Abstract
The endosteal niche is critical for the maintenance of hematopoietic stem cells (HSCs). However, it consists of a heterogeneous population in terms of differentiation stage and function. In this study, we characterized endosteal cell populations and examined their ability to maintain HSCs. Bone marrow endosteal cells were subdivided into immature mesenchymal cell-enriched ALCAM−Sca-1+ cells, osteoblast-enriched ALCAM+Sca-1−, and ALCAM–Sca-1− cells. We found that all 3 fractions maintained long-term reconstitution (LTR) activity of HSCs in an in vitro culture. In particular, ALCAM+Sca-1− cells significantly enhanced the LTR activity of HSCs by the up-regulation of homing- and cell adhesion–related genes in HSCs. Microarray analysis showed that ALCAM−Sca-1+ fraction highly expressed cytokine-related genes, whereas the ALCAM+Sca-1− fraction expressed multiple cell adhesion molecules, such as cadherins, at a greater level than the other fractions, indicating that the interaction between HSCs and osteoblasts via cell adhesion molecules enhanced the LTR activity of HSCs. Furthermore, we found an osteoblastic markerlow/− subpopulation in ALCAM+Sca-1− fraction that expressed cytokines, such as Angpt1 and Thpo, and stem cell marker genes. Altogether, these data suggest that multiple subsets of osteoblasts and mesenchymal progenitor cells constitute the endosteal niche and regulate HSCs in adult bone marrow.
Introduction
During postnatal life, bone marrow (BM) supports both self-renewal and differentiation of hematopoietic stem cells (HSCs) in specialized niches. These niches, which are composed of cellular components located around stem cells, facilitate the signaling networks that control the balance between self-renewal and differentiation.1–5 Long-term HSCs (LT-HSCs) are retained in a quiescent state in the BM, where they are anchored to specialized niches along the endosteum (the border between the bone and the BM) and in perivascular sites adjacent to the endothelium. Cytokines, chemokines, adhesion molecules, proteolytic enzymes, neurotransmitters, and transcription factors regulate the balance between quiescence and activation (proliferation and migration) of HSCs.4,6 In particular, Angiopoietin-1 (Angpt1),5 Kit-ligand (Kitl),7,8 chemokine (C-X-C motif) ligand 12 (Cxcl12),9–12 thrombopoietin (Thpo),13,14 Wnt,15 Jagged1 (Jag1),16 osteopontin (OPN),17,18 and N-cadherin (Cdh2)19–21 are known to be involved in the niche regulation of HSCs in BM, and cooperative regulation among cytokine signals and cell adhesion molecules is required for the maintenance of HSCs. We previously reported that Tie2/Angpt1 and Mpl/Thpo signaling between HSCs and the endosteal (osteoblastic) niche cells plays a critical role in the enhancement of cell-to-cell and cell-to-extracellular matrix interactions of HSCs with niche cells and in the maintenance of cell-cycle quiescence of HSCs. Therefore, the functionality of the endosteal niche cells is critical for maintaining a sufficient reserve of LT-HSCs.14,22 Because the cells in the endosteum are a heterogeneous population in terms of their degree of differentiation and accompanying functions,23,24 the precise cellular and molecular contribution of endosteal cell populations to the HSC-supportive microenvironment is still unclear.
In addition to osteoblasts, other mesenchymal cells, including mesenchymal stem/progenitor cells (MSCs/MPCs), reticular cells, adipocytes, and other stromal cells, also have been implicated in regulation of HSC maintenance in the endosteal niche.11,25,26 In addition, osteoclasts play a critical role in progenitor mobilization.10 Furthermore, it has been shown that the vascular endothelial network is distributed throughout BM and extends into the outermost region close to the endosteal surfaces, raising the possibility that HSCs, osteoblasts, and the perivascular niche have a close relationship in the regulation of hematopoiesis, bone formation, and vascular remodeling.27,28
Activated leukocyte cell-adhesion molecule (ALCAM) is a member of the immunoglobulin superfamily and mediates homophilic and heterophilic (ALCAM-CD6) adhesion.29 ALCAM is expressed on HSCs,30 metastasizing melanoma,31 neuronal cells,32 endothelial cells,30 hematopoiesis-supporting osteoblastic cell lines,33,34 and MSCs.35–37 We previously reported that ALCAM was expressed in the perichondrium in mouse embryonic limb.35 ALCAM+ perichondrial cells exhibited multilineage differentiation capacity, and ALCAM-ALCAM homophilic interactions supported the maintenance of MSCs. We also reported that ALCAM was involved in the maintenance of HSCs and primitive hematopoietic progenitor cells.30,35 In addition, ALCAM plays a critical role in angiogenesis.30 Therefore, it is reasonable to hypothesize that ALCAM-mediated homophilic cell adhesion among HSCs, MSCs, and vascular endothelial cells is associated with the mechanism of HSC maintenance in adult BM niches.
In this study, we identified subpopulations of adult BM endosteal cells by the expression of ALCAM and Sca-1. Nonhematopoietic and nonendothelial CD45−CD31−Ter119− endosteal cells were subdivided into 3 fractions: ALCAM+Sca-1−, ALCAM−Sca-1+, and ALCAM−Sca-1− cells. We found that osteoblasts were enriched in the Sca-1− population, whereas immature mesenchymal cells were enriched in the ALCAM−Sca-1+ fraction. All 3 fractions maintained long-term reconstitution (LTR) activity, and ALCAM+Sca-1− cells in particular showed robust supporting activity for HSCs. Microarray analysis showed that cytokine-related genes were highly expressed in the ALCAM−Sca-1+ fraction. In contrast, compared with other fractions, the ALCAM+Sca-1− fraction showed greater levels of expression of genes related to cell adhesion. Furthermore, by using single-cell gene expression analysis, we found a subpopulation in the ALCAM+Sca-1− fraction that expressed genes encoding cytokines but not osteoblastic markers (osteoblast markerlow/−). In addition, some of these osteoblast markerlow/− ALCAM+Sca-1− cells expressed pluripotency markers. These data suggest that multiple subsets of osteoblasts and MPCs constitute the endosteal niche in the adult BM.
Methods
Mice
C57BL/6 mice were purchased from Japan SLC. C57BL/6 mice congenic for the Ly5 locus (B6-Ly5.1) were purchased from Sankyo-Lab Service. Animals were cared for in accordance with the guidelines of the Keio University School of Medicine. All animal experiments were approved by the Keio University School of Medicine ethical review board.
Antibodies
The following antibodies (Abs) were used for fluorescence-activated cell-sorting (FACS) analysis and cell sorting: anti-CD45 (30-F11; BD Biosciences), -CD31 (MEC 13.3; BD Biosciences), -Ter119 (eBioscience), –Sca-1 (E13-161.7; BD Biosciences), -PDGFRα (APA5, a gift from Dr Takakura, Osaka University), -ALCAM (R&D Systems), -CD45.1 (A20; BD Biosciences), -CD45.2 (104; BD Biosciences), -CD3 (500A2; BD Biosciences), -B220 (RA3-6B2; BD Biosciences), –Mac-1 (M1/70; BD Biosciences), -Gr-1 (RB6-8C5; BD Biosciences), -CD4 (RM4-5; BD Biosciences), -CD8 (53-6.7; BD Biosciences), -CD41 (MWReg30; BD Biosciences), and –c-Kit (2B8; BD Biosciences). A mixture of CD4, CD8, B220, Ter119, Mac-1, and Gr-1 was used as the lineage mix. The following Abs were used for immunohistochemical staining of BM sections: anti-CD45, -CD31, -ALCAM, –Sca-1, and -osteocalcin (OCN; Takara Bio Inc).
Cell preparation and sorting
Endosteal cells were isolated by the use of published procedures, with minor modifications.38 It has been reported that the collagenase treatment of bone is applicable for the isolation of osteoblasts from marrow-depleted bone.39 In addition, collagenase treatment increased the yield of the MSC population.40 After we flushed out the BM cells of femurs and tibias, the bones were minced with scissors. Then, bone fragments were incubated at 37°C with a type I collagenase (3 mg/mL; Worthington) in Dulbecco modified Eagle medium with 10% fetal calf serum and gently agitated for 90 minutes at 37°C. The dissociated cells were collected, and bone-associated mononuclear cells (MNCs) were isolated with the use of density centrifugation with Lymphoprep (Axis-Shield). Subsequently, the cells were stained with phycoerythrin-conjugated anti-CD45, CD31, Ter119, fluorescein isothiocyanate–conjugated anti-Sca1, biotinylated anti-ALCAM, and streptavidin-allophycocyanin. Propidium iodide was used for identifying and excluding dead cells. In the case of the sorting of the endosteal cell population, CD45−CD31−Ter119− endosteal cells were enriched by magnetic cell sorting (Miltenyi Biotec) by the use of antiphycoerythrin microbeads (Miltenyi Biotec) before FACS sorting. The stained cells were analyzed and sorted by FACSVantage (BD Biosciences). Because the contaminating subpopulations could result in the different populations being less distinctive, we reanalyzed the purity of the isolated endosteal cell fractions by FACS and confirmed that the purity of each endosteal fraction was reproducibly greater than 98%.
Immunohistochemical staining
Preparation of BM sections and the procedure for fluorescent immunohistochemistry were described previously.22 Fluorescent images were obtained by the use of a confocal laser-scanning microscope, FV1000 (Olympus).
Cell culture
Fractionated cells were maintained in Mesenchymal Stem Cell Basal Medium (Lonza). For osteogenic induction, cells were cultured for 7 days in osteogenic induction medium (Lonza). Osteoblastic differentiation was evaluated by alkaline phosphatase (ALP) staining. Calcium deposits were stained with 1% alizarin red. For adipogenic induction, cells were cultured in adipogenic induction medium (Lonza), and after 1 week of cultivation, the medium was changed to adipogenic maintenance medium (Lonza) for an additional 7 days. Adipogenic differentiation was assessed by the accumulation of lipid vesicles by the use of an oil red O staining. Cells were counterstained with hematoxylin. Chondrogenic potential was assayed for by culturing cells in Differentiation Basal Medium Chondrogenic (Lonza), supplemented with Chondrogenic SingleQuots kit (Lonza), and 10 ng/mL of transforming growth factor-β3 (TGF-β3; Lonza) for 2 weeks. After cultivation, cells were stained with alcian blue. The high-density pellet culture using ALCAM−Sca-1+ cells was performed as described previously.41 After 3 weeks of culture, the pellet was fixed and stained with alcian blue.
Colony assay and BM reconstitution assay
For the colony assay, BM-derived lineage−Sca-1+c-Kit+ cells (LSK cells, 1 × 104 cells) and fractionated bone-derived cells were cocultured in Myelocult for 5 days. Cultured cells were trypsinized, and CD45.1 cells were sorted. Colony-forming assays were performed with methylcellulose medium (Methocult M3434; StemCell Technologies). The number of colonies containing more than 50 cells was scored on day 7, and colony morphology was evaluated under a microscope.
For the BM reconstitution assay, BM-derived LSK cells (Ly5.1+, 1 × 104 cells) and fractionated bone-derived cells were cocultured in Myelocult. CD45.1+ cells were sorted after 2 days of coculture and transplanted into lethally irradiated Ly5.2 mice (200 cells/mice) along with 2 × 105 competitor cells (Ly5.2). Percentages of donor-derived cells (Ly5.1) were analyzed monthly by FACS.
Microarray analysis
Total RNA was extracted from fractionated bone-derived cells by the use of TRIzol Reagent (Invitrogen). Biotin-labeled cRNA was prepared from total RNA with the Affymetrix 2-cycle target-labeling assay (Affymetrix). After biotin-labeled cRNA was purified, it was fragmented and hybridized to the Affymetrix Mouse Genome Array 430 2.0 (Affymetrix). Hybridization, washing, and staining with streptavidin-phycoerythrin conjugate and scanning were performed essentially as recommended by Affymetrix. After scanning, raw data were normalized by the use of GeneChip Operating Software (Affymetrix, Version 1.4) and the MAS5.0 algorithm. To determine correlations in the expression data, Pearson correlation coefficients were calculated with Microsoft Excel (Microsoft), and hierarchical clustering of each subset was performed in R (Version 2.9.0, http://www.r-project.org/) with the Bioconductor pvclust package (http://www.bioconductor.org/). Differentially expressed genes were selected by the use of 2 criteria: (1) present call at least one subset and (2) a greater than 3-fold difference between subsets. These genes were clustered by the use of the Bioconductor stats package. The microarray data have been deposited in Gene Expression Omnibus (National Center for Biotechnology Information) and are accessible through GEO Series accession number GSE17597 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17597).
Quantitative real-time PCR analysis
Quantitative real-time PCR (Q-PCR) was performed on an ABI 7500 Fast Real-Time PCR System with TaqMan Fast Universal PCR master mix (Applied Biosystems). The TaqMan gene expression assay mixes used in the Q-PCR assay are listed in the supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Data were analyzed with the use of 7500 Fast System SDS software, Version 1.3.1. All experiments were performed in triplicate.
Q-PCR array analysis
To analyze gene expression in hematopoietic cells after coculture with endosteal populations, RNA was isolated from CD45+ cells (1 × 103 cells/sample) after 2 days of coculture with each endosteal population and reverse transcribed with RT for PCR Kit (Clontech). cDNA was amplified for specific target amplification (STA) by the use of pooled 0.2× TaqMan Gene Expression Assays mix (Applied Biosystems). The thermal cycling conditions used for STA were 18 cycles of 95°C for 15 seconds and 60°C for 4 minutes. To analyze age-dependent changes in gene expression in endosteal populations, RNA was isolated from ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ cells (5000 cells/each fraction) derived from BM from 2-, 4-, 6-, and 8-week-old mice, and reverse transcribed with the use of RT for PCR Kit. After cDNA synthesis, STA (14 cycles) was performed. For single-cell Q-PCR analysis, ALCAM+Sca-1− and ALCAM−Sca-1+ cells were sorted directly into the mixture of CellsDirect 2× Reaction Mix (Invitrogen), 0.2× TaqMan Gene Expression Assays, and SuperScript III RT/Platinum Taq Mix (Invitrogen). Reverse transcription (50°C for 15 minutes) and STA (22 cycles) were serially performed.
Gene expression was analyzed by the use of the BioMark 48·48 Dynamic Array (Fluidigm). Preamplified cDNA was diluted with TE buffer (1:5) and used for Q-PCR array analysis. Data were analyzed by the use of BioMark Real-Time PCR Analysis Software Version 2.0 (Fluidigm). TaqMan Gene Expression Assays used in this study are listed in the supplemental data.
Statistical analysis
The significance of differences among groups was determined by use of the 2-tailed Student t test, Tukey test, or Dunnett test.
Results
Isolation of cell fractions adhering to the endosteum
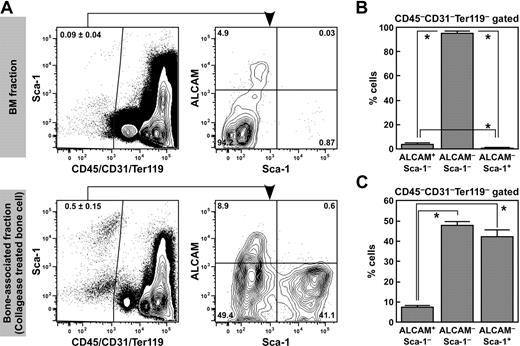
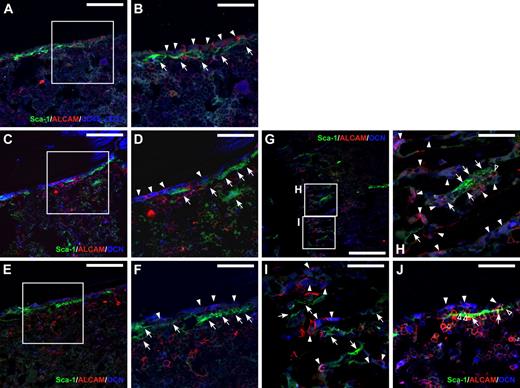
For isolation of bone-associated cells from the endosteum, we treated femur and tibia bone fragments with collagenase after flushing out the BM. Nonhematopoietic and nonendothelial CD45−CD31−Ter119− endosteal cells were enriched in the cells isolated from collagenase-treated bone fragments (0.5% ± 0.15%) rather than in the BM cells (0.09% ± 0.04%; Figure 1A). Next, we analyzed the expression of ALCAM and Sca-1 in the CD45−CD31−Ter119− endosteal cell population. The CD45−CD31−Ter119− population consisted of Sca-1+ and Sca-1− fractions. The Sca-1− population was further subdivided into ALCAM+Sca-1− and ALCAM−Sca-1− fractions (Figure 1A). The proportions of ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ cells were 3.6% plus or minus 1.5%, 95.6% plus or minus 1.6%, and 0.66% plus or minus 0.3%, respectively, in the BM CD45−CD31−Ter119− population (Figure 1B), and 0.0013% plus or minus 0.00065%, 0.038% plus or minus 0.005%, and 0.00011% plus or minus 0.00013%, respectively, in the total BMMNCs. In contrast, the frequency of ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ cells were 7.3% plus or minus 0.9%, 47.7% plus or minus 1.8%, and 42.1% plus or minus 3.5%, respectively, in the bone-associated (collagenase-treated bone-derived) CD45−CD31−Ter119− population (Figure 1C), and 0.024% plus or minus 0.006%, 0.16% plus or minus 0.04%, and 0.14% plus or minus 0.03%, respectively, in the total bone-associated MNCs. These data suggest that both ALCAM+Sca-1− and ALCAM−Sca-1+ cells were enriched in collagenase-treated bone-associated cells. Immunohistochemical staining for ALCAM, Sca-1, and CD45/CD31 or OCN demonstrated that ALCAM+ endosteal cells and Sca-1+ endosteal cells were localized to the endosteal surface of BM (Figure 2). The number of ALCAM−Sca-1+ cells was greater than ALCAM+Sca-1− cells in the diaphysis. The number of ALCAM+Sca-1− cells increased in the metaphyseal region. In addition, we found that some of the ALCAM+Sca-1+ hematopoietic cells adhered to OCN+ALCAM+Sca-1− cells and OCN−ALCAM−Sca-1+ cells (Figure 2J).
Isolation and frequency of endosteal cell populations. (A) Representative FACS profile of ALCAM and Sca-1 expression in CD45−CD31−Ter119− cells in BM cells (top) and the bone-associated fraction (cells isolated from bone fragments using collagenase treatment, bottom). CD45−CD31−Ter119− cells were gated, and the expression of Sca-1 and ALCAM was analyzed. CD45−CD31−Ter119− cells were subdivided into 3 fractions: ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ cells. (B) Frequency of ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ populations in the CD45−CD31−Ter119− fraction in BM cells. Data represent means ± SD (*P < .01, n = 6). (C) Frequency of ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ populations in the CD45−CD31−Ter119− fraction in bone-associated cells. Data represent means ± SD (*P < .01, n = 6).
Isolation and frequency of endosteal cell populations. (A) Representative FACS profile of ALCAM and Sca-1 expression in CD45−CD31−Ter119− cells in BM cells (top) and the bone-associated fraction (cells isolated from bone fragments using collagenase treatment, bottom). CD45−CD31−Ter119− cells were gated, and the expression of Sca-1 and ALCAM was analyzed. CD45−CD31−Ter119− cells were subdivided into 3 fractions: ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ cells. (B) Frequency of ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ populations in the CD45−CD31−Ter119− fraction in BM cells. Data represent means ± SD (*P < .01, n = 6). (C) Frequency of ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ populations in the CD45−CD31−Ter119− fraction in bone-associated cells. Data represent means ± SD (*P < .01, n = 6).
Localization of ALCAM+ and Sca-1+ cells in BM. (A and B) Immunohistochemical staining of ALCAM (red), Sca-1 (green), and CD45, CD31 (blue) in adult mouse BM. CD45−/CD31− ALCAM+ cells (arrowheads) and flat-shaped Sca-1+ cells (arrows) were localized to the endosteum. (B) Greater magnification of the enclosed box in panel A. Scale bar: 100 μm (A), 50 μm (B). (C-J) Immunohistochemical staining of ALCAM (red), Sca-1 (green), and OCN (blue) in adult mouse BM. Multiple views of the trabecular bone surface are shown. (C-F) Diaphysial region. (G-I) Metaphyseal region. A portion of OCN-positive osteoblastic cells expressed ALCAM on the bone surface in the trabecular area (arrowheads). In addition, flat-shaped Sca-1+ cells were also localized to the endosteal surface (arrows). Of note, there were no bone-lining nonhematopoietic cells that expressed both ALCAM and Sca-1. (D and F) Greater magnification of the enclosed box in panels C and E, respectively. (H and I) Greater magnification of the enclosed box in panel G. (J) Some of ALCAM+Sca-1+ hematopoietic cells (open arrowheads) adhered to the OCN+ALCAM+Sca-1− cells (arrows) and OCN−ALCAM−Sca-1+ cells (arrow). Scale bar: 200 μm (G), 100 μm (C,E), and 50 μm (D,F,H-J).
Localization of ALCAM+ and Sca-1+ cells in BM. (A and B) Immunohistochemical staining of ALCAM (red), Sca-1 (green), and CD45, CD31 (blue) in adult mouse BM. CD45−/CD31− ALCAM+ cells (arrowheads) and flat-shaped Sca-1+ cells (arrows) were localized to the endosteum. (B) Greater magnification of the enclosed box in panel A. Scale bar: 100 μm (A), 50 μm (B). (C-J) Immunohistochemical staining of ALCAM (red), Sca-1 (green), and OCN (blue) in adult mouse BM. Multiple views of the trabecular bone surface are shown. (C-F) Diaphysial region. (G-I) Metaphyseal region. A portion of OCN-positive osteoblastic cells expressed ALCAM on the bone surface in the trabecular area (arrowheads). In addition, flat-shaped Sca-1+ cells were also localized to the endosteal surface (arrows). Of note, there were no bone-lining nonhematopoietic cells that expressed both ALCAM and Sca-1. (D and F) Greater magnification of the enclosed box in panels C and E, respectively. (H and I) Greater magnification of the enclosed box in panel G. (J) Some of ALCAM+Sca-1+ hematopoietic cells (open arrowheads) adhered to the OCN+ALCAM+Sca-1− cells (arrows) and OCN−ALCAM−Sca-1+ cells (arrow). Scale bar: 200 μm (G), 100 μm (C,E), and 50 μm (D,F,H-J).
It has recently been reported that the alpha-type platelet-derived growth factor receptor (PDGFRα)-positive Sca-1+CD45−Ter119− population contains a large proportion of multipotent MSCs.40,42 Therefore, we also examined the expression patterns of PDGFRα and Sca-1 in CD45−CD31−Ter119− cells and established that there were 3 fractions: PDGFRα+Sca-1− (5.21% ± 0.58%), PDGFRα−Sca-1− (39.2% ± 2.89%), and PDGFRα+Sca-1+ (45.3% ± 2.62%; supplemental Figure 1).
Differentiation potential of endosteal cell fractions
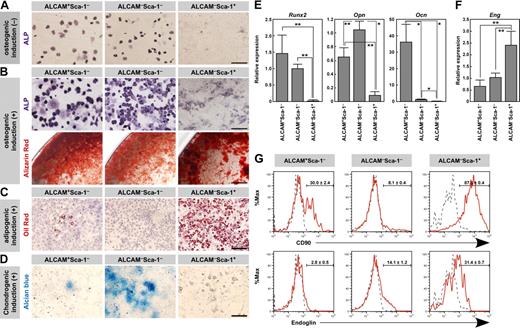
Next, we examined the differentiation potential of these 3 kinds of fractionated cells into the osteogenic, adipogenic, and chondrogenic lineages. ALP staining of freshly isolated cells showed that the Sca-1− cells expressed ALP, whereas Sca-1+ cells did not (Figure 3A). These data suggest that Sca-1− populations contained osteoblasts. After 7 days of culture, alizarin red-positive calcium deposition was observed in Sca-1− populations in the presence of osteogenic induction medium (Figure 3B). Calcium mineralization was not observed in the Sca-1− populations without osteogenic induction (data not shown). Although the primary ALCAM−Sca-1+ cells did not express ALP, this fraction efficiently differentiated into the osteoblastic lineage after osteogenic induction, as evidenced by the positive staining with ALP and alizarin red (Figure 3B). As for adipogenic differentiation, oil red O staining showed that a large number of cells after adipogenic induction contained lipid droplets in ALCAM−Sca-1+ cells (Figure 3C). In contrast, lipid accumulation was observed in few ALCAM+Sca-1− cells and in no ALCAM−Sca-1− cells (Figure 3C). Chondrogenic phenotype was evaluated by staining with alcian blue. In the presence of TGF-β3, the alcian blue-stained extracellular matrix was observed in ALCAM−Sca-1− cells (Figure 3D). ALCAM−Sca-1− population expressed a chondrocyte marker gene, type X collagen, without chondrogenic induction (see the result of Q-PCR array analysis shown in Figure 7), indicating that chondrocytic cells were contained in this fraction. Alcian blue-stained extracellular matrix was not observed in ALCAM−Sca-1+ cells (Figure 3D). We then tested the chondrogenic differentiation of ALCAM−Sca-1+ cells in the pellet culture to increase the cell density. Although alcian blue-stained intracytoplasmic vesicles were observed, ALCAM−Sca-1+ cells did not show the typical chondrocyte morphology and alcian blue-stained extracellular matrix was not observed in the pellet in culture with TGF-β3 (supplemental Figure 2).
Osteo-, adipo-, and chondrogenic potential of fractionated endosteal cells. (A) ALP staining of freshly isolated ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ populations. Scale bar: 50 μm. (B) Differentiation of osteoblasts from ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ populations after osteogenic induction. Osteoblastic differentiation was determined by ALP staining (top) and calcium deposition (alizarin red staining, bottom) after osteogenic induction. Scale bar: 50 μm (upper), 0.5 mm (lower). (C) Differentiation of adipocytes (oil red O-positive cells) from each population after adipogenic induction. Scale bar: 200 μm. (D) Alcian blue staining after chondrogenic induction. Scale bar: 200 μm. (E) qPCR analysis of the osteoblastic markers Runx2, Opn, and Ocn in fractionated endosteal populations. Data represent means ± SD (*P < .01, **P < .05). Representative data from 3 independent experiments are shown. (F) qPCR analysis of the MPC marker Eng in fractionated endosteal populations. Data represent means ± SD (**P < .05). Representative data from 3 independent experiments are shown. (G) Representative results of flow cytometric analysis of the expression of CD90 (top) and Endoglin (bottom) in ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ populations. Red line, specific Abs; gray dotted line, isotype control. Data represent means ± SD.
Osteo-, adipo-, and chondrogenic potential of fractionated endosteal cells. (A) ALP staining of freshly isolated ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ populations. Scale bar: 50 μm. (B) Differentiation of osteoblasts from ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ populations after osteogenic induction. Osteoblastic differentiation was determined by ALP staining (top) and calcium deposition (alizarin red staining, bottom) after osteogenic induction. Scale bar: 50 μm (upper), 0.5 mm (lower). (C) Differentiation of adipocytes (oil red O-positive cells) from each population after adipogenic induction. Scale bar: 200 μm. (D) Alcian blue staining after chondrogenic induction. Scale bar: 200 μm. (E) qPCR analysis of the osteoblastic markers Runx2, Opn, and Ocn in fractionated endosteal populations. Data represent means ± SD (*P < .01, **P < .05). Representative data from 3 independent experiments are shown. (F) qPCR analysis of the MPC marker Eng in fractionated endosteal populations. Data represent means ± SD (**P < .05). Representative data from 3 independent experiments are shown. (G) Representative results of flow cytometric analysis of the expression of CD90 (top) and Endoglin (bottom) in ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ populations. Red line, specific Abs; gray dotted line, isotype control. Data represent means ± SD.
Next we examined the gene expression of the osteoblastic markers Runt-related transcription factor 2 (Runx2), Osteopontin (Opn), and Ocn and the MSC-associated gene, Endoglin (Eng). Consistent with the osteogenic potential, Runx2 was expressed in the ALCAM+Sca-1− and ALCAM−Sca-1− cells (Figure 3E). There was no significant difference in the Runx2 expression level between ALCAM+Sca-1− and ALCAM−Sca-1− cells. The Opn expression level was greater in ALCAM−Sca-1− cells than in ALCAM+Sca-1− cells. However, the ALCAM+Sca-1− fraction showed a significantly greater level of Ocn expression than the ALCAM−Sca-1− fraction, indicating that the ALCAM+Sca-1− fraction contained more differentiated osteoblasts than the ALCAM−Sca-1− fraction. In contrast, ALCAM+Sca-1− cells expressed both Runx2 and Ocn. It was reported that Runx2 promotes osteoblast differentiation at an early stage but inhibits osteoblast differentiation at a later stage.43 Therefore, it is possible that Runx2 in ALCAM+Sca-1− cells inhibited terminal differentiation of ALCAM+Sca-1− cells. ALCAM−Sca-1+ cells showed greater expression levels of Eng than the Sca-1− fractions (Figure 3F).
Next we examined the expression of CD90 and Endoglin by FACS. As shown in Figure 3G, most ALCAM−Sca-1+ cells expressed CD90 (87.6% ± 0.4%), whereas ALCAM−Sca-1− cells showed a lower frequency of CD90+ cells (8.1% ± 0.4%). Interestingly, both CD90+ and CD90− populations were observed in ALCAM+Sca-1− cells (30.0% ± 2.4% of ALCAM+ Sca-1− cells expressed CD90). Similar to the result of Q-PCR analysis, ALCAM−Sca-1+ cells showed greater expression levels of Endoglin (31.4% ± 0.7%) compared with Sca-1− populations (ALCAM+Sca-1− cells: 2.8% ± 0.5%, ALCAM−Sca-1− cells: 14.1% ± 1.2%).
Hematopoiesis-supporting activity of fractionated endosteal cells
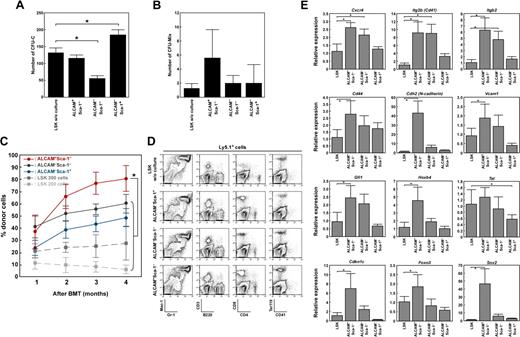
Next, we investigated the ability of these fractionated endosteal cells to support hematopoiesis. First, we examined the capacity of ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ cells to maintain colony formation in BM LSK cells. The colony-forming units in culture (CFU-C) were increased in coculture with ALCAM−Sca-1+ cells, whereas CFU-C was decreased in coculture with ALCAM−Sca-1− cells (Figure 4A). However, there was no significant difference in the number of CFU-Mix after coculture with each endosteal population (Figure 4B). To further examine whether these fractionated cells were involved in maintenance of HSCs, an LTR assay was performed. The LSK cells cocultured with either of the endosteal fractions showed greater LTR activity with greater chimerisms of donor cells in the peripheral blood (Figure 4C) and reconstituted the myeloid, lymphoid, erythroid, and megakaryocytic lineages (Figure 4D).
Endosteal populations maintain the LTR activity of HSCs. (A and B) LSK cells were cultured on a feeder layer of ALCAM+Sca-1−, ALCAM−Sca-1−, or ALCAM−Sca-1+ cells. After 5 days of culture, cells were harvested and cultured in methylcellulose medium. (A) Number of CFU-C. (B) Number of CFU-Mix. Data represent means ± SD (*P < .01). (C and D) LSK cells (Ly5.1+) were cultured on feeder layers of ALCAM+Sca-1−, ALCAM−Sca-1−, or ALCAM−Sca-1+ cells for 2 days. Then, CD45.1+ cells were sorted, and 200 cells were transplanted into recipient mice (Ly5.2+) along with 2 × 105 competitor cells. (C) Percentages of donor-derived (Ly5.1+) cells in recipient mice 1 to 4 months after BM transplantation. Approximately 200 or 300 LSK cells without coculture were transplanted into recipient mice as a control. Data represent means ± SD (n = 5, *P < .01). (D) Representative FACS profiles of donor-derived (Ly5.1+) myeloid, B, T, erythroid, and megakaryocytic lineages in recipient mice. (E) LSK cells were cultured on feeder layers of ALCAM+Sca-1−, ALCAM−Sca-1−, or ALCAM−Sca-1+ cells. After 2 days of coculture, CD45+ cells were sorted, and the expression of Cxcr4, Itg2b (Cd41), Itgb2, Cd44, Cdh2, Vcam1, Gfi1, Hoxb4, Tel, Cdkn1c, Foxo3, and Sox2 was analyzed by Q-PCR array. Data represent means ± SD (*P < .05). Hprt1 was used for an endogenous control.
Endosteal populations maintain the LTR activity of HSCs. (A and B) LSK cells were cultured on a feeder layer of ALCAM+Sca-1−, ALCAM−Sca-1−, or ALCAM−Sca-1+ cells. After 5 days of culture, cells were harvested and cultured in methylcellulose medium. (A) Number of CFU-C. (B) Number of CFU-Mix. Data represent means ± SD (*P < .01). (C and D) LSK cells (Ly5.1+) were cultured on feeder layers of ALCAM+Sca-1−, ALCAM−Sca-1−, or ALCAM−Sca-1+ cells for 2 days. Then, CD45.1+ cells were sorted, and 200 cells were transplanted into recipient mice (Ly5.2+) along with 2 × 105 competitor cells. (C) Percentages of donor-derived (Ly5.1+) cells in recipient mice 1 to 4 months after BM transplantation. Approximately 200 or 300 LSK cells without coculture were transplanted into recipient mice as a control. Data represent means ± SD (n = 5, *P < .01). (D) Representative FACS profiles of donor-derived (Ly5.1+) myeloid, B, T, erythroid, and megakaryocytic lineages in recipient mice. (E) LSK cells were cultured on feeder layers of ALCAM+Sca-1−, ALCAM−Sca-1−, or ALCAM−Sca-1+ cells. After 2 days of coculture, CD45+ cells were sorted, and the expression of Cxcr4, Itg2b (Cd41), Itgb2, Cd44, Cdh2, Vcam1, Gfi1, Hoxb4, Tel, Cdkn1c, Foxo3, and Sox2 was analyzed by Q-PCR array. Data represent means ± SD (*P < .05). Hprt1 was used for an endogenous control.
Intriguingly, regardless of the feeder cells used for coculture, the numbers of hematopoietic cells did not increase in the coculture (data not shown), but the chimerisms of donor cells were much greater than those of freshly isolated LSK cells (Figure 4C). In particular, CD45+ cells cocultured with ALCAM+Sca-1− cells showed significantly greater LTR activity compared with those cocultured with the other fractions. Next we investigated the expression of HSC markers and genes related to homing, lodgment, and self-renewal in cocultured LSK cells. We found that cells cocultured with ALCAM+Sca-1− fractions showed significantly greater expression levels of homing- and cell adhesion–related genes, such as chemokine (C-X-C motif) receptor 4 (Cxcr4), integrin alpha 2b (Itg2b), integrin beta 2 (Itgb2), CD44 antigen (Cd44), Cdh2, and vascular cell adhesion molecule 1 (Vcam1), compared with cells cocultured with ALCAM−Sca-1+ cells (Figure 4E). In addition, ALCAM+Sca-1− cells significantly up-regulated the expression levels of growth factor–independent 1 (Gfi1), homeo box B4 (Hoxb4), cyclin-dependent kinase inhibitor 1C (P57, Cdkn1c), forkhead box O3 (Foxo3), and SRY-box containing gene 2 (Sox2) in LSK cells (Figure 4E). These data suggest that, during culture, ALCAM+Sca-1− cells enhanced the LTR activity of HSCs or enriched the cell population that originally had high LTR activity.
Gene expression profiles of endosteal cells
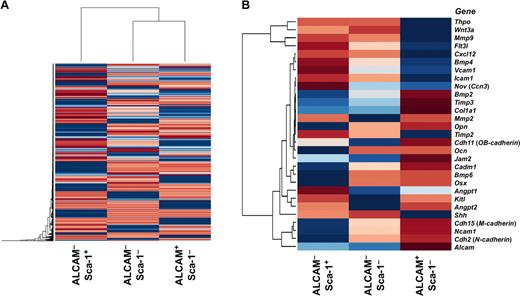
Next we performed microarray analysis by using ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ cells (Figure 5A-B). Microarray analysis showed that cytokine-related and cell adhesion-related genes were expressed differentially among these endosteal cell fractions. Compared with the other fractions, ALCAM+Sca-1− cells tended to express highly osteoblastic markers and genes related to cell-to-cell and cell-to-extracellular adhesion, such as Cdh2, Cadherin 11 (OB-cadherin, Cdh11), and Alcam (Figure 5B). In contrast, ALCAM−Sca-1+ cells highly expressed growth factor– and cytokine-related genes that are known to be involved in the regulation of both quiescence and proliferation of LT-HSCs, such as Angpt1, fms-related tyrosine kinase 3 ligand (Flt3l), Cxcl12, bone morphogenetic protein 4 (Bmp4), and Kitl (Figure 5B). Similarly, we also analyzed gene expression in fractionated cells according to the expression of PDGFRα and Sca-1 in the CD45−CD31−Ter119− cell population. The coefficients of correlation suggested that PDGFRα+Sca-1−, PDGFRα−Sca-1−, and PDGFRα+Sca-1+ cells corresponded to ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ cells, respectively (supplemental Figure 3). Indeed, the expression levels of cell adhesion-related genes were greater in PDGFRα+Sca-1− cells, as were levels of cytokine-related genes in PDGFRα+Sca-1+ cells (data not shown).
Microarray analysis of fractionated endosteal cells. (A) Hierarchical clustering analysis of gene expression in ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ populations. (B) Heat map of genes implicated in the control of HSCs in BM niches. The ALCAM+Sca-1− population tended to express genes involved in cell-to-cell and cell-to-ECM (extracellular matrix) interactions between HSCs and niche cells. In contrast, the ALCAM−Sca-1+ population highly expressed growth factors, cytokines and chemokines known to be involved in the regulation of quiescence and proliferation of HSCs.
Microarray analysis of fractionated endosteal cells. (A) Hierarchical clustering analysis of gene expression in ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ populations. (B) Heat map of genes implicated in the control of HSCs in BM niches. The ALCAM+Sca-1− population tended to express genes involved in cell-to-cell and cell-to-ECM (extracellular matrix) interactions between HSCs and niche cells. In contrast, the ALCAM−Sca-1+ population highly expressed growth factors, cytokines and chemokines known to be involved in the regulation of quiescence and proliferation of HSCs.
Single-cell level gene expression analysis in fractionated endosteal cells
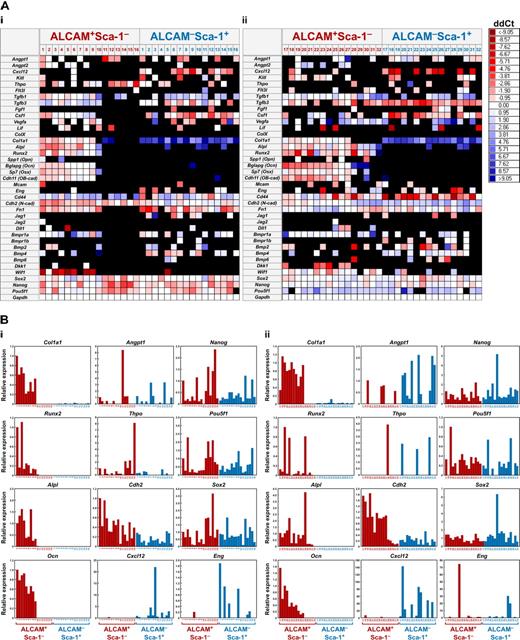
Next, to characterize the ALCAM+Sca-1− and ALCAM−Sca-1+ cells in greater detail, we performed single-cell Q-PCR array analysis by using a nanofluidic platform (Figure 6). We analyzed 39 genes in 72 single cells in each population. As a general trend of gene expression pattern in each population, osteoblastic markers and Cdh2 were highly expressed in ALCAM+Sca-1− cells, and the cells highly expressing cytokine-related genes were enriched in ALCAM−Sca-1+ cells, which is consistent with the results of the differentiation assay (Figure 3) and microarray analysis (Figure 5). At the single-cell level, 22.5% of ALCAM+Sca-1− cells expressed at least one gene related to cytokines such as Angpt1, Thpo, or Clxcl12. Furthermore, interestingly, we found a subpopulation in ALCAM+Sca-1− cells that showed very low or no expression of osteoblastic markers (26 of 72 cells). Eleven of these 26 osteoblastic markerlow/− ALCAM+Sca-1− cells expressed at least one cytokine-related gene that is involved in the niche regulation of LT-HSCs. Furthermore, 9 of the 26 osteoblastic markerlow/− cells expressed at least one pluripotent stem cell marker such as Sox2, POU domain, class 5, transcription factor 1 (Pof5f1, Oct3/4), or Nanog homeobox (Nanog) at relatively high levels compared with other ALCAM+Sca-1− or ALCAM−Sca-1+ cells. These data indicate that ALCAM+Sca-1− cells are a heterogeneous population that contains an osteoblastic markerlow/− subpopulation, some of which might be immature niche cells expressing cytokine- or pluripotent marker–related genes.
Single-cell Q-PCR array. (A) Heat maps of the representative gene expression patterns of selected genes in ALCAM+Sca-1− and ALCAM−Sca-1+ cells at the single-cell level. The horizontal axis in the heat map is a list of genes tested, including internal controls, and the vertical axis represents cell samples (16 independent cells in each fraction/experiment). The color in each cross point indicates delta-delta Ct (ddCt). Representative data from 2 (of 5) independent experiments are shown. (B) Expression levels of collagen type I alpha 1 (Col1a1), Runx2, alkaline phosphatase (Alp1), Ocn, Angpt1, Thpo, Cdh2, Cxcl12, Nanog, Pou5f1, Sox2, and Eng in each single cell.
Single-cell Q-PCR array. (A) Heat maps of the representative gene expression patterns of selected genes in ALCAM+Sca-1− and ALCAM−Sca-1+ cells at the single-cell level. The horizontal axis in the heat map is a list of genes tested, including internal controls, and the vertical axis represents cell samples (16 independent cells in each fraction/experiment). The color in each cross point indicates delta-delta Ct (ddCt). Representative data from 2 (of 5) independent experiments are shown. (B) Expression levels of collagen type I alpha 1 (Col1a1), Runx2, alkaline phosphatase (Alp1), Ocn, Angpt1, Thpo, Cdh2, Cxcl12, Nanog, Pou5f1, Sox2, and Eng in each single cell.
Developmental changes in gene expression in endosteal cells
Next, we examined changes in gene expression in ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ cells isolated from 2-, 4-, 6-, and 8-week-old mice. We analyzed the expression levels of osteoblastic markers, cell adhesion molecules, cytokines, proteolytic enzymes, and extracellular matrices implicated in the control of HSCs (Figure 7). The expression levels of Angpt1, Kitl, and Flt3l were constant, whereas Cxcl12, TGF-β3 (Tgfb3), and fibronectin 1 (Fn1) expression levels increased with age. Thpo was highly expressed in ALCAM−Sca-1− cells from 2 weeks, and expression was down-regulated with age. In contrast, the expression of Thpo in ALCAM−Sca-1+ cells was up-regulated with age. Jag1 was highly expressed in 2- and 4-week-old ALCAM−Sca-1+ cells relative to the 6- or 8-week-old adult ALCAM−Sca-1+ cells.
Dynamic array analysis of endosteal populations isolated from 2-, 4-, 6-, and 8-week-old BM. To analyze changes in gene expression in ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ cells isolated from the BM of 2-, 4-, 6-, and 8-week-old mice, gene expression was examined by dynamic array. (A) Representative gene expression pattern of selected genes. The horizontal axis is a list of genes tested, including internal controls, and the vertical axis represents the quadruplicate cell samples. The color in each cross point indicates ddCt. (B) Changes in gene expression levels of Angpt1, Kitl, Angpt2, Flt3l, Tgfb3, Vegfa, Cxcl12, Fn1, Mmp9, Thpo, Jag1, and Mmp13. Gapdh was used for an endogenous control. Data represent means ± SD. Statistical differences were analyzed among each population and between each age (*P < .05).
Dynamic array analysis of endosteal populations isolated from 2-, 4-, 6-, and 8-week-old BM. To analyze changes in gene expression in ALCAM+Sca-1−, ALCAM−Sca-1−, and ALCAM−Sca-1+ cells isolated from the BM of 2-, 4-, 6-, and 8-week-old mice, gene expression was examined by dynamic array. (A) Representative gene expression pattern of selected genes. The horizontal axis is a list of genes tested, including internal controls, and the vertical axis represents the quadruplicate cell samples. The color in each cross point indicates ddCt. (B) Changes in gene expression levels of Angpt1, Kitl, Angpt2, Flt3l, Tgfb3, Vegfa, Cxcl12, Fn1, Mmp9, Thpo, Jag1, and Mmp13. Gapdh was used for an endogenous control. Data represent means ± SD. Statistical differences were analyzed among each population and between each age (*P < .05).
In addition to these findings, we found that vascular endothelial growth factor A (Vegfa) in ALCAM+Sca-1− and ALCAM−Sca-1− cells and angiopoietin 2 (Angpt2) in ALCAM+Sca-1− cells were expressed at high levels in younger mice. The expression of these angiogenic factors was down-regulated with development. Moreover, we found that matrix metallopeptidase 9 (Mmp9) and Mmp13 were specifically expressed in 2- and 4-week-old ALCAM−Sca-1− cells. High levels of Vegfa, Angpt2, Mmp9, and Mmp13 expression in young mice might be associated with vascular remodeling in the BM. In addition, MMP9 is known to induce the shedding of membrane-anchored growth factors such as Kitl,7,44 whereas it has been reported that soluble Kitl induces proliferation of HSCs.7 We hypothesized that endosteal cells induce HSC proliferation through the activation of vascular remodeling and production of soluble forms of growth factors during postnatal development.
Discussion
In the BM, HSCs maintain their stem cell activity through interactions with their niches.5 It has been shown that quiescent HSCs reside in the endosteal area known as the endosteal niche,16,45 indicating that cells in the endosteum have a critical function in the maintenance of quiescence of HSCs. HSCs also reside adjacent to sinusoidal blood vessels in the BM.11,46 This is called a perivascular niche and is also involved in HSC maintenance. The endosteal niche comprises bone-lining cells, which are heterogeneous in their function and degree of differentiation.23,24 In this study, we subdivided CD45−CD31−Ter119− cells into 3 fractions according to their ALCAM and Sca-1 expression (Figure 1). The expression of osteoblastic markers and differentiation potential of these 3 fractions revealed that osteoblasts were enriched in the Sca-1− populations.
Recent reports on the localization of MSCs and skeletal progenitor cells indicated that they exist within BM, especially in the perivascular area.26,40 In our study, the frequency of bone-associated ALCAM−Sca-1+ cells was much greater than the anticipated frequency of MSCs in BM. The frequencies were 42.1% plus or minus 3.5% in the bone-associated CD45−CD31−Ter119− population, 0.14% plus or minus 0.03% in the total MNCs, and 0.017% plus or minus 0.001% in the total BM cells. Although the ALCAM−Sca-1+ population showed greater expression of MSC-associated genes and differentiated into osteoblasts and adipocytes, the chondrogenic potential of ALCAM−Sca-1+ cells was vanishingly low compared with that of the previously identified MSC population40,41 in the presence of TGF-β3. We assumed that the bone-associated ALCAM−Sca-1+ cells were a progenitor population that had restricted differentiation potential compared with BM-localized primitive MSCs. However, it is still unclear whether ALCAM−Sca-1+ cells are bipotential (osteo- and adipogenic potential) MPCs or a heterogeneous population of progenitor cells from the osteoblast and adipocyte lineage. A clonal assay will be required to clarify the differentiation potential of ALCAM−Sca-1+ cells.
ALCAM is known to be expressed in human MSCs,36,37 and we previously reported that ALCAM was expressed in the mouse embryonic perichondrial cells that have multilineage differentiation potential.35 ALCAM-ALCAM–homophilic interactions supported the maintenance of MSCs in the embryo. In this study, we found that ALCAM was expressed mainly in the osteoblast-enriched mouse adult BM endosteum. It is possible that the function of ALCAM changes with developmental stages, and it would be interesting to clarify the developmentally related function of ALCAM in the regulation of MSCs and osteoblasts. In addition, in earlier studies, MSCs were obtained by in vitro culture of nonhematopoietic cells. However, we previously reported that freshly isolated ALCAM− cells became to express ALCAM in culture,35 and therefore the differences in characteristics of the ALCAM+ cell populations between the present data and those of previous studies by other groups36,37 may be the result of differences in the cell isolation method.
It was hypothesized that osteoblasts act as important regulators of HSC activity.16,22,45,47 Therefore, the functional analysis has been required for elucidation of the role of prospectively isolated osteoblasts in HSC maintenance. In this study, we found that all 3 endosteal cell fractions directly obtained from bone maintained LTR activity in vitro. Notably, compared with other fractions, ALCAM+Sca-1− cells showed significantly greater potential for maintenance of the LTR activity of HSCs. Recently, Mayack and Wagers39 isolated osteoblasts by using anti-OPN Ab and showed that OPN+ osteoblasts have the potential to maintain LTR activity of HSCs. In this study, we found that the Opn expression level in ALCAM+Sca-1− cells was lower than in ALCAM−Sca-1− cells (Figure 3E and 7A). In addition, the single-cell Q-PCR array showed that the number of Opn-expressing cells was low in ALCAM+Sca-1− cells (Figure 6A). These data suggest that ALCAM+Sca-1− cells were a population distinct from the previously identified OPN+ osteoblastic population. Q-PCR array analysis revealed that LSK cells cocultured with ALCAM+Sca-1− cells had significantly greater levels of Cd44, Cdh2, Vcam1, Gfi1, Hoxb4, Cdkn1c, Foxo3, and Sox2 expression than LSK cells cocultured with other fractions. These data suggest either that the cell-cell interaction between HSCs and osteoblasts mediated by cell adhesion molecules including ALCAM enhanced the LTR activity of HSCs, or that, during coculture, ALCAM+Sca-1− cells enriched the cell population that originally had high LTR activity. To elucidate definitively the function of ALCAM, it would be interesting to determine the relevance of the adhesive interaction between HSC and niche cells to HSC maintenance.
Microarray analysis (Figure 5) revealed that cytokine- and cell adhesion–related genes were expressed in distinct endosteal cell fractions. ALCAM−Sca-1+ cells tended to express highly cytokine-related genes, and therefore, this population may regulate HSCs through the production of cytokines that affect both HSC proliferation and quiescence. However, ALCAM+Sca-1− cells expressed genes for multiple cell adhesion molecules, indicating that this population physically regulates quiescence of HSCs via cell adhesion molecules.
Interestingly, by using single-cell gene expression analysis with Q-PCR array, we identified an osteoblastic markerlow/− subpopulation in ALCAM+Sca-1− fraction that expresses genes for both cell adhesion molecules, such as Cdh2, and cytokines, such as Angpt1 and Thpo (Figure 6). Notably, the osteoblastic markerlow/− subpopulation in ALCAM+Sca-1− cells includes cells that express relatively high levels of pluripotent markers. It would be interesting to elucidate the differentiation potential of osteoblastic markerlow/− ALCAM+Sca-1− cells and their function in the maintenance of LT-HSCs. In addition, more detailed fractionation of endosteal cells based on single-cell gene expression may provide detailed characterization of niche cell components.
Because the cell cycle of HSCs shifts from a cycling to a quiescent state during postnatal BM development,22 the function of the BM niche shifts from expansion to maintenance of quiescence of HSCs. We found that osteoblast populations from the bones of younger mice (2-4 weeks old) expressed high levels of Vegfa, Angpt2, and MMPs (Figure 7), indicating that the endosteal cells induce vascular remodeling in the developing BM. We hypothesized that the endosteal population affects HSC expansion through the stimulation of vascular niche function.
Manipulation of niche components and their signaling pathways is of special interest for regenerative medicine.20,48 For ex vivo expansion of HSCs, our data indicate that it would be necessary to mimic the niche of young BM. The molecules expressed in the adult BM niche are targets for the protection of HSCs from various stressors. In addition, our study raises the possibility that antagonizing the activity of niche-related factors and cell adhesion molecules could potentially repress the interaction of leukemic stem cells with their niche and thereby decrease their resistance to chemotherapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Nobuyuki Takakura (Osaka University) for providing the anti-PDGFRα Ab (APA5). We also thank Dr Tokuhiro Kimura (Keio University) for helpful advice.
This work was supported by a Grant-in-Aid for Specially Promoted Research and a Grant-in-Aid for Young Scientists (S) from The Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, by the “High-Tech Research Center” Project for Private Universities: matching fund subsidy from MEXT, and by the Takeda Science Foundation.
Authorship
Contribution: Y.N., F.A., H.I., K.H., Y.G., Y.M., and H.Y. performed the experiments; I.K. performed the statistical analysis for microarray experiments; Y.N. and F.A. analyzed the results and made the figures; F.A and T.S. designed the research; and Y.N., F.A., H.I., and T.S. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Fumio Arai, 35 Shinano-machi, Shinjuku-ku, Tokyo, 160-8582, Japan; e-mail: farai@sc.itc.keio.ac.jp; or Toshio Suda, 35 Shinano-machi, Shinjuku-ku, Tokyo, 160-8582, Japan; e-mail: sudato@sc.itc.keio.ac.jp.
References
Author notes
Y.N. and F.A. contributed equally to this work.