Abstract
Sequence variants at the TERT-CLPTM1L locus in chromosome 5p have been recently associated with disposition for various cancers. Here we show that this locus including the gene encoding the telomerase reverse-transcriptase TERT at 5p13.33 is rarely but recurrently targeted by somatic chromosomal translocations to IGH and non-IG loci in B-cell neoplasms, including acute lymphoblastic leukemia, chronic lymphocytic leukemia, mantle cell lymphoma and splenic marginal zone lymphoma. In addition, cases with genomic amplification of TERT locus were identified. Tumors bearing chromosomal aberrations involving TERT showed higher TERT transcriptional expression and increased telomerase activity. These data suggest that deregulation of TERT gene by chromosomal abnormalities leading to increased telomerase activity might contribute to B-cell lymphomagenesis.
Introduction
Chromosomal translocations to the immunoglobulin heavy chain (IGH) or light chain (IGL or IGK) loci, and less frequently to non-IG loci represent well known mechanisms of oncogene activation in B-cell neoplasms.1,2 Many of the genes deregulated by chromosomal translocations in these neoplasms are involved in important cellular processes like cell-cycle control (CCND1, CCND3, BCL6, and CDK6), apoptosis (BCL2), proliferation (MYC), or signal transduction (BCL3 and MALT1).1
Telomere maintenance across cell divisions is essential for tumor cell growth and immortalization.3 Here, we provide evidence that the telomerase reverse-transcriptase (TERT) gene at 5p13.33, which encodes for the rate-limiting catalytic protein subunit of the telomerase,4-6 is deregulated by chromosomal translocations to IG and non-IG loci in precursor and mature B-cell malignancies.
Methods
Patient samples
The features of B-cell malignancies showing aberrations in the TERT region (5p1) are listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The study was performed as part of the “Molecular Mechanisms in Malignant Lymphoma” Network Project for which approval from the Institutional Review Board of the Medical Faculity of the Christian-Albrechts-University Kiel has been obtained.
FISH
Fluorescence in situ hybridization (FISH) was performed on fixed cells from bone marrow, peripheral blood, or lymph node cell suspensions as described previously.7 Generation of FISH probes as well as FISH procedures are described in supplemental Methods. A list of FISH probes used is shown in supplemental Table 2.
Array CGH to custom-designed arrays
The microarrays were designed using the eArray software from Agilent (https://earray.chem.agilent.com/earray/) and the 4 × 44k format. The experimental procedures were performed according to the manufacturer's instructions. Arrays were scanned with the GenePix4000B Scanner (Axon Instruments) and log ratios obtained with the comparative genomic hybridization (CGH) Analytics Version 3.5.14 software (Agilent). All microarray data are available to be viewed at ArrayExpress under accession number E-MEXP-2675 (http://www.ebi.ac.uk/miamexpress/).
Quantitative reverse transcription PCR
RNA was extracted from tumor cell–containing samples and control tissue, using the RNeasy Mini Kit (QIAGEN) and transcribed into cDNA with the QuantiTect Rev. Transcription Kit (QIAGEN). TERT transcripts were amplified and detected as described elsewhere.8
TRAP assay
The PCR-based telomeric repeat amplification protocol (TRAP) assay was performed as described previously.9,10 Protein from tumor cell containing samples and control tissue was isolated according to Kim et al.11
Additional information about materials and methods is available in supplemental Methods.
Results and discussion
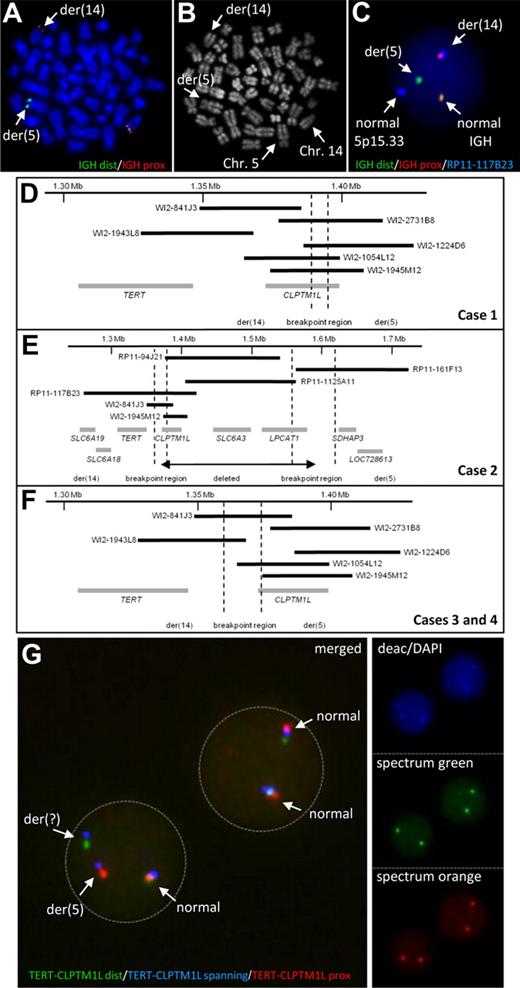
By FISH screening of B-cell neoplasms for translocations affecting the IGH locus we observed a cytogenetically cryptic translocation t(5;14)(p15;q32) involving the IGH locus in 3 cases of chronic lymphocytic leukemia (CLL) and one case of precursor B-cell acute lymphoblastic leukemia (ALL; Figure 1A-C). The breakpoints in 5p15.33 in all 4 cases mapped to a region containing the TERT (telomerase reverse transcriptase; chr5: 1.306 Mb to 1.348 Mb) and CLPTM1L (cleft lip and palate transmembrane 1–like; chr5: 1.371 Mb to 1.398 Mb) genes (Figure 1D-F, supplemental Table 1).
Characterization of chromosomal aberrations affecting the TERT-CLPTM1L locus. IGH FISH (A) and R-banding analysis (B) of the same metaphase of case 1 showing the cytogenetic cryptic t(5;14). (C) Interphase FISH showing a fusion of IGH and the TERT-CLPTM1L locus in case 1. (D-F) Schematic maps of the breakpoint regions in cases with t(5;14)(p15;q32) based on FISH mapping. Black bars indicate the hybridized BAC and Fosmid clones; gray bars represent the genes in the region. The breakpoint regions were determined by the respective FISH hybridization patterns. (G) FISH using the TERT-CLPTM1L 3-color break-apart assay on MCL case 10 showing break in the clone RP11-117B23 (blue signal). The R-banding image with Chromomycin A3 (C2659, Sigma-Aldrich) and Methyl green (M8884, Sigma-Aldrich) was obtained using a 63×/1.40 numeric aperture oil objective in a Zeiss Axioskop Imager M1 fluorescence microscope (Zeiss) with R-banding filter sets (Zeiss) and documented using the IKAROS imaging system version 5.2.11 (MetaSystems). FISH images were acquired using a 63×/1.40 numeric aperture oil objective in a Zeiss Axioskop 2 fluorescence microscope (Zeiss) equipped with the appropriate filter sets (AHF) and documented using a CV-M300 camera (JAI Corporation) and the ISIS imaging system Version 5.2.11 (MetaSystems). dist indicates distal; and prox, proximal.
Characterization of chromosomal aberrations affecting the TERT-CLPTM1L locus. IGH FISH (A) and R-banding analysis (B) of the same metaphase of case 1 showing the cytogenetic cryptic t(5;14). (C) Interphase FISH showing a fusion of IGH and the TERT-CLPTM1L locus in case 1. (D-F) Schematic maps of the breakpoint regions in cases with t(5;14)(p15;q32) based on FISH mapping. Black bars indicate the hybridized BAC and Fosmid clones; gray bars represent the genes in the region. The breakpoint regions were determined by the respective FISH hybridization patterns. (G) FISH using the TERT-CLPTM1L 3-color break-apart assay on MCL case 10 showing break in the clone RP11-117B23 (blue signal). The R-banding image with Chromomycin A3 (C2659, Sigma-Aldrich) and Methyl green (M8884, Sigma-Aldrich) was obtained using a 63×/1.40 numeric aperture oil objective in a Zeiss Axioskop Imager M1 fluorescence microscope (Zeiss) with R-banding filter sets (Zeiss) and documented using the IKAROS imaging system version 5.2.11 (MetaSystems). FISH images were acquired using a 63×/1.40 numeric aperture oil objective in a Zeiss Axioskop 2 fluorescence microscope (Zeiss) equipped with the appropriate filter sets (AHF) and documented using a CV-M300 camera (JAI Corporation) and the ISIS imaging system Version 5.2.11 (MetaSystems). dist indicates distal; and prox, proximal.
The IGH locus harbors strong transcriptional enhancers, which can activate oncogenes by translocation on both der(14) and der(5) chromosomes12 (supplemental Figure 1). FISH mapping showed the TERT gene juxtaposed to the IGH locus indicating TERT translocation to der(14) in all 4 cases. The signal patterns indicated CLPTM1L to be disrupted by the translocation in case 1 (Figure 1D). In case 2, the breakpoint was associated with an interstitial deletion resulting in loss of CLPTM1L (Figure 1E). This deletion was confirmed by high-resolution array CGH, which also allowed fine-mapping the translocation breakpoint to a repeat-rich region centromeric of TERT (1.354 Mb to 1.366 Mb; supplemental Figure 2A). The 5p15.33 breakpoints of cases 3 and 4 were shown to be between TERT and CLPTM1L (Figure 1F).
To investigate the frequency of rearrangements in the TERT-CLPTM1L region we screened additional cases by FISH (supplemental Figure 3, supplemental Table 1). Among 34 lymphoid neoplasms, in which conventional cytogenetics revealed an aberration in 5p1, 5 cases showed breaks and one case showed amplification at the TERT-CLPTM1L region. Four of these 6 cases were diagnosed as mantle cell lymphomas (MCLs). Thus, we extended the screen to 123 primary MCLs without known 5p1 abnormality and identified 2 additional cases, one with break (Figure 1G) and one with amplification at the TERT-CLPTM1L region (supplemental Table 1).
None of the additional 6 cases with translocations detected by FISH showed juxtaposition to any of the 3 IG loci. Cytogenetically, in 4 of the 6 variant translocations (ie, non-IG), 7p11, 9q31∼33, 10q25 and 19p13 were identified as partners of 5p15. In a splenic marginal zone lymphoma (SMZL, case 5) with t(5;7)(p15.33;p11), FISH and tiling array CGH again confirmed a breakpoint centromeric to TERT with loss of CLPTM1L similar to that in the t(5;14)-positive CLL mentioned above (Figure 1E, supplemental Figure 2B). Except for the SMZL case, the remaining 5 additional cases showed the same breakpoint in 5p15.33 as in cases 3 and 4 (Figure 1F).
The inactivation of CLPTM1L by deletion and disruption in 3/10 cases with break at the TERT-CLPTM1L region and also the fact that in precursor B-cell ALL with IGH translocation oncogene activation has always been assigned to the der14 chromosome13 (supplemental Figure 1) strongly supports TERT to be the gene targeted by these aberrations.
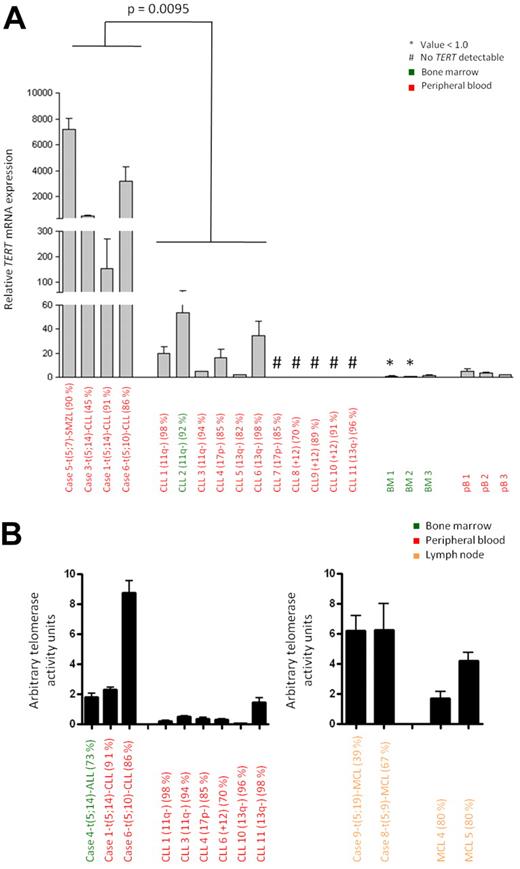
To investigate the effect of the identified chromosomal aberrations on TERT expression, we performed qRT-PCR in cases with t(5;14)/TERT-IGH juxtaposition (n = 3; Figure 2A for CLL, supplemental Figure 5 for ALL) and 5p15.33 variant translocations (n = 5; Figure 2A, supplemental Figure 4 for MCL) where suitable material was available. Significantly increased TERT mRNA expression was observed in all 3 CLLs and the SMZL with a break in the TERT-CLPTM1L region compared with eleven CLLs without aberration at the TERT-CLPTM1L region as detected by FISH (Figure 2A). Similarly, the 3 MCLs with break in 5p15.33 showed higher TERT mRNA expression compared with 5 MCLs without break in the TERT-CLPTM1L region and controls (supplemental Figure 4). The ALL with t(5;14) displayed high TERT expression but this was also detected in 6 ALL cases without such a change (supplemental Figure 5). These findings show that independent of the presence of centromeric (CLPTM1L) deletions or the translocation partner, rearrangements affecting the TERT-CLPTM1L region are associated with high TERT expression.
TERT expression and telomerase activity of B-cell malignancies with and without aberration in the TERT-CLPTM1L locus. (A) Relative TERT mRNA expression determined by qRT-PCR. Cases 1, 3, 5, and 6 with 5p15.33 break by FISH were compared with CLL 1-11 showing no 5p15.33 aberration by FISH. The P value was estimated using the Mann-Whitney test. Samples with undetectable TERT mRNA expression were excluded from statistical analysis. (B) Telomerase activity measured by TRAP assay. Cases 1, 4, and 6 and cases 8 and 9 with 5p15.33 aberration by FISH were compared with CLL cases 1, 3, 4, 6, 10 and 11 and MCL cases 4 and 5 without aberration in this region by FISH. The relevant cytogenetic aberrations and the tumor cell contents are given in brackets.
TERT expression and telomerase activity of B-cell malignancies with and without aberration in the TERT-CLPTM1L locus. (A) Relative TERT mRNA expression determined by qRT-PCR. Cases 1, 3, 5, and 6 with 5p15.33 break by FISH were compared with CLL 1-11 showing no 5p15.33 aberration by FISH. The P value was estimated using the Mann-Whitney test. Samples with undetectable TERT mRNA expression were excluded from statistical analysis. (B) Telomerase activity measured by TRAP assay. Cases 1, 4, and 6 and cases 8 and 9 with 5p15.33 aberration by FISH were compared with CLL cases 1, 3, 4, 6, 10 and 11 and MCL cases 4 and 5 without aberration in this region by FISH. The relevant cytogenetic aberrations and the tumor cell contents are given in brackets.
We next assessed whether the up-regulation of TERT mRNA was also associated with increased telomerase activity. Using the TRAP assay, cases of CLL (cases 1 and 6) and MCL (cases 8 and 9) with chromosomal breakpoints at the TERT-CLPTM1L locus showed markedly higher telomerase activity compared with appropriate disease and tissue controls (Figure 2B, supplemental Table 3).
Taken together, we show that the TERT-CLPTM1L region in 5p15 is recurrently targeted by chromosomal translocations in B-cell neoplasms. Based on the finding that CLPTM1L is recurrently lost or disrupted by the mentioned aberrations, it does not seem to contribute to the process of lymphomagenesis. Interestingly, germ line sequence variants in the TERT-CLPTM1L region were recently shown to be associated with predisposition for several cancer types.14-17 All 6 additional translocations and both amplifications identified herein were of somatic origin as cells without the aberration were always present. The pattern of IGH translocations as well as the increased TERT expression and telomerase activation in cases with 5p15 translocations strongly suggest TERT to be the candidate gene involved in lymphomagenesis. Telomerase activity, which is clearly detectable in up to 90% of human tumors, including hematologic neoplasias, but not in most normal somatic cells, is in part explained by changes in chromatin structure or amplification of the TERT locus.11,18-20 Moreover activation of TERT transcription through viral integration into the TERT promoter has been shown to be associated with B-cell lymphoma and other tumors.21,22 Remarkably, on the cytogenetic level the t(5;14)/IGH-TERT was the sole abnormality in one case of CLL, suggesting TERT deregulation to be an early event in lymphomagenesis. This is in accordance with the recent observation that mice with constitutive expression of TERT in thymocytes and peripheral T cells are prone to develop T-cell lymphomas, which are more invasive than those of wild-type mice and may be explained by the recent finding that TERT acts, independent of telomerase activity, as a cofactor in the Wnt pathway.23,24 Remarkably, TERT translocations might not be restricted to lymphatic neoplasms as a recent study by Zhao et al also identified a translocation breakpoint within CLPTM1L in the breast cancer cell line HCC1954 similar to one case of CLL (case 1) with translocation t(5;14)(p15;q32) described herein.25
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors gratefully thank Dorit Schuster, Magret Ratjen, Ursula Schnaidt, Reina Zühlke-Jenisch, Astrid Schneider, Simone Hartmann, and Claudia Becher for their technical assistance.
This work was supported by the network Project of the Deutsche Krebshilfe “Molecular Mechanisms in Malignant Lymphomas” 70-3173-TR3.
Authorship
Contribution: I.N. and R.S. designed experiments; L.H., E.C.B., R.D.G., D.H., A.M., J.A.M.C., S.S., and M.J.S.D. provided tumor samples; I.N., M.S., J.I.M.S., L.H., T.A., O.A., S.G., W.K., and H.T. generated and/or analyzed experimental data; and I.N., J.I.M.S., and R.S. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Inga Nagel, Institute of Human Genetics, Christian-Albrechts University Kiel & University Hospital Schleswig-Holstein, Campus Kiel, Schwanenweg 24, 24105 Kiel, Germany; e-mail: inagel@medgen.uni-kiel.de.