primer sets and SYBR Green technology (Applied Biosystems). MicroRNA was reverse transcribed using microRNA-specific primers and the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). Expression was measured using the TaqMan MircoRNA Assay (Applied Biosystems). Changes were quantified using the ΔΔCT method.
Western blotting
Samples were boiled at 100°C for 5 minutes, resolved, blotted on polyvinylidene difluoride membranes, and probed using either goat anti–human EGFL7 (R-12) antibody (Santa Cruz Biotechnology Sc34416, 1:1000), hamster anti–mouse Notch1 (Millipore MAB5414, 1:1000), goat anti–mouse Notch4 (E11; Santa Cruz Biotechnology Sc32634, 1:1000), or mouse anti–β-tubulin (Sigma-Aldrich T4026, 1:1000). Antibody complexes were detected using horseradish peroxidase–conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) and the PICO substrate kit (Pierce Chemical). Embryonic EGFL7 protein expression was performed using 25 μg of cytosolic protein isolated from whole embryos.
Coimmunoprecipitation assays
HEK293 cells were transiently transfected with pcDNA4(MycHis)Egfl7 and pHyTC-N4Fc or pHyTC-N1Fc13 using Lipofectamine 2000 (Invitrogen). The pcDNA4(MycHis) Egfl7 plasmid contains a MycHis-tagged Egfl7 in the pcDNA4 vector. The pHyTC-N4Fc construct encodes the signal peptide and EGF repeats of murine Notch4 fused to human Fc. After 48 hours, cells were lysed in immunoprecipitation (IP) buffer (50mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.5, 150mM NaCl, 1% Triton X-100, 10% glycerol, 1.5mM MgCl2, 1mM ethylenediaminetetraacetic acid, protease inhibitor cocktail; Sigma-Aldrich). Protein A-agarose beads (Invitrogen) were added to the cleared lysates and incubated at 4°C for 1 hour. Beads were discarded and lysates were incubated either alone, with the NOTCH4, or mouse anti-c-MYC antibodies (Millipore MAB8865) overnight at 4°C. Protein-antibody complexes were collected by incubating with protein A-agarose beads for 1 hour at 4°C, followed by centrifugation and 3 washes with IP buffer. HUVECs were infected with an adenovirus encoding human EGFL7. Cells were lysed as described for HEK293 cells, and EGFL7 was immunoprecipated using the MYC antibody. For pull-downs using embryonic lysates, 3 embryos were pooled, snap frozen, transferred to IP buffer, and homogenized. Lysates were subjected to the same IP protocol as described for HEK293 cells.
Immunohistochemistry
Embryos and yolk sacs were dissected and fixed in 4% paraformaldehyde. For whole-mount immunostaining, an antibody against CD31 (MEC13.3, BD Biosciences PharMingen 553370, 1:100) was used and developed with a horseradish peroxidase–conjugated secondary donkey anti–rat IgG (Jackson ImmunoResearch Laboratories, 1:1000), followed by treatment with diaminobenzidine (Vector). For embryonic sections, embryos were incubated in 30% sucrose overnight at 4°C after paraformaldehyde fixation and embedded in OCT compound for sectioning (thickness, 12 μm). Sections were incubated with the rat anti–mouse CD31 antibody (1:100) and biotinylated-secondary rabbit anti–rat IgG (Vector, 1:1000). The signal was amplified using the ABC Elite Kit (Vector) and developed using NovaRed Substrate Kit (Vector). Nuclei were counterstained using Mayer Hematoxylin solution (Sigma-Aldrich). For immunofluorescence (IF), sections were incubated with the rat anti–mouse CD31 antibody (1:100) and a goat anti–mouse smooth muscle actin antibody (Abcam, Ab21027, 1:1000). Primary antibodies were detected using species-specific AlexaFluor- and Cy3-coupled secondary antibodies (Invitrogen). Cell nuclei were visualized using 4,6-diamidino-2-phenylindole. Eyes were isolated from P5 pups, fixed in 4% paraformaldehyde, and retinas dissected. Retinal vasculature was highlighted using fluorescein isothiocyanate-labeled Bandeiraea simplicifolia (BS-1) isolectin (Sigma-Aldrich).
Constructs and cell culture
A human EGFL7myc cDNA was engineered into the pCCLIRESGFP vector provided by Stefano Rivella (WCMC). MISSION shRNA lentiviral plasmids TRCN0000053658 (pLKO58), TRCN0000053659 (pLKO59), TRCN0000053661 (pLKO61), and TRCN0000053662 (pLKO62), and the control pLKO1 were purchased from Sigma-Aldrich. HUVECs were isolated by standard protocol from human umbilical cords14 and maintained in complete medium (EGM-2 Bullet kit; Lonza Walkersville). HUVECs were infected with lentiviral vectors to generate stable populations.15 EGFL7 expression was determined by quantitative reverse-transcribed polymerase chain reaction (RT-PCR) and Western blot analysis using an antibody against the c-MYC tag (Santa Cruz Biotechnology; 9E10). Experiments were performed with at least 3 independently generated HUVEC lines.
Proliferation assay
HUVECs were suspended in complete medium and seeded on rat type I collagen (BD Biosciences) in a 24-well plate (1 × 104 cells/well). Four days later, WST-8 assay (Dojindo) was performed and cell number was determined.
HUVEC monolayer wounding assay
HUVECs were seeded in a 24-well plate (1 × 105 cells/well). The next day, the confluent monolayer was stripped with a pipette tip. The stripped region was photographed at 0, 8, and 24 hours. The TScratch program16 was used to determine the percentage area filled at the different time points.
Capillary sprouting assay
HUVEC lines were added to Cytodex 3 beads (GE Healthcare) in complete medium and suspended in 2.5 mg/mL fibrinogen (500 beads) in a 24-well plate. The bead/fibrinogen solution was mixed with 0.625 units of thrombin (Sigma-Aldrich) and incubated for 10 to 15 minutes at 37°C. Thirty minutes after polymerization, Detroit 551 skin fibroblasts (ATCC) were seeded on top of the fibrin clots in EGM-2 medium and medium was changed every other day. Wells were visualized by microscopy on day 7.
Luciferase reporter assay
HUVECs were seeded in 24-well plates in complete medium (2 × 105 cells/well). The next day, the pGL3 vector (Promega), containing 11-CSL binding consensus sites, upstream of a SV40 promoter, was introduced using Lipofectamine 2000 (Invitrogen), along with a plasmid containing the control Renilla luciferase gene. Two days after lipofection, luciferase and Renilla activity was determined with Dual-Luciferase Reporter Assay System (Promega).
Endothelial morphogenesis coculture assay
HUVECs were coinfected with adenoviruses encoding full-length murine Notch4 and GFP at a multiplicity of infection of 30 and 5, respectively.13 As a control, the Notch4 adenovirus was replaced with a LacZ adenovirus. Two days later, each lentiviral-infected HUVEC line (1.5 × 105 cells; EGFL7-overexpressing, EGFL7 knockdown, and control lines) was seeded on fibrin gels in a 24-well plate. Three hours later, each adenovirus-infected line (2 × 104 cells) was added on top of the monolayer in complete medium. Seven days later, HUVEC morphogenesis was determined by measuring the number of GFP+ cells with processes per field.
Yeast-2-hybrid and yeast-3-hybrid systems
Protein-protein interactions were studied in yeast using MATCHMAKER GAL4 TWO HYBRID System 3 and the pBRIDGE 3-hybrid vector system (BD Biosciences). Interactions were monitored by the ability of the transformed yeast to grow on defined medium and activate α-galactosidase activity.
Protein sequence analysis, quantification, and statistics
Analysis of EGFL7 protein sequence was done using Prosite (http://ca.expasy.org). Vascular measurements and imaging were done using Volocity Imaging Software (Version 5.2.1; Improvision). Error bars correspond to SEM, unless otherwise stated. Differences between wild-type and transgenic mice are regarded as significant when P is less than .05 using an unpaired Student t test.
Results
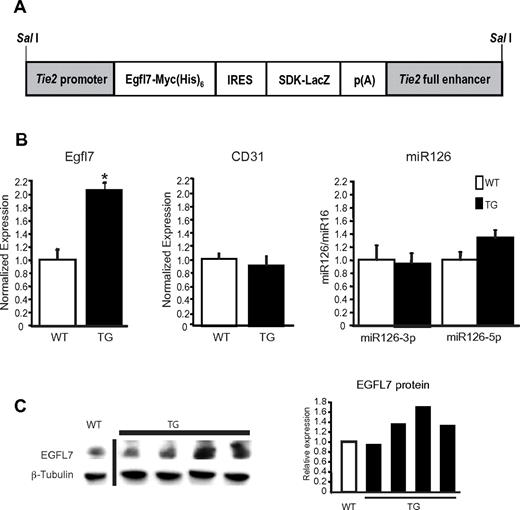
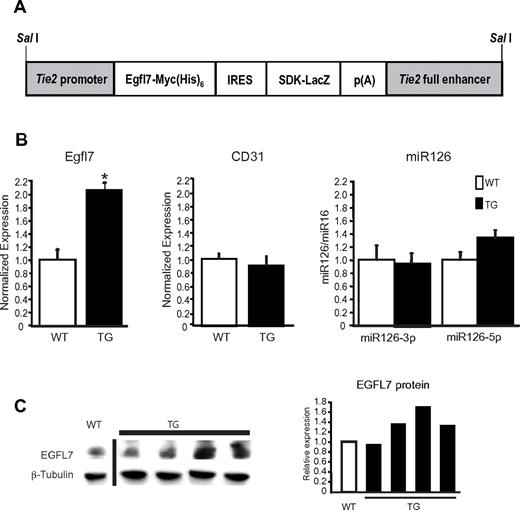
To determine the role of Egfl7 in vivo in the vascular system, we generated Tie2-Egfl7 transgenic mice. These mice carry an Egfl7 transgene under the control of the Tie2 promoter, resulting in overexpression specifically in the endothelium and cells of the early hematopoietic lineage (Figure 1A).12,17 Two transgenic lines were generated, both of which were viable and fertile. Line 2 expressed higher Egfl7 levels than line 1 and exhibited a more pronounced phenotype. We therefore used line 2 for all subsequent analyses. EGFL7 expression was detected in the endothelium of many vessels, including the head, dorsal aorta, cardinal, and umbilical veins in both wild-type and transgenic embryos at E12.5 by immunohistochemistry (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). EGFL7 was coexpressed with collagen IV, consistent with its localization to the extracellular matrix (supplemental Figure 1B). Exogenous EGFL7 was correctly expressed, as observed by immunohistochemistry using an antibody against the MYC tag (supplemental Figure 1A). To measure Egfl7 overexpression, mRNA was isolated from whole embryos at E12.5 and expression was measured by quantitative RT-PCR (Figure 1B). Although expression varied among transgenic embryos, a subset showed a significant 2-fold increase in Egfl7 expression (Figure 1B left graph), whereas endothelial cell number, as measured by CD31 transcripts, was unaffected (Figure 1B middle graph). EGFL7 protein levels also varied, with some transgenics exhibiting a 1.3- to 1.7-fold increase of EGFL7 (Figure 1C). miR126 expression, however, was not significantly altered between genotypes (Figure 1B right graph). Thus, the phenotypes in the Tie2-Egfl7 mice can be ascribed to overexpression of Egfl7 and not miR126.
Egfl7 mRNA and protein expression in Tie2-Egfl7 transgenic embryos. (A) Schematic organization of Tie2-Egfl7 transgene construct used to generate mice that overexpress Egfl7. (B) Average Egfl7, CD31, and miR126 expression in whole embryos at E12.5, as measured by quantitative RT-PCR. The levels of Egfl7 were normalized to endothelial cell number using CD31 expression. Values are represented as fold difference relative to wild-type: WT, n = 6; TG, n = 4. *P < .01. (C) Detection of EGFL7 and β-tubulin protein in cytosolic fraction isolated from wild-type and transgenic embryos at E12.5. Quantification of protein levels is shown in the graph, and values are made relative to wild-type expression level. White bar represents wild-type; and black bars, transgenic. A vertical line has been inserted to indicate a repositioned gel lane.
Egfl7 mRNA and protein expression in Tie2-Egfl7 transgenic embryos. (A) Schematic organization of Tie2-Egfl7 transgene construct used to generate mice that overexpress Egfl7. (B) Average Egfl7, CD31, and miR126 expression in whole embryos at E12.5, as measured by quantitative RT-PCR. The levels of Egfl7 were normalized to endothelial cell number using CD31 expression. Values are represented as fold difference relative to wild-type: WT, n = 6; TG, n = 4. *P < .01. (C) Detection of EGFL7 and β-tubulin protein in cytosolic fraction isolated from wild-type and transgenic embryos at E12.5. Quantification of protein levels is shown in the graph, and values are made relative to wild-type expression level. White bar represents wild-type; and black bars, transgenic. A vertical line has been inserted to indicate a repositioned gel lane.
Embryonic Egfl7 overexpression results in partial lethality and defects in cardiac morphogenesis
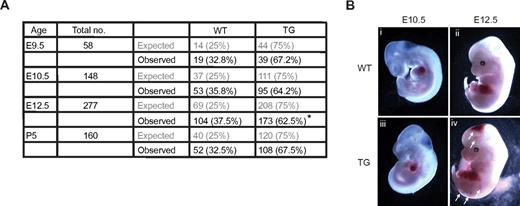
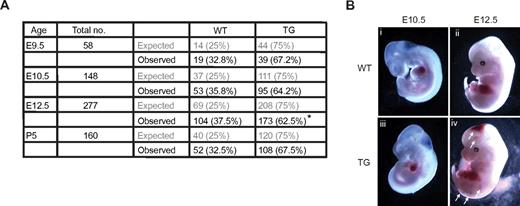
The effect of overexpressing Egfl7 in the endothelium was determined by analysis of embryos from Tie2-Egfl7 hemizygous intercrosses. Mutant phenotypes were incompletely penetrant, possibly reflecting a dosage-dependent effect. The number of transgenics observed was lower than expected (Figure 2A). This was significant at E12.5 but not at any other stage, suggesting that some of the Tie2-Egfl7 transgenic embryos died between E10.5 and E12.5. Hemorrhaging was observed at various sites in the head and trunk in 13% (14 of 105) of transgenic embryos at E12.5 (Figure 2Biv arrows) but was not observed earlier (Figure 2Biii) or in wild-type mice (Figure 2Bi-ii).
Overexpressing Egfl7 results in partial embryonic lethality and hemorrhaging. (A) The expected and observed number of embryos and pups obtained from Tie2-Egfl7 hemizygous intercrosses at E9.5, E10.5, E12.5, and P5. *P = .004. (B) Gross phenotype of wild-type (i-ii) and Tie2-Egfl7 transgenic embryos (iii-iv) at E10.5 and E12.5. Arrows indicate hemorrhaging in Tie2-Egfl7 mice at E12.5 (iv). Original magnification ×20 (E10.5), ×12 (E12.5). Images were captured on Discovery V20 Stereomicroscope (Carl Zeiss).
Overexpressing Egfl7 results in partial embryonic lethality and hemorrhaging. (A) The expected and observed number of embryos and pups obtained from Tie2-Egfl7 hemizygous intercrosses at E9.5, E10.5, E12.5, and P5. *P = .004. (B) Gross phenotype of wild-type (i-ii) and Tie2-Egfl7 transgenic embryos (iii-iv) at E10.5 and E12.5. Arrows indicate hemorrhaging in Tie2-Egfl7 mice at E12.5 (iv). Original magnification ×20 (E10.5), ×12 (E12.5). Images were captured on Discovery V20 Stereomicroscope (Carl Zeiss).
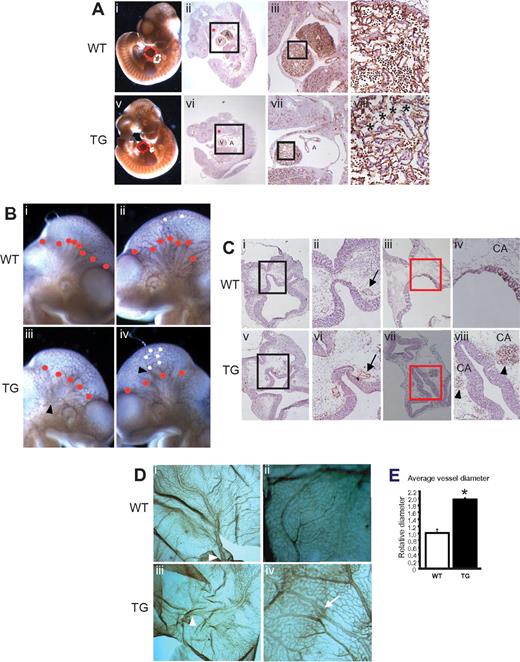
Egfl7 overexpression caused a reduction in the size of the atrium and ventricle (Figure 3Av), as observed by whole-mount CD31 immunostaining and parasagittal sections of Tie2-Egfl7 embryos at E10.5 (Figure 3Avi-vii). The hearts of the transgenics, but not controls, also showed a reduction in ventricular traberculation (Figure 3Aiv,viii). In summary, endothelial overexpression of Egfl7 results in hemorrhaging and cardiac defects. The heart abnormalities may also, in part, explain the loss of embryos observed between E10.5 and E12.5.
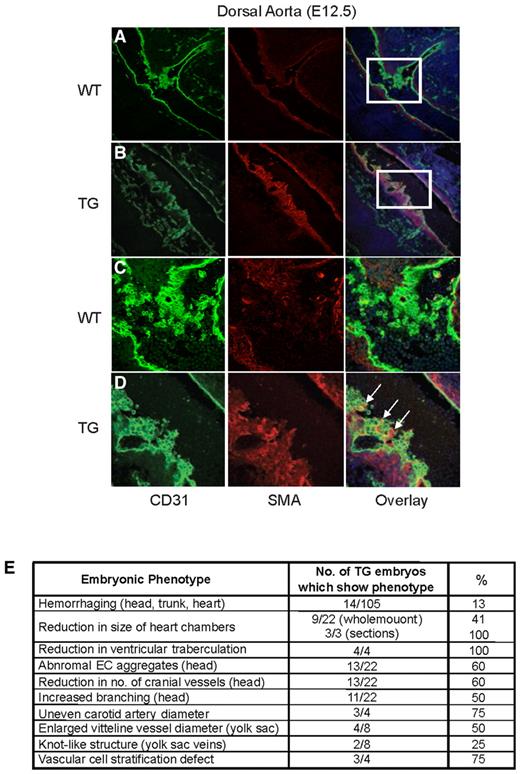
Embryonic Egfl7 overexpression causes defects in the developing heart and in head and yolk sac vasculature. (A) CD31 immunostaining of whole-mount wild-type (i) and transgenic (v) embryos at E10.5. White and red lines represent the atrium (A) and ventricle (V), respectively. Original magnification ×30. CD31 immunostaining of parasagittal sections of wild-type (ii-iv) and Tie2-Egfl7 transgenic embryos (vi-viii). High-magnification images of black-boxed areas in subpanels ii and vi (iii,vii). High-magnification images of boxed areas in subpanels iii and vii (iv,viii). Original magnification ×1.25 (ii,vi), ×4 (iii,vii), and ×20 (iv,viii). Black asterisks represent reduction in traberculation; and red asterisks, branchial arch. (B) CD31 immunostaining of whole-mount embryos at E10.5. Tie2-Egfl7 transgenic embryos (iii-iv) showed abnormal endothelial aggregates (arrowheads), a decrease in the number of major cranial vessels (red dots), and increased branching (white dots) compared with wild-type littermates (i-ii). Original magnification ×60. (C) CD31 immunostaining of cross sections from wild-type (i-iv) and Tie2-Egfl7 transgenic embryos (v-viii) at E10.5. High-magnification images of black-boxed areas in subpanels i and v (ii,vi). High-magnification images of the boxed areas in subpanels iii and vii (iv,viii). Arrows indicates carotid artery (CA); and arrowheads, hemorrhaging from CA. Original magnification ×4 (i,iii,v,vii) and ×10 (ii,iv,vi,viii). (D) CD31 immunostaining of E12.5 wild-type (i-ii) and transgenic (iii-iv) yolk sacs. Arrowheads indicate vitelline vessels; and arrow, knot-like structure in vein. Quantification of vitelline vessel diameter in wild-type (□, n = 2) and transgenic (■, n = 4) yolk sacs. (v) *P < .03. Original magnification ×25.5 (i,iii) and × 41 (ii,iv). Whole-mount images were captured on Discovery V20 Stereomicroscope (Carl Zeiss) and immunohistochemistry images on the Axioplan 2 Imaging Upright Microscope (Carl Zeiss).
Embryonic Egfl7 overexpression causes defects in the developing heart and in head and yolk sac vasculature. (A) CD31 immunostaining of whole-mount wild-type (i) and transgenic (v) embryos at E10.5. White and red lines represent the atrium (A) and ventricle (V), respectively. Original magnification ×30. CD31 immunostaining of parasagittal sections of wild-type (ii-iv) and Tie2-Egfl7 transgenic embryos (vi-viii). High-magnification images of black-boxed areas in subpanels ii and vi (iii,vii). High-magnification images of boxed areas in subpanels iii and vii (iv,viii). Original magnification ×1.25 (ii,vi), ×4 (iii,vii), and ×20 (iv,viii). Black asterisks represent reduction in traberculation; and red asterisks, branchial arch. (B) CD31 immunostaining of whole-mount embryos at E10.5. Tie2-Egfl7 transgenic embryos (iii-iv) showed abnormal endothelial aggregates (arrowheads), a decrease in the number of major cranial vessels (red dots), and increased branching (white dots) compared with wild-type littermates (i-ii). Original magnification ×60. (C) CD31 immunostaining of cross sections from wild-type (i-iv) and Tie2-Egfl7 transgenic embryos (v-viii) at E10.5. High-magnification images of black-boxed areas in subpanels i and v (ii,vi). High-magnification images of the boxed areas in subpanels iii and vii (iv,viii). Arrows indicates carotid artery (CA); and arrowheads, hemorrhaging from CA. Original magnification ×4 (i,iii,v,vii) and ×10 (ii,iv,vi,viii). (D) CD31 immunostaining of E12.5 wild-type (i-ii) and transgenic (iii-iv) yolk sacs. Arrowheads indicate vitelline vessels; and arrow, knot-like structure in vein. Quantification of vitelline vessel diameter in wild-type (□, n = 2) and transgenic (■, n = 4) yolk sacs. (v) *P < .03. Original magnification ×25.5 (i,iii) and × 41 (ii,iv). Whole-mount images were captured on Discovery V20 Stereomicroscope (Carl Zeiss) and immunohistochemistry images on the Axioplan 2 Imaging Upright Microscope (Carl Zeiss).
Embryonic Egfl7 overexpression causes defects in the vasculature of the head and yolk sac
Defects were observed in the head vasculature of Tie2-Egfl7 embryos at E10.5 (Figure 3B-C). Specifically, abnormal endothelial cell aggregates (Figure 3Biii-iv arrowheads) and a decrease in the number of major cranial vessels (Figure 3Biii-iv red dots) were seen in 60% (13 of 22) of the transgenic embryos. Half (11 of 22) of the transgenic mice showed increased branching at the distal ends of the vessels (Figure 3Biv white dots). Interestingly, the arterial vessels were most severely affected. The diameter of the carotid artery was uneven and appeared collapsed in 75% (3 of 4) of transgenic embryos (Figure 3Cvi arrow). In addition, hemorrhaging from this vessel was seen at both E10.5 (Figure 3Cviii arrowheads) and E12.5 (data not shown). These abnormalities were not observed in wild-types (Figure 3Ci-iv).
The yolk sac vasculature was also affected (Figure 3D). At E12.5, transgenic vitelline vessels exhibited a 2-fold increase in diameter compared with controls (Figure 3Diii arrowhead, 3Di). Knot-like structures were observed in the veins of 25% (2 of 8) of transgenic yolk sacs analyzed (Figure 2Div arrow) but not in wild-types (Figure 2Dii). These findings indicate that elevated levels of Egfl7 result in abnormal vessel formation and suggest that Egfl7 plays a key role in the patterning of the head and yolk sac vessels.
Elevated levels of Egfl7 cause defects in vascular cell stratification
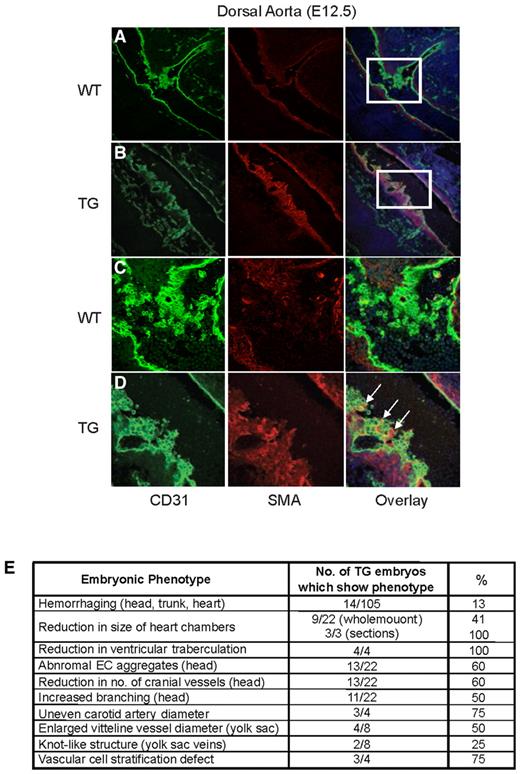
To determine whether defects in vessel stability contributed to the observed hemorrhaging, mural cell recruitment was analyzed. Immunofluorescence analysis of E12.5 transgenic embryos revealed stratification defects in cells surrounding the dorsal aorta (Figure 4B,D). In wild-type embryos, smooth muscle actin α (SMA+) cells were positioned adjacent to the CD31+ cell layer and the 2 cell types existed separately (Figure 4A,C). In contrast, SMA+ cells were interspersed with CD31+ cells in 75% (3 of 4) of transgenic embryos analyzed. This was only observed at branch points (Figure 4B,D, arrows), suggesting that Egfl7 overexpression causes stratification defects, specifically at sites of vascular growth and remodeling.
Tie2-Egfl7 transgenic mice exhibit defects in vascular cell stratification at branch points of the dorsal aorta. Immunofluorescence staining of the dorsal aorta at E12.5. CD31+ cells are shown in green (Alexa Flour 488), and SMA+ cells are shown in red (Cy3). Images were captured on the TCS AOBS SP2 microscope (Leica). (A-B) Wild-type and transgenic dorsal aorta, respectively. Original magnification 20×/0.7NA, water objective. (C-D) High-magnification image of boxed areas in panels A and B. Original magnification 63×/1.2NA, water objective. (E) Summary of phenotypes observed in Egfl7 transgenic embryos.
Tie2-Egfl7 transgenic mice exhibit defects in vascular cell stratification at branch points of the dorsal aorta. Immunofluorescence staining of the dorsal aorta at E12.5. CD31+ cells are shown in green (Alexa Flour 488), and SMA+ cells are shown in red (Cy3). Images were captured on the TCS AOBS SP2 microscope (Leica). (A-B) Wild-type and transgenic dorsal aorta, respectively. Original magnification 20×/0.7NA, water objective. (C-D) High-magnification image of boxed areas in panels A and B. Original magnification 63×/1.2NA, water objective. (E) Summary of phenotypes observed in Egfl7 transgenic embryos.
In summary, embryonic endothelial Egfl7 overexpression results in partial lethality and hemorrhaging. This is accompanied by defects in cardiac morphogenesis, vessel patterning, and vascular cell stratification (summarized in Figure 4E).
Postnatal Egfl7 overexpression results in retinal vascular defects
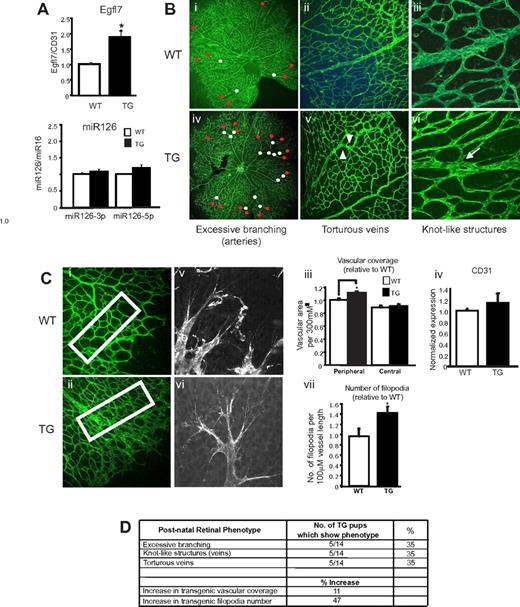
To test whether Eglf7 overexpression also affected postnatal angiogenesis, retinal vascular development was examined. EGFL7 protein was localized to endothelial sprouts in the P5 retina (supplemental Figure 2A-B). We observed a similar expression pattern for NOTCH4 (supplemental Figure 2C-D). In transgenic retinas, Egfl7 mRNA was significantly increased by 2-fold (Figure 5A), whereas miR126 expression (Figure 5A) and EGFL7 protein localization were unaffected (supplemental Figure 2B). Immunofluoresence analysis using fluorescein isothiocyanate–labeled BS-1 lectin revealed subtle defects in transgenic arterial and venous vessel morphology, including increased arterial branching (Figure 5Biv red dots). In control retinas, arteries branched into one or 2 vessels (Figure 5Bi). In contrast, as many as 5 branches were observed to originate from one transgenic artery (Figure 5Biv). The number of branch points along the arterial vessel length was also elevated (Figure 5Biv, white dots). These arterial defects were apparent in 35% (5 of 14) of the transgenics but not in wild-types. We also noted venous abnormalities in those transgenic retinas with arterial defects. Specifically, tortuosity (Figure 5Bv arrowheads) and knot-like structures (Figure 5Bvi arrow) were observed. Interestingly, the knot-like structures resembled those in the vasculature of transgenic yolk sacs (Figure 3Div arrow). A significant 11% increase was observed in the vascular coverage of the transgenic peripheral plexus compared with wild-type controls (Figure 5Ci-iii). Quantitative RT-PCR analysis revealed that CD31 expression was also increased by 20% in transgenics (Figure 5Civ). In addition, the number of filopodia was approximately 50% higher (Figure 5Cv-vii). Radial expansion of the developing vasculature and vessel length, however, was unaffected (supplemental Figure 3). These findings, summarized in Figure 5D, indicate that overexpressing Egfl7 in the postnatal retina induces a hyperangiogenic response and results in abnormal vessel patterning. The retinal defects are not completely penetrant, possibly because of a dosage-dependent effect of Egfl7.
Overexpressing Egfl7 in the postnatal retina results in arterial and venous defects and an increase in vascular coverage. (A) Quantitative RT-PCR analysis of Egfl7 and miR126 expression in P5 retinas isolated from wild-type (□, n = 4) and transgenic (■, n = 6) mice. Egfl7 expression is normalized to endothelial cell number using CD31 expression. *P < .03. (B) Wild-type (i-iii) and transgenic (iv-vi) retinal vasculature at P5, stained with fluorescein isothiocyanate–labeled BS-1 lectin. Red dots represent branching at the distal ends of the vessel; white dots, branching along the vessel length; arrowheads, tortuous veins; and arrows, venous knot-like structures. Original magnification 10×/0.4NA, water objective (i,iv), 20×/0.7NA, water objective (ii,v), and 63×/1.2NA, (iii,vi). (C) Vascular coverage in wild-type (i) and transgenic (ii) retinas at P5. Boxed regions correspond to areas where coverage was measured. (iii) Average vascular coverage at the peripheral and central plexus for wild-type (□, n = 8) and transgenic (■, n = 14) retinas. *P < .05. (iv) CD31 mRNA expression in wild-type (□, n = 4) and transgenic (■, n = 6) retinas. Filopodia in wild-type (v) and transgenic (vi) retinas. (vii) Average filopodia number per 100-μM vessel length for wild-type (□, n = 5) and transgenic (■, n = 9) retinas. *P < .05. Values for vascular coverage and filopodia number are represented as fold difference relative to wild-type. Original magnification 20× (i,ii) and 40×/1.3NA, water objective (v,vi). (D) Summary of phenotypes observed in Egfl7 transgenic retinal vasculature at P5. Images of the retinal vasculature were captured on the TCS AOBS SP2 microscope (Leica), and filopodia was imaged using the LSM 5Live DuoScan microscope (Carl Zeiss).
Overexpressing Egfl7 in the postnatal retina results in arterial and venous defects and an increase in vascular coverage. (A) Quantitative RT-PCR analysis of Egfl7 and miR126 expression in P5 retinas isolated from wild-type (□, n = 4) and transgenic (■, n = 6) mice. Egfl7 expression is normalized to endothelial cell number using CD31 expression. *P < .03. (B) Wild-type (i-iii) and transgenic (iv-vi) retinal vasculature at P5, stained with fluorescein isothiocyanate–labeled BS-1 lectin. Red dots represent branching at the distal ends of the vessel; white dots, branching along the vessel length; arrowheads, tortuous veins; and arrows, venous knot-like structures. Original magnification 10×/0.4NA, water objective (i,iv), 20×/0.7NA, water objective (ii,v), and 63×/1.2NA, (iii,vi). (C) Vascular coverage in wild-type (i) and transgenic (ii) retinas at P5. Boxed regions correspond to areas where coverage was measured. (iii) Average vascular coverage at the peripheral and central plexus for wild-type (□, n = 8) and transgenic (■, n = 14) retinas. *P < .05. (iv) CD31 mRNA expression in wild-type (□, n = 4) and transgenic (■, n = 6) retinas. Filopodia in wild-type (v) and transgenic (vi) retinas. (vii) Average filopodia number per 100-μM vessel length for wild-type (□, n = 5) and transgenic (■, n = 9) retinas. *P < .05. Values for vascular coverage and filopodia number are represented as fold difference relative to wild-type. Original magnification 20× (i,ii) and 40×/1.3NA, water objective (v,vi). (D) Summary of phenotypes observed in Egfl7 transgenic retinal vasculature at P5. Images of the retinal vasculature were captured on the TCS AOBS SP2 microscope (Leica), and filopodia was imaged using the LSM 5Live DuoScan microscope (Carl Zeiss).
EGFL7 shows hallmarks of a Notch antagonist in primary endothelial cells
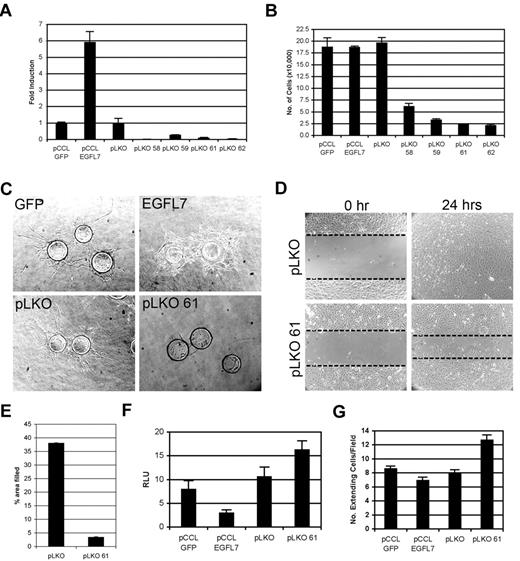
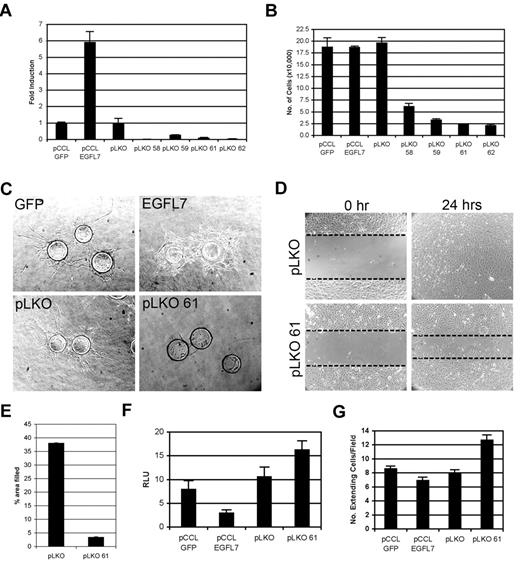
The hyperangiogenic response observed in the Tie2-Egfl7 retinas is similar to that found when Notch function is disrupted during retinal angiogenesis.18-21 We therefore characterized EGFL7 function in endothelial cells and determined its effect on Notch signaling by in vitro angiogenesis assays using HUVECs. EGFL7 expression was modulated by lentiviruses either encoding human EGFL7 (pCCLEGFL7) or shRNA against 4 different regions of EGFL7 (pLKO58, 59, 61, and 62). Quantitative RT-PCR analysis demonstrated a 6-fold increase in ectopic EGFL7 expression in pCCLEGFL7 cells and a 70% to 99% knockdown in the EGFL7 shRNA lines (Figure 6A). The level of miR126 was similar in all cell lines tested (data not shown).
EGFL7 functions as an antagonist of Notch in HUVECs. (A) Human EGFL7 expression in lentivirally generated HUVEC lines, as measured by quantitative RT-PCR. Empty lentiviruses pCCL GFP and pLKO served as controls for EGFL7 overexpression and knockdown, respectively. Data are represented as fold induction relative to the lentiviral controls. (B) Proliferation assay of HUVEC lines grown in complete medium for 4 days. (C) Capillary sprouting assay. Control (GFP and pLKO), EGFL7 overexpressing (EGFL7), and knockdown (pLKO 61) HUVEC lines were coated on a cytodex bead, beads embedded in fibrin gels, and visualized on day 7. (D) HUVEC monolayer wounding assay at 0 and 24 hours. Dotted lines highlight the edges of the monolayer. (E) Quantitation of HUVEC migration in monolayer wounding assay, represented as percentage of area filled at 8 hours. (F) Transactivation of Notch/CSL-luciferase reporter in EGFL7 overexpressing and knockdown HUVEC lines. Data represented as relative luciferase units (RLU). (G) Notch4-dependent HUVEC morphogenesis assay. Control HUVECs (pCCL-GFP or pLKO) or EGFL7 overexpressing (EGFL7) or knockdown (pLKO61) cells were grown as monolayer on a fibrin gel. GFP- or Notch4/GFP-expressing HUVECs were overlaid on top of the monolayer. At day 7, cocultures were visualized and the number of GFP+ cells undergoing morphogenesis, as seen by the extending of processes into the surrounding matrix per field, was determined. Experiments were performed in triplicate, and error bars represent SD.
EGFL7 functions as an antagonist of Notch in HUVECs. (A) Human EGFL7 expression in lentivirally generated HUVEC lines, as measured by quantitative RT-PCR. Empty lentiviruses pCCL GFP and pLKO served as controls for EGFL7 overexpression and knockdown, respectively. Data are represented as fold induction relative to the lentiviral controls. (B) Proliferation assay of HUVEC lines grown in complete medium for 4 days. (C) Capillary sprouting assay. Control (GFP and pLKO), EGFL7 overexpressing (EGFL7), and knockdown (pLKO 61) HUVEC lines were coated on a cytodex bead, beads embedded in fibrin gels, and visualized on day 7. (D) HUVEC monolayer wounding assay at 0 and 24 hours. Dotted lines highlight the edges of the monolayer. (E) Quantitation of HUVEC migration in monolayer wounding assay, represented as percentage of area filled at 8 hours. (F) Transactivation of Notch/CSL-luciferase reporter in EGFL7 overexpressing and knockdown HUVEC lines. Data represented as relative luciferase units (RLU). (G) Notch4-dependent HUVEC morphogenesis assay. Control HUVECs (pCCL-GFP or pLKO) or EGFL7 overexpressing (EGFL7) or knockdown (pLKO61) cells were grown as monolayer on a fibrin gel. GFP- or Notch4/GFP-expressing HUVECs were overlaid on top of the monolayer. At day 7, cocultures were visualized and the number of GFP+ cells undergoing morphogenesis, as seen by the extending of processes into the surrounding matrix per field, was determined. Experiments were performed in triplicate, and error bars represent SD.
In cultured human endothelial cells, activation of Notch signaling has been shown to inhibit endothelial cell proliferation, new sprout formation, and migration.22-27 EGFL7 knockdown strongly suppressed endothelial cell proliferation in all lines tested (Figure 6B). Therefore, we selected one HUVEC line, pLKO61, for further experiments. EGFL7 overexpression had no effect on proliferation (Figure 6B).
The ability of the HUVEC lines to form a capillary network was evaluated using a capillary sprouting assay26,28 (Figure 6C). HUVECs overexpressing EGFL7 failed to form the uniform capillary network observed in control cells but instead developed into an overgrown monolayer without dominant sprouts (Figure 6C). In contrast, EGFL7 knockdown resulted in a failure of the HUVECs to sprout (Figure 6C).
The function of EGFL7 in HUVEC migration was examined using a monolayer-wounding assay (Figure 6D). Cells were grown to confluence, a region was stripped away, and migration into the stripped area was monitored at 0, 8, and 24 hours. Knockdown of EGFL7 significantly reduced HUVEC migration, as seen by a failure of the pLKO61 HUVECs to completely migrate into the wounded area after 24 hours (Figure 6D). At 8 hours, migration of the knockdown cells was reduced by more than 85% relative to the control (Figure 6E). We found that blocking Notch signaling with the γ-secretase inhibitor, compound E, partially rescued migration of EGFL7 knockdown cells (supplemental Figure 4). This further supports our findings that loss of EGFL7 correlated with an increase in endogenous Notch signaling.
In summary, EGFL7 overexpression in HUVECs leads to a dysregulated and hyperangiogenic response, similar to that found when Notch signaling is suppressed. Knockdown of EGFL7 suppresses endothelial cell proliferation, angiogenic sprouting, and migration, reminiscent of Notch activation.
To further explore the hypothesis that Egfl7 affects endothelial cell behavior via modulation of Notch, the effect of altering the level of EGFL7 on Notch signaling was determined (Figure 6F). The lentiviral-infected HUVEC lines were transfected with a Notch-dependent CSL-luciferase reporter plasmid. EGFL7 overexpression resulted in a 2-fold decrease in luciferase reporter activity (Figure 6F). In contrast, reporter activity in pLKO61-infected HUVECs was increased by 50% (Figure 6F). Next, the effect of altering EGFL7 expression on Notch-dependent HUVEC morphogenesis was tested (Figure 6G). We have previously shown that overexpressing NOTCH4 in HUVECs promotes formation of cellular processes.13 Here, NOTCH4/GFP-overexpressing HUVECs were added to a HUVEC monolayer infected with pCCLEGFL7, pLKO61, or an empty vector control. In the absence of ectopic NOTCH4, the morphology of the control GFP-expressing cells remained unchanged, independent of the level of EGFL7 expression in the monolayer (data not shown). Ectopic expression of EGFL7 in the monolayer cells reduced the number of NOTCH4/GFP HUVECs with altered morphology, whereas EGFL7 knockdown resulted in an increase in the number of NOTCH4/GFP HUVECs that sent out processes (Figure 6G). Together, these data suggest that in primary endothelial cells EGFL7 functions to antagonize Notch signaling and is consistent with the notion that Egfl7 overexpression in vivo may disrupt Notch function during retinal angiogenesis.
EGFL7 interacts with Notch receptors in vitro and in vivo
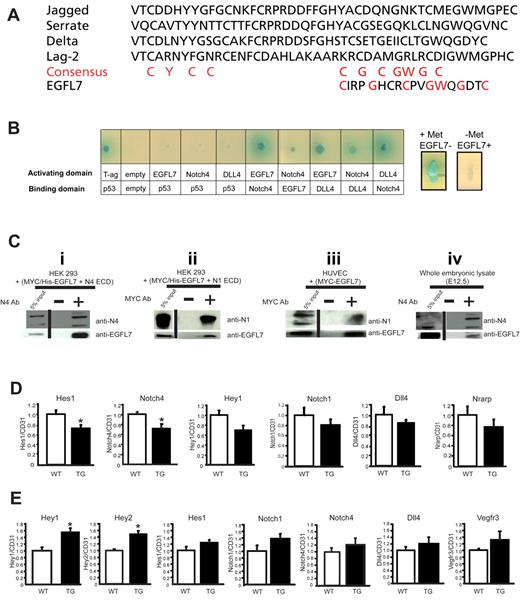
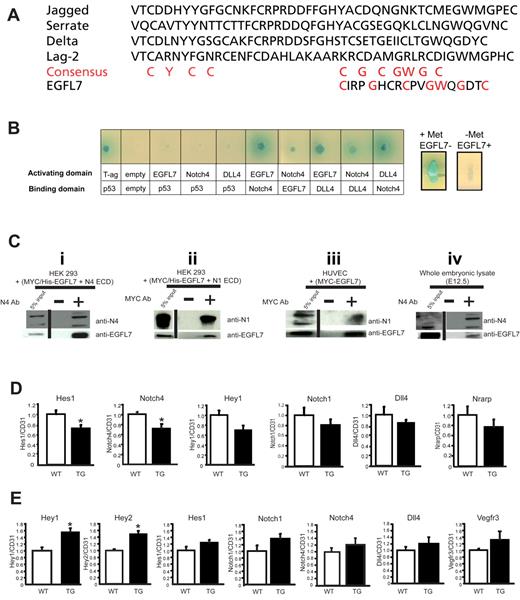
We have previously shown that EGFL7 contains a putative DSL domain.2 Alignment of the DSL-like sequence of EGFL7 with the DSL domain of known Notch ligands revealed that the spacing of critical cysteine residues in the carboxyl-terminal half of this motif is conserved (Figure 7A). Furthermore, NOTCH4 and EGFL7 share a similar expression pattern (supplemental Figure 2).2,29 To determine whether EGFL7 interacts with Notch, we used a yeast-2-hybrid system (Figure 7B). Full-length Egfl7, Dll4, or the extracellular domain (ECD) of Notch4 was fused to either the DNA-binding domain or the transcriptional activation domain of Gal4. The Notch ligand DLL4 was used as a control. Protein-protein interactions were monitored by the ability of transformed yeast to grow in defined medium and express α-galactosidase. We found that EGFL7 interacted with NOTCH4, and this interaction appeared to be at least as robust as the control NOTCH4-DLL4 interaction (Figure 7B left panel). Interestingly, EGFL7 also interacted with DLL4 (Figure 7B left panel). To test whether EGFL7 and DLL4 compete for NOTCH4 binding, a yeast-3-hybrid assay was performed (Figure 7B right panels). Induced EGFL7 overexpression disrupted the NOTCH4-DLL4 interaction, as observed by the loss of α-galactosidase expression (Figure 7B right panels). This finding suggests that EGFL7 and DLL4 bind to a similar region on NOTCH4.
EGFL7 interacts with Notch receptors and regulates Notch target gene expression in vivo. (A) Alignment of the DSL domain of Jagged1, Serrate, Delta, and Lag-2 with the putative DSL domain in EGFL7. Red letters represent the consensus sequence. (B) Yeast-2-hybrid assay (left panel): EGFL7 interacts with NOTCH4 and DLL4. Full-length EGFL7, DLL4, or the extracellular domain of NOTCH4 were fused to either the DNA-binding domain or the transcriptional activation domain of GAL4, and protein-protein interactions were monitored by the ability of the transformed yeast to grow on defined medium, and expression of α-galactosidase. Yeast-3-hybrid assay (right panels): EGFL7 abolishes NOTCH4-DLL4 interaction. The Egfl7 ORF was cloned downstream of a methionine repressible promoter (Met25) and transformed into a yeast strain expressing Notch4-GAL activating domain and DLL4-GAL4 DNA-binding domain fusions. Expression of α-galactosidase was then assayed on X-gal selection plates with or without methionine. (Ci-ii) Coimmunoprecipitation assays with protein extracts prepared from HEK293 cells transfected with plasmids encoding MYC/His-tagged-Egfl7 and Notch4 ECD or Notch1 ECD. (i) A NOTCH4 antibody was used to immunoprecipitate NOTCH4 ECD, and protein complexes were probed for NOTCH4 and EGFL7 by Western blot. (ii) A MYC antibody was used to immunoprecipitate EGFL7, and protein complexes were probed for NOTCH1 and EGFL7 by Western blot. (iii) Coimmunoprecipitation assay with protein extracts prepared from HUVECs infected with an adenovirus encoding MYC-tagged-Egfl7. A MYC antibody was used to immunoprecipitate EGFL7, and protein complexes were probed for NOTCH1 and EGFL7 by Western blot. (iv) Coimmunoprecipitation assays using protein lysates prepared from E12.5 embryos. An antibody against NOTCH4 was used to immunoprecipitate NOTCH4, and protein complexes were probed for NOTCH4 and EGFL7 by Western blot. Vertical lines have been inserted to indicate a repositioned gel lane. (D) Notch target gene expression in wild-type (□, n = 4) and Tie2-Egfl7 transgenic (■, n = 6) retinas. Gene expression was measured by quantitative RT-PCR and normalized to endothelial cell number using CD31 expression. (E) Notch target gene expression in wild-type (white bars, n = 6) and Tie2-Egfl7 transgenic embryos (black bars, n = 4). Data are represented as fold change compared with wild-type. *P < .05.
EGFL7 interacts with Notch receptors and regulates Notch target gene expression in vivo. (A) Alignment of the DSL domain of Jagged1, Serrate, Delta, and Lag-2 with the putative DSL domain in EGFL7. Red letters represent the consensus sequence. (B) Yeast-2-hybrid assay (left panel): EGFL7 interacts with NOTCH4 and DLL4. Full-length EGFL7, DLL4, or the extracellular domain of NOTCH4 were fused to either the DNA-binding domain or the transcriptional activation domain of GAL4, and protein-protein interactions were monitored by the ability of the transformed yeast to grow on defined medium, and expression of α-galactosidase. Yeast-3-hybrid assay (right panels): EGFL7 abolishes NOTCH4-DLL4 interaction. The Egfl7 ORF was cloned downstream of a methionine repressible promoter (Met25) and transformed into a yeast strain expressing Notch4-GAL activating domain and DLL4-GAL4 DNA-binding domain fusions. Expression of α-galactosidase was then assayed on X-gal selection plates with or without methionine. (Ci-ii) Coimmunoprecipitation assays with protein extracts prepared from HEK293 cells transfected with plasmids encoding MYC/His-tagged-Egfl7 and Notch4 ECD or Notch1 ECD. (i) A NOTCH4 antibody was used to immunoprecipitate NOTCH4 ECD, and protein complexes were probed for NOTCH4 and EGFL7 by Western blot. (ii) A MYC antibody was used to immunoprecipitate EGFL7, and protein complexes were probed for NOTCH1 and EGFL7 by Western blot. (iii) Coimmunoprecipitation assay with protein extracts prepared from HUVECs infected with an adenovirus encoding MYC-tagged-Egfl7. A MYC antibody was used to immunoprecipitate EGFL7, and protein complexes were probed for NOTCH1 and EGFL7 by Western blot. (iv) Coimmunoprecipitation assays using protein lysates prepared from E12.5 embryos. An antibody against NOTCH4 was used to immunoprecipitate NOTCH4, and protein complexes were probed for NOTCH4 and EGFL7 by Western blot. Vertical lines have been inserted to indicate a repositioned gel lane. (D) Notch target gene expression in wild-type (□, n = 4) and Tie2-Egfl7 transgenic (■, n = 6) retinas. Gene expression was measured by quantitative RT-PCR and normalized to endothelial cell number using CD31 expression. (E) Notch target gene expression in wild-type (white bars, n = 6) and Tie2-Egfl7 transgenic embryos (black bars, n = 4). Data are represented as fold change compared with wild-type. *P < .05.
To test for direct interaction between EGFL7 and NOTCH4, we investigated whether the 2 proteins could coimmunoprecipitate (Figure 7C). Plasmids expressing a MYC/his-tagged Egfl7 cDNA (pcDNA4(MycHis)Egfl7) and the ECD of Notch4 (pHyTC-N4Fc) were cotransfected into HEK293 cells. NOTCH4-containing complexes were immunoprecipitated with a NOTCH4-specific antibody, and coimmunoprecipitated EGFL7 was detected on a Western blot using an antibody against EGFL7 (Figure 7Ci). These data showed that NOTCH4 and EGFL7 directly interact. When we repeated this experiment using the ECD of NOTCH1 (pHyTC-N1Fc) and an anti-MYC antibody to pull-down MYC-tagged EGFL7, we found that EGFL7 and NOTCH1 also formed a complex (Figure 7Cii). In addition, we observed an interaction between MYC-tagged EGFL7 and endogenous NOTCH1 when EGFL7 was overexpressed in HUVECs (Figure 7Ciii). This is consistent with a recent report that detected the same interaction in HUVECs and adult mouse brain.5
To determine whether EGFL7 and NOTCH4 interact in vivo, coimmunoprecipitation experiments were repeated using protein lysate prepared from whole wild-type E12.5 embryos. Endogenous NOTCH4 was immunoprecipitated by the NOTCH4 antibody and endogenous EGFL7 was readily detectable by Western blot (Figure 7Civ). Binding was also observed using protein lysate prepared from E10.5 embryos (data not shown). These findings confirm that EGFL7 and NOTCH4 are capable of interacting during embryonic development in vivo.
Egfl7 overexpression results in altered Notch target gene expression in vivo
To determine whether EGFL7 binding to Notch receptors affected downstream signaling in vivo, Notch target gene expression was measured in P5 retinas by quantitative RT-PCR and normalized to endothelial cell number using CD31 expression (Figure 7D). Hes1 and Notch4 expression was significantly down-regulated by 30% in the Tie2-Egfl7 transgenic mice (Figure 7D). Hey1, Notch1, Dll4, and Nrarp transcripts were also reduced by 20% to 30% (Figure 7D). These results support the notion that Egfl7 acts as a Notch antagonist during postnatal angiogenesis and are consistent with the phenotypes observed in the retina and in HUVECs.
To test whether EGFL7 overexpression also affected the levels of Notch targets during embryogenesis, we measured gene expression in E12.5 wild-type and transgenic embryos (Figure 7E). Interestingly, expression of the transcription factors Hey1 and Hey2 showed a 60% and 40% significant increase, respectively, in Tie2-Egfl7 embryos (Figure 7E). Expression of several other Notch targets, including Hes1, Notch1, Notch4, Dll4, and Vegfr3 (Figure 7E), was also up-regulated, whereas the levels of Vegfr2 and EphrinB2 were unaltered (supplemental Figure 5). Thus, in contrast to the postnatal retina, embryonic overexpression of Eglf7 results in an increase in the level of some, but not all, Notch target genes.
Discussion
The expression of Egfl7 at sites of endothelial progenitors2,7 led us to hypothesize that this factor has a critical role in the developing vasculature. Here, we provide evidence that Egfl7 has distinct functions during mammalian vascular development and that at least some of these functions are mediated through its modulation of the Notch signaling pathway. A 2-fold increase of Egfl7 expression results in partial embryonic lethality and is accompanied by a wide range of angiogenic defects during both embryonic and postnatal vascular development. The phenotypes observed are not completely penetrant, and this may reflect a dosage-dependent effect of Egfl7. It is probable that those mice homozygous for the transgene will exhibit a more pronounced phenotype. Severity of the phenotype decreases as development proceeds, and this correlates with the loss of transgenic embryos between E10.5 and E12.5.
The in vivo gain-of-function findings are corroborated by our studies carried out in primary endothelial cells. EGFL7 overexpression in HUVECs results in a hyperangiogenic response, whereas knockdown causes a reduction in cell proliferation, sprouting, and migration. Egfl7 knockdown in zebrafish also causes vascular abnormalities, including defects in the extension and junctional arrangements of endothelial cells and tubulogenesis.3,8 We have previously shown that shRNA-mediated knockdown of Egfl7 during embryonic stem cell differentiation results in abnormal endothelial cords and the formation of CD31+ cell aggregates.30 Interestingly, Egfl7 loss of function in mice does not appear to cause any obvious abnormalities.10 The different results obtained from Egfl7 knockdown in vitro and knockout in vivo could be explained by compensating factors in the developing embryos or differences in genetic background strains. It is also possible that Egfl7 loss of function in mice causes a subtle and transient phenotype, early in vascular development, similar to what is observed during embryonic stem cell differentiation.30
We showed that EGFL7 interacts with endothelial-specific NOTCH. Several pieces of evidence suggest that Egfl7 acts as a Notch antagonist in both the postnatal retina and in human primary endothelial cells. First, EGFL7 knockdown in HUVECs results in cellular responses similar to Notch activation.22-28 Second, EGFL7 inhibits Notch signaling in a CSL-luciferase reporter assay and blocks Notch-dependent morphogenesis. Third, Egfl7 overexpression significantly down-regulates expression of Notch target genes in the postnatal retina. This results in a hyperangiogenic response that is reminiscent of Notch inhibition. In support, a similar antagonistic role for Egfl7 on the Notch pathway was proposed in cultured adult neural stem cells.5 The mutant phenotype in the transgenic postnatal retina was less severe than previously reported for inhibition of Notch signaling.18-21,31 It is possible that EGFL7 predominantly functions as an antagonist of NOTCH4 and, to a lesser extent, NOTCH1. This is consistent with the similar localization of EGFL7 and NOTCH4 on sprouting retinal vessels and the preferential down-regulation of Notch4 transcripts in the transgenic retina. In addition, Notch1 inhibition alone is sufficient to disrupt mammalian retinal angiogenesis, whereas Notch4 inhibition is not necessary.31 This may explain the subtlety of our retinal phenotype.
In contrast to the postnatal retina, embryonic overexpression of Egfl7 significantly increases the levels of the Notch target transcription factors Hey1 and Hey2, suggesting that Egfl7 may act as an agonist of Notch signaling during embryogenesis. To reconcile this difference, we propose that the mode of EGFL7-Notch interactions depends on the microenvironment within a particular vascular bed, similar to the way autocrine and paracrine vascular endothelial growth factor signaling differs in their ability to regulate vascular homeostasis and angiogenesis.32 Specifically, it is probable that the presence of EGFL7 in a secreted- or extracellular matrix-bound form, the availability of Notch receptors and ligands, as well as the post-translational modifications that have been reported for JAGGED1- and DLL4-Notch interactions18 will dictate the way EGFL7 affects the Notch pathway.
Although many Notch targets are up-regulated in Tie2-Egfl7 embryos, these embryos did not recapitulate the phenotypes associated with Notch gain-of-function mutations, including an enlarged dorsal aorta and arterialization of vessels.29,33,34 This may be explained by the level at which Egfl7 is overexpressed. The 2-fold overexpression in Tie2-Egfl7 embryos is lower than the levels of overexpression observed for the Dll4 transgenic model34 or in mice that express a constitutively active form of Notch4.33 It is also conceivable that Egfl7 up-regulates Hey1 and Hey2 expression independent of Notch, possibly through basic fibroblast growth factor.35 Recently, Egfl7 has been shown to promote cell migration in hepatocellular carcinoma cells via the epidermal growth factor receptor pathway.36 Therefore, Egfl7 may also modulate these pathways during angiogenesis.
By what mechanism does Egfl7 modulate the Notch pathway? In general, Notch ligands are membrane-bound and interact with Notch receptors on an opposing cell. Truncated, secreted versions of the ligands are capable of acting as inhibitors of Notch signaling.37-39 Notch ligands can also inhibit Notch in a cell autonomous manner, known as cis-inhibition, when receptor and ligand are coexpressed in the same cell.40 Our data suggest that, in the postnatal retina and in primary endothelial cells, Egfl7 acts as a Notch antagonist and it does so by binding to the Notch receptor and/or its corresponding ligand, thus preventing ligand-receptor interaction (supplemental Figure 6). In support, we have shown that EGFL7 is not only capable of binding to the Notch receptors and DLL4 but is also capable of disrupting the receptor-ligand interaction. This could be achieved both cell autonomously and nonautonomously, and data obtained from the CSL-dependent Notch reporter assay and the endothelial morphogenesis coculture assay suggest that either is plausible. In the embryo, it is possible that extracellular matrix-bound EGFL7 interacts with Notch receptors on opposing cells and exerts its agonistic effect in this way.
In conclusion, by overexpressing Egfl7 in vivo and manipulating EGFL7 levels in HUVEC, we uncovered a critical role for this factor in vascular development. We also showed, for the first time, that EGFL7 physically interacts with endothelial NOTCH and that it is capable of modulating Notch signaling in vivo.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Sergey Kuprianov (TSRI Mouse Genetics Core) for Tie2-Egfl7 mice, MSKCC Molecular Cytology for assistance with imaging, Stefano Rivella (WCMC) for the pCCLIRESGFP, Christopher Kitajewski for reagents, David Nichol for illustration, and Andrea Victor for generating the pCCLEGFL7 plasmid and animal husbandry.
This work was supported by a Tri-Starr Stem Cell Scholar Postdoctoral Fellowship (D.N.) and the National Institutes of Health (grants K01DK744629 and R01CA136673, C.S.; grant R01HL082098, H.S.).
National Institutes of Health
Authorship
Contribution: D.N., C.S., J.K., and H.S. designed the research and analyzed the data; D.N., C.S., M.J.F., K.B., and A.S. performed the research; and D.N. and H.S. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Heidi Stuhlmann, Department of Cell and Developmental Biology, Weill Cornell Medical College, 1300 York Ave, New York, NY 10065; e-mail: hes2011@med.cornell.edu.