Defining the molecular mechanisms underpinning fetal (γ) globin gene silencing may provide strategies for reactivation of γ-gene expression, a major therapeutic objective in patients with β-thalassemia and sickle cell disease (SCD). We have previously demonstrated that symmetric methylation of histone H4 Arginine 3 (H4R3me2s) by the protein arginine methyltransferase PRMT5 is required for recruitment of the DNA methyltransferase DNMT3A to the γ-promoter, and subsequent DNA methylation and gene silencing. Here we show in an erythroid cell line, and in primary adult erythroid progenitors that PRMT5 induces additional repressive epigenetic marks at the γ-promoter through the assembly of a multiprotein repressor complex containing the histone modifying enzymes SUV4-20h1, casein kinase 2α (CK2α), and components of the nucleosome remodeling and histone deacetylation complex. Expression of a mutant form of PRMT5 lacking methyltransferase activity or shRNA-mediated knockdown of SUV4-20h1 resulted in loss of complex binding to the γ-promoter, reversal of both histone and DNA repressive epigenetic marks, and increased γ-gene expression. The repressive H4K20me3 mark induced by SUV4-20h1 is enriched on the γ-promoter in erythroid progenitors from adult bone marrow compared with cord blood, suggesting developmental specificity. These studies define coordinated epigenetic events linked to fetal globin gene silencing, and provide potential therapeutic targets for the treatment of β-thalassemia and SCD.
Introduction
The developmental switch in human β-like globin gene subtype from fetal (γ) to adult (β) that begins at birth heralds the onset of the hemoglobinopathies, β-thalassemia and sickle cell disease (SCD). The observation that increased adult γ-globin gene expression (in the setting of hereditary persistence of fetal hemoglobin [HPFH] mutations) significantly ameliorates the clinical severity of β-thalassemia and SCD has prompted the search for therapeutic strategies to reverse γ-globin gene silencing.1 To date, this has been achieved through pharmacologic induction, using compounds that broadly influence epigenetic modifications, including DNA methylation and histone deacetylation.2,–4 The development of more targeted therapies is dependent on the identification of the molecular mechanisms underpinning fetal globin gene silencing. These mechanisms have remained elusive, despite exhaustive study of the HPFH mutations,5 and considerable progress in many other aspects of globin gene regulation.
Central to silencing of the γ-genes is DNA methylation, which marks critical CpG dinucleotides flanking the γ-gene transcriptional start site in adult bone marrow erythroid cells.6,7 We have recently shown that these marks are established as a consequence of recruitment of the DNA methyltransferase, DNMT3A to the γ-promoter by the protein arginine methyltransferase PRMT5.8 PRMT5 induces the repressive histone mark, H4R3me2s, which serves as a template for direct binding of DNMT3A, and subsequent DNA methylation. Loss of PRMT5 binding, or its enzymatic activity leads to demethylation of the CpG dinucleotides and γ-gene activation.8
Here we demonstrate that, in addition to the H4R3me2s mark and DNA methylation, PRMT5 binding to the γ-promoter, and its enzymatic activity are essential for assembly of a multiprotein complex on the γ-promoter, which induces a range of coordinated repressive epigenetic marks. Disruption of this complex leads to reactivation of γ-gene expression. These studies provide a platform for the development of targeted therapies for β-thalassemia and SCD.
Methods
Cell culture
K562 cells were grown as described previously.9 Human bone marrow (BM) and cord blood (CB) were collected after approval by the Melbourne Health and Royal Women's Hospital Human Research Ethics Committees. CD34+ cells isolated from fresh adult BM and CB were cultured in Iscove modified Dulbecco medium supplemented with 15% (vol/vol) fetal calf serum, stem cell factor (SCF; 100 ng/mL), interleukin-3 (10 ng/mL), and FMS-like tyrosine kinase-3 ligand (500 ng/mL) for 7 days, followed by erythropoietin (EPO; 5 U/mL) alone for 5 days to induce erythroid differentiation. To induce high levels of γ-gene expression in CB, the culture conditions described by Bhanu et al were used.10 Briefly, cells were grown in Dulbecco modified Eagle medium containing 30% (vol/vol) fetal bovine serum, 1% bovine serum albumin (wt/vol), β-mercaptoethanol (10−5M), dexamethasone (10−6M), transferring (0.3 mg/mL), glutamine (2mM), EPO (4 U/mL), SCF (50 ng/mL) and transforming growth factor-β1 (1.25 ng/mL). For low fetal hemoglobin conditions for adult BM, the culture conditions described by Sankaran et al.11 Briefly, cells were grown in StemSpan SFEM medium with 1× CC100 cytokine mix for 6 days and then reseeded into the same medium supplemented with SCF (20 ng/mL), EPO (1 U/mL), interleukin-3 (5 ng/mL), dexamethasone (2 × 10−6M), and β-estradiol (10−6M). Cell-surface marker analyses with CD71 and Glycophorin A indicated that cultured cells were greater than 90% erythroid lineage.
Protein interaction studies
Immunoprecipitation and immunoblotting were performed as described previously.12 Antibodies used in the immunoprecipitations: FLAG (Sigma-Aldrich), PRMT5, CK2α, SUV4-20h1, SUV39h1, MBD2, MeCP2 (Abcam), Mi2, mSin3A, DNMT1, DNMT3A, DNMT3B, MBD3, Brg-1, HP1, tubulin, GATA-1 (Santa Cruz Biotechnology), HDAC1 (Upstate Biotechnology). Many of these were also used in chromatin-immunoprecipitation (ChIP) assays.
Size exclusion chromatography
Size exclusion chromatography was performed on a calibrated Superose 12 HR 10/30 gel filtration column (Amersham/Pharmacia), and 500-μL fractions were collected. Fractions were concentrated by trichloroacetic acid precipitation, electrophoresed on a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to a polyvinylidene difluoride membrane, and analyzed by Western blot with selected antibodies.
ChIP analysis
ChIP assays were performed as described previously.8 Chromatin fractions from K562 cells, or erythroid progenitors from CB or BM were immunoprecipitated with specific antibodies (Abs). No antibody and normal rabbit immunoglobulin G (IgG) served as the controls. Antibodies used were: H4R3me2s, H4S1ph, H4K20me3, H3K9me3, H3K27me3, SUV4-20h1, CK2α (Abcam), H4K5ac, H4K8ac, H4K12ac, H4K16ac (Upstate Biotechnology), and RNA polII (Santa Cruz Biotechnology). The relative enrichment was calculated using the method described in Wysocka et al.13 The percentage of ChIP DNA was calculated relative to the input DNA. In all assays it ranged from 0.35% to 0.71%. In comparative experiments, the most substantial enrichment was assigned the value 1, and all other conditions were normalized to this value. Each experiment was performed at least twice independently.
shRNA and retroviral infection
The shRNA-mediated knockdown of PRMT5 was described previously.8 The shRNA target sequences for SUV4-20h1 and CK2α were inserted into the XhoI/HpaI sites in the Lentilox 3.7 vector (ATCC). The oligonucleotide sequences were: SUV4-20h1 kd target sequence: GGCTCTAAGAGACATTGAA; and CK2α kd target sequence: GGCCCTATCTGTCTCCTGA.
Virus production in 293T cells and infection of K562 cells or erythroid progenitors from BM were performed as described.9 Transduced cells were selected for green fluorescent protein expression by fluorescence-activated cell sorting.
Bisulfite sequence analysis
Bisulfite sequence analysis was performed as described previously.14 Primers to amplify the bisulfite treated γ-promoter are described previously.8 Polymerase chain reaction (PCR) was performed with HiFi Taq polymerase (Roche) as follows: 30 cycles, 94°C for 20 seconds, 55°C for 20 seconds, and 68°C for 35 seconds. PCR products were cloned into pCRII (Invitrogen) followed by nucleotide sequencing using the Big-Dye Termination method (ABI). The significance of the differences between cell lines was calculated using the Fisher exact test, with all P values less than .05.
Quantitative RT-PCR
Total RNA was isolated from cells with Trizol reagent (Invitrogen). cDNA was generated using the reverse transcription system (Promega). Quantitative reverse-transcriptase PCR (Q-RT-PCR) primers were described previously.8 The identities of the amplified bands were confirmed by sequencing. Q-RT-PCR was done in a Rotorgene 2000 (Corbett Research) or an LC480 (Roche), in a final volume of 20 μL. Reaction mixtures contained 1× reaction buffer, 2.5mM MgCl2, 0.05mM deoxynucleotides (Roche), 0.1μM gene-specific primers, 1 U of Taq polymerase (Fisher Biotech), a 1:10 000 dilution of SYBR Green I (Molecular Probes,) and 2 μL of sample or standard. Cycling conditions were 94°C for 15 seconds, 57°C for 30 seconds, and 72°C for 30 seconds. Standard curves for hypoxanthine-guanine phosphoribosyltransferase and γ-globin were generated from the various K562 cell lines, BM, and CB cells. The relative quantity of the transcripts was calculated for all individual cell lines. Each reaction was done in duplicate.
Results
PRMT5 induces multiple repressive histone modifications
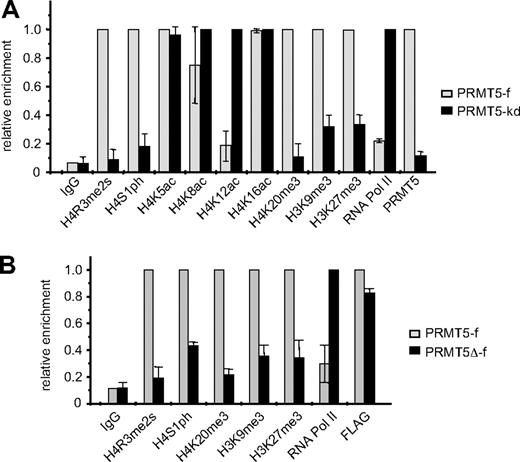
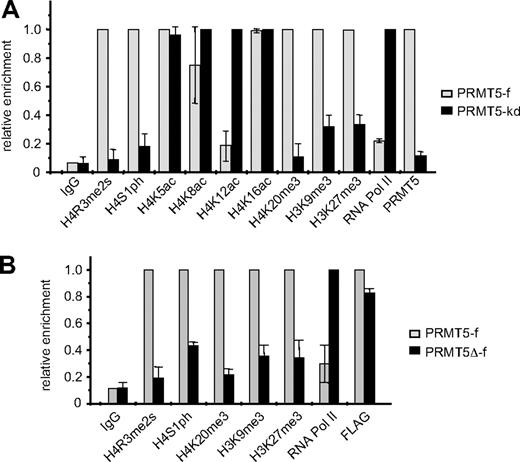
We have previously established K562 cell lines in which FLAG-tagged PRMT5 expression is enforced (PRMT5-f; to < 2-fold greater than the endogenous levels), or in which expression of PRMT5 is markedly reduced (> 90%) via expression of a specific shRNA (PRMT5-kd).8 These lines exhibit contrasting γ-globin gene expression profiles, with silencing in the PRMT5-f cells, and up-regulation in the PRMT5-kd cells. Commensurate with this, they also exhibit opposite levels of the repressive H4R3me2s mark, which is enriched at the proximal γ-promoter in PRMT5-f cells, and lost in the PRMT5-kd line.8 To determine whether other repressive histone modifications were induced at the γ-promoter in response to increased PRMT5 expression, we performed ChIP analyses on our PRMT5-f and PRMT5-kd cells using a range of specific antibodies (Figure 1A). Two other histone H4 modifications that have been linked to gene silencing, phosphorylation of H4S1 (H4S1ph),15 and tri-methylation of H4K20 (H4K20me3)16 were also differentially localized to the γ-promoter in the 2 lines, with high levels observed in the PRMT5-f cells, and a marked reduction in the PRMT5-kd line. Tri-methylation of H3K9, which has previously been shown to be required for H4K20me3,16 and tri-methylation of H3K27, which has been linked to DNA methylation and gene silencing,17,18 were also increased in the PRMT5-f compared with the PRMT5-kd lines, although these changes were less striking and so we focused on the H4S1ph and H4K20me3 modifications. The alterations in repressive marks were reflected in localization of RNA polymerase II (RNA polII) to the promoter, which was markedly reduced in the PRMT5-f cells compared with the knockdown cells. Assessment of histone H4 acetylation in the 2 cell lines revealed a substantial reduction in H4K12ac, and a modest loss of H4K8ac in the PRMT5-f cells, but no alterations at lysine 5 or 16.
Comparison of histone modifications at the γ-promoter in PRMT5-f and PRMT5-kd K562 cells. (A) Enrichment of various histone modifications, RNA polII and PRMT5 at the γ-promoter was measured by chromatin immunoprecipitation (ChIP) in K562 cells expressing PRMT5-f, or in PRMT5-kd cells. (B) Enrichment of various histone modifications, RNA polII and FLAG-tagged proteins at the γ-promoter was measured by ChIP in K562 cells expressing PRMT5-f or PRMT5Δ-f. The error bars correspond to the SD. The SD is between the means of 3 independent experiments.
Comparison of histone modifications at the γ-promoter in PRMT5-f and PRMT5-kd K562 cells. (A) Enrichment of various histone modifications, RNA polII and PRMT5 at the γ-promoter was measured by chromatin immunoprecipitation (ChIP) in K562 cells expressing PRMT5-f, or in PRMT5-kd cells. (B) Enrichment of various histone modifications, RNA polII and FLAG-tagged proteins at the γ-promoter was measured by ChIP in K562 cells expressing PRMT5-f or PRMT5Δ-f. The error bars correspond to the SD. The SD is between the means of 3 independent experiments.
We examined whether these repressive histone marks were dependent on the methyltransferase activity of PRMT5 by performing ChIP analyses on the K562 cells expressing PRMT5Δ-f, a FLAG-tagged mutant form of PRMT5, in which 5 amino acids in the S-adenosyl-L-methionine binding motif had been deleted19 (Figure 1B). The mutant form of PRMT5 retained the ability to bind robustly to the γ-promoter (as indicated by the FLAG ChIP), but not to induce the H4R3me2s mark. Concomitantly, we observed a distinct reduction of H4S1ph, H4K20me3, H3K9me3 and H3K27me3 at the γ-promoter, coincident with increased occupancy by RNA PolII in the PRMT5Δ-f–expressing cells. These findings indicated that the methyltransferase activity of PRMT5, and not just its physical occupation of the promoter, was integral for the subsequent generation of multiple repressive histone marks.
Recruitment of a multiprotein repressor complex to the γ-promoter is dependent on PRMT5
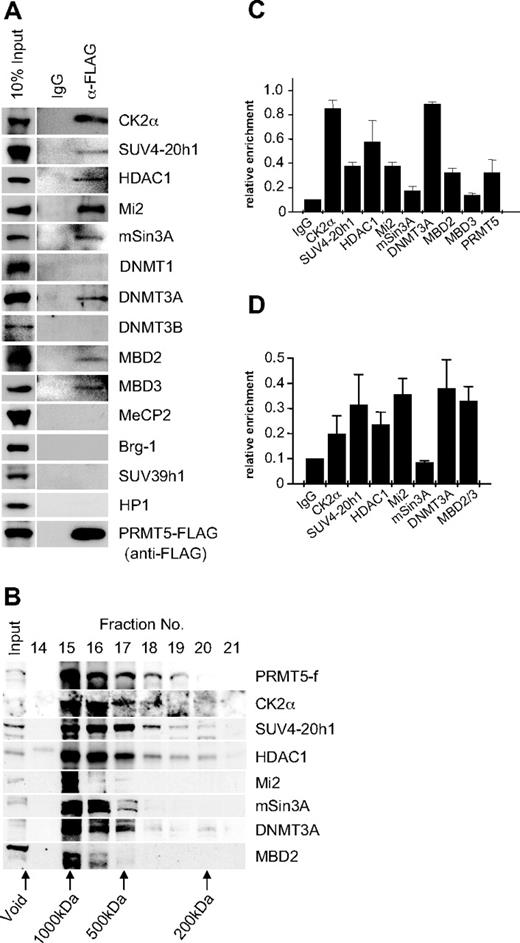
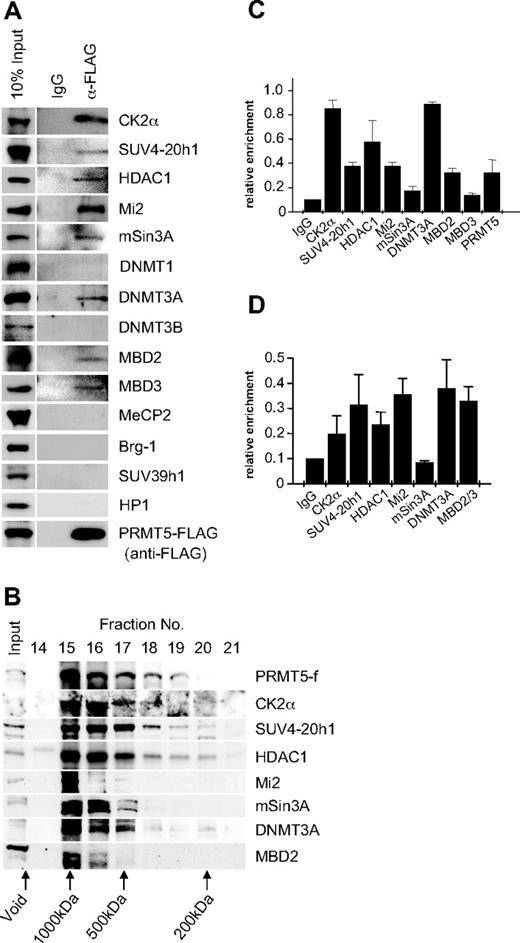
PRMT5 has been linked to transcriptional repression through the formation of 3 multiprotein complexes, 1 containing mSin3A, HDAC1, and HDAC2, another containing SWI/SNF components Brg1 and Brm, and a third containing MBD2/MBD3 and components of the nucleosome remodeling and histone deacetylation (NuRD) complex.20,,–23 To determine whether these factors associated with PRMT5 in K562 cells, we performed immunoprecipitations with extract from the PRMT5-f cells using the anti-FLAG antisera, and blotted the precipitates with antibodies to a range of candidate protein partners (Figure 2A). We identified a putative complex in which PRMT5 associates with the repressor proteins Mi2, mSin3A, MBD2 and HDAC1, as well as MBD3. Brg-1 was not associated with PRMT5 in this context. We also identified the DNA methyltransferase DNMT3A in association with PRMT5, consistent with our earlier findings that demonstrated a direct protein/protein interaction between these 2 enzymes. Two other DNA methyltransferases, DNMT3B and DNMT1, were not associated with PRMT5 in this context. Casein kinase 2α (CK2α) and SUV4-20h1, the enzymes linked to the repressive markers H4S1ph and H4K20me3s identified in our ChIP analysis, were also found to coimmunoprecipitate with PRMT5.15,16 However, SUV39h1 and HP1 did not coimmunoprecipitate with PRMT5 in this context.
Recruitment of multiple repressor proteins to the γ-promoter is dependent on PRMT5. (A) Western blot analysis of proteins immunoprecipitated from K562 cells expressing PRMT5-f with α-FLAG antibodies (Abs). A mock immunoprecipitation with normal rabbit immunoglobulin G (IgG) was used as a negative control. Abs used are indicated on the right. (B) Western blot analysis of extract from K562 cells expressing PRMT5-f fractionated by Superose 12 gel filtration. Column fractions were concentrated and analyzed using the antibodies indicated. (C) ChIP analysis on chromatin derived from PRMT5-f–expressing K562 cells with the stated Abs. (D) ChIP-ReChIP analysis on chromatin derived from FLAG immunoprecipitates from PRMT5-f K562 cells with the stated Abs. The precipitated DNA was amplified with primers specific for the γ-promoters. Enrichment was calculated relative to normal rabbit IgG. The error bars correspond to the SD. Each experiment was performed twice independently.
Recruitment of multiple repressor proteins to the γ-promoter is dependent on PRMT5. (A) Western blot analysis of proteins immunoprecipitated from K562 cells expressing PRMT5-f with α-FLAG antibodies (Abs). A mock immunoprecipitation with normal rabbit immunoglobulin G (IgG) was used as a negative control. Abs used are indicated on the right. (B) Western blot analysis of extract from K562 cells expressing PRMT5-f fractionated by Superose 12 gel filtration. Column fractions were concentrated and analyzed using the antibodies indicated. (C) ChIP analysis on chromatin derived from PRMT5-f–expressing K562 cells with the stated Abs. (D) ChIP-ReChIP analysis on chromatin derived from FLAG immunoprecipitates from PRMT5-f K562 cells with the stated Abs. The precipitated DNA was amplified with primers specific for the γ-promoters. Enrichment was calculated relative to normal rabbit IgG. The error bars correspond to the SD. Each experiment was performed twice independently.
The predicted molecular mass of the complex based on the coimmunoprecipitation studies was approximately 770 kDa. To confirm that the proteins coimmunoprecipitating with PRMT5 formed a multiprotein complex in vivo, we performed gel filtration chromatography with cellular extract from K562 cells expressing PRMT5-f, and analyzed fractions by Western blot (Figure 2B). PRMT5 eluted in the earliest protein-containing fractions, defining it as a component of a complex with a molecular mass of approximately 750 kDa, substantially less than the exclusion limit of the matrix (Mr 2 × 106). All other PRMT5 associated proteins defined in our coimmunoprecipitation studies that were tested exhibited peak levels in the fractions containing PRMT5. We confirmed the localization of the complex components on the proximal γ-promoter by ChIP in K562 cells expressing PRMT5-f (Figure 2C). Binding of complex components extended to regions of the promoter and γ-gene immediately adjacent to the proximal promoter, but was not observed at more distal sequences, with the exception of DNMT3A, which still displayed a binding peak on the proximal γ-promoter (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). We confirmed that these proteins formed a multiprotein complex assembled on the γ-promoter using a ChIP-reChIP strategy, in which chromatin immunoprecipitated with FLAG antisera was reimmunoprecipitated with antisera to specific complex components (Figure 2D).
PRMT5 methyltransferase activity is essential for complex assembly
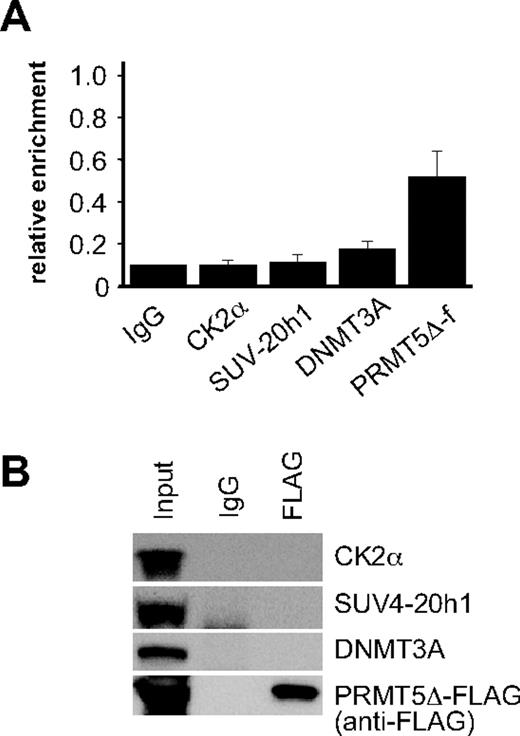
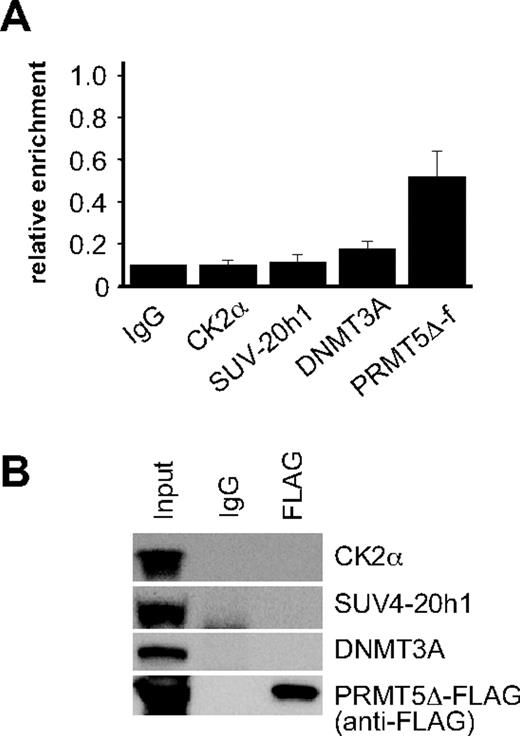
To ascertain whether assembly of the repressor complex at the γ-promoter was dependent on the methyltransferase activity of PRMT5, we performed ChIP assays on PRMT5Δ-f–expressing cells using antisera to the FLAG epitope, and selected complex components (Figure 3A). Binding of PRMT5Δ-f to the γ-promoter was readily detected indicating that the mutant form of PRMT5 binds to this site in the absence of enzymatic activity. Despite this, we were unable to detect CK2α or SUV4-20h1 binding, and occupancy of the γ-promoter by DNMT3A was markedly reduced. Consistent with this, we were also unable to demonstrate interactions between PRMT5Δ-f and selected complex components by coimmunoprecipitation (Figure 3B). These findings were in keeping with our analysis of the repressive histone marks in PRMT5Δ-f–expressing cells (Figure 1B), and indicated that the methyltransferase activity of PRMT5, and not just its binding to the γ-promoter, was essential for assembly of the repressor complex.
Assembly of the multiprotein repressor complex is dependent on PRMT5 enzymatic activity. (A) ChIP analysis on chromatin derived from PRMT5Δ-f– expressing K562 cells with the stated Abs. The precipitated DNA was amplified with primers specific for the γ-promoters. Enrichment was calculated relative to normal rabbit IgG. The error bars correspond to the SD. Each experiment was performed 3 times independently. Binding of CK2α, SUV4-20h1 and DNMT3A were not significant (P > .05) using Fisher exact test. (B) Western blot analysis of proteins immunoprecipitated from PRMT5Δ-f–expressing K562 cells with anti-FLAG Ab. A mock IP with normal rabbit IgG was used as a negative control. Abs used are indicated on the right.
Assembly of the multiprotein repressor complex is dependent on PRMT5 enzymatic activity. (A) ChIP analysis on chromatin derived from PRMT5Δ-f– expressing K562 cells with the stated Abs. The precipitated DNA was amplified with primers specific for the γ-promoters. Enrichment was calculated relative to normal rabbit IgG. The error bars correspond to the SD. Each experiment was performed 3 times independently. Binding of CK2α, SUV4-20h1 and DNMT3A were not significant (P > .05) using Fisher exact test. (B) Western blot analysis of proteins immunoprecipitated from PRMT5Δ-f–expressing K562 cells with anti-FLAG Ab. A mock IP with normal rabbit IgG was used as a negative control. Abs used are indicated on the right.
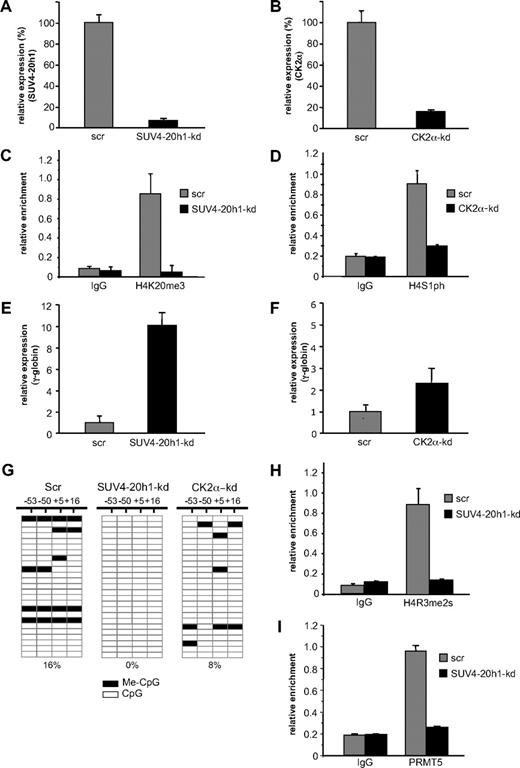
SUV4-20h1 is also critical for γ-globin gene repression
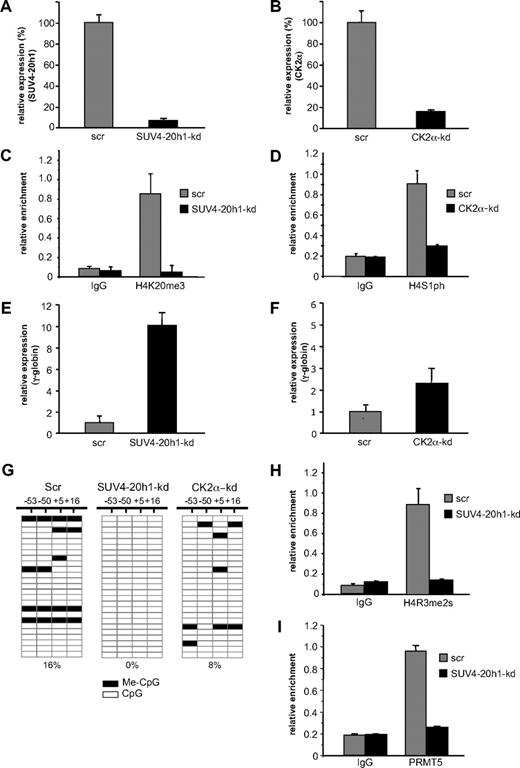
Our data with PRMT5-kd and PRMT5Δ-f cells demonstrated that PRMT5-induced symmetric di-methylation of histone H4R3 acted as a trigger to induce a range of repressive epigenetic marks, culminating in DNA methylation and gene silencing. In addition to the H4R3me2s mark, the differential between PRMT5-kd and PRMT5Δ-f cells was most evident with the H4K20me3 and H4S1ph marks, and our identification of the enzymes responsible for these histone modifications in the PRMT5-dependent complex suggested that they might also be central to γ-gene silencing. To evaluate this, we used an shRNA-mediated approach to achieve a knockdown of SUV4-20h1 to less than 10% (Figure 4A), and of CK2α to approximately 20% (Figure 4B), with cells expressing scrambled shRNAs (scr) serving as controls. The reduced level of SUV4-20h1 resulted in a marked reduction of the H4K20me3 mark on the γ-promoter (Figure 4C), and the H4S1p mark was reduced 3-fold in the CK2α-kd cells (Figure 4D). As a result of the reduction of these repressive marks, γ-gene expression increased 10-fold in the SUV4-20h1-kd cells (Figure 4E), with a more modest 2.5-fold increase observed in the CK2α-kd cells (Figure 4F). These findings suggested that SUV4-20h1 played an important role in γ-gene silencing, in concert with PRMT5. We have previously shown that loss of PRMT5 led to a marked reduction in methylation of 4 CpG dinucleotides immediately flanking the γ-gene transcriptional start site that are essential for gene silencing.6,–8 We therefore used bisulfite sequencing to assess the methylation status of these CpGs in the SUV4-20h1-kd, CK2α-kd, and scrambled control cells (Figure 4G). Consistent with the observation that globin gene expression is not detectable in a large proportion of uninduced K562 cells, we observed methylation of the 4 CpG dinucleotides in 16% of clones derived from scrambled control cells. In contrast, no methylation was detectable on any of these CpG dinucleotides in the SUV4-20h1-kd cells. A more modest reduction in DNA methylation was observed in the CK2α-kd cells, commensurate with the level of γ-gene induction observed in these cells. To determine whether PRMT5 recruitment and establishment of the H4R3me2s mark at the γ-promoter were influenced by SUV4-20h1, we performed ChIP analysis on the SUV4-20h1-kd cells with anti-H4R3me2s (Figure 4H) and anti-PRMT5 (Figure 4I) antibodies. Consistent with a coordinate role for the 2 methylases in γ-gene silencing, the levels of both PRMT5 binding and the H4R3me2s mark were substantially reduced in the SUV4-20h1-kd cells compared with scr control.
SUV4-20h1 mediates transcriptional silencing of the γ-gene. (A) Expression of SUV4-20h1 and (B) CK2α was analyzed in K562 cells expressing specific shRNAs or scrambled control sequences (scr). (C) H4K20me3 levels at the γ-promoter in SUV4-20h1-kd and scr control cells, and (D) H4S1ph levels at the γ-promoter in CK2α-kd and scr control cells were measured by ChIP analysis. The error bars correspond to the SD. Each experiment was performed twice independently. Normal rabbit IgG served as the control. (E) Analysis of γ-globin gene expression in SUV4-20h1-kd and scr control cells, and (F) CK2α-kd and scr control cells. (G) DNA methylation at the human γ-gene in SUV4-20h1-kd, CK2α-kd and scr control cells. Each row shows the methylation status of individual CpG dinucleotides derived from sequence analysis of 25 representative (of at least 40) individual cloned polymerase chain reaction (PCR) products of the γ-genes after bisulfite modification from knockdown or scr control K562 cells. The differences between the 2 knockdown lines and the scrambled control were highly significant (P < .05). The numbers at the top represent the positions of the CpG dinucleotides relative to the transcriptional start site of the γ-gene. (H) ChIP of the H4R3me2s level and (I) PRMT5 binding at the γ-promoter in SUV4-20h1-kd and scr control cells. Normal rabbit IgG served as the control. The error bars correspond to the SD. Each experiment was performed 3 times independently.
SUV4-20h1 mediates transcriptional silencing of the γ-gene. (A) Expression of SUV4-20h1 and (B) CK2α was analyzed in K562 cells expressing specific shRNAs or scrambled control sequences (scr). (C) H4K20me3 levels at the γ-promoter in SUV4-20h1-kd and scr control cells, and (D) H4S1ph levels at the γ-promoter in CK2α-kd and scr control cells were measured by ChIP analysis. The error bars correspond to the SD. Each experiment was performed twice independently. Normal rabbit IgG served as the control. (E) Analysis of γ-globin gene expression in SUV4-20h1-kd and scr control cells, and (F) CK2α-kd and scr control cells. (G) DNA methylation at the human γ-gene in SUV4-20h1-kd, CK2α-kd and scr control cells. Each row shows the methylation status of individual CpG dinucleotides derived from sequence analysis of 25 representative (of at least 40) individual cloned polymerase chain reaction (PCR) products of the γ-genes after bisulfite modification from knockdown or scr control K562 cells. The differences between the 2 knockdown lines and the scrambled control were highly significant (P < .05). The numbers at the top represent the positions of the CpG dinucleotides relative to the transcriptional start site of the γ-gene. (H) ChIP of the H4R3me2s level and (I) PRMT5 binding at the γ-promoter in SUV4-20h1-kd and scr control cells. Normal rabbit IgG served as the control. The error bars correspond to the SD. Each experiment was performed 3 times independently.
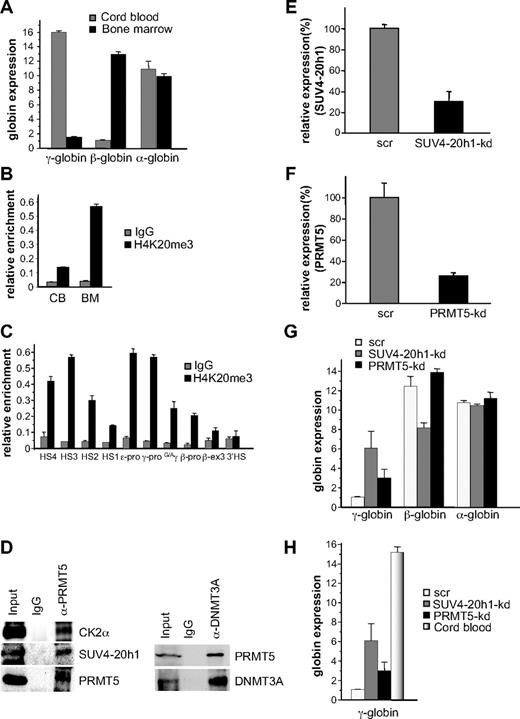
Role of SUV4-20h1 in developmental globin gene silencing
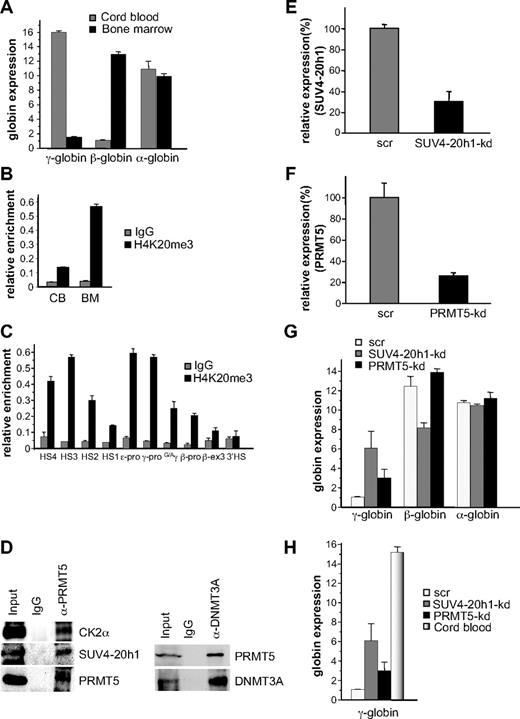
Based on our expression and DNA methylation data, it appeared that SUV4-20h1 played a more central role in γ-gene silencing than CK2α. We therefore determined whether the H4K20me3 mark was evident at the human γ-globin promoter in a developmentally specific pattern in primary human cells, we isolated erythroid progenitors from CB and adult BM and cultured them in conditions that supported high levels of cellular expansion. ChIP analysis demonstrated a 2-fold increase in the H4K20me3 mark at the γ-promoter in adult BM erythroid progenitors compared with progenitors derived from CB (supplemental Figure 2). Our previous analysis of globin expression in cultured CB progenitors had demonstrated almost equivalent levels of γ- and β-globin expression,8 which may have accounted for the relatively modest differential in the H4K20me3 mark between these cells and BM progenitors. To address this, we cultured CB progenitors in conditions that promoted high levels of γ-gene expression,10 and BM progenitors in low fetal hemoglobin conditions11 (Figure 5A), and examined the H4K20me3 mark in these cells. In this setting, we observed a 4-fold difference in the H4K20me3 mark at the γ-promoter in adult BM erythroid progenitors compared with progenitors derived from CB (Figure 5B). We then examined the distribution of the H4K20me3 mark across the β-globin locus in these BM erythroid progenitors using primer pairs spanning regions of the locus control region (LCR) hypersensitive sites (HS1-4), the γ-, ϵ- and β-globin promoters, the intergenic region between the Gγ- and Aγ-genes, exon 3 of the β-gene, and the 3′HS (Figure 5C). Enrichment was observed at the γ- and ϵ-promoters compared with the β-promoter in these adult cells. We also observed increased levels at the HSs of the LCR, particularly at HS4 and HS3. To confirm the presence of the multiprotein complex in BM erythroid progenitors we immunoprecipitated extract from these cells with PRMT5 antibodies, and blotted the precipitates with antibodies to selected complex components (Figure 5D left panel). Binding of PRMT5 to CK2α and SUV4-20h1 was readily detected, but we were unable to detect DNMT3A using this approach (not shown). However, coimmunoprecipitation of DNMT3A with PRMT5 in these extracts was demonstrated using the reverse order of antibodies (Figure 5D right panel).
SUV4-20h1 induced epigenetic modification of the γ-globin genes is developmentally specific. (A) Quantitative reverse-transcriptase (Q-RT)–PCR analysis of γ-, β-, and α-globin gene expression in cord blood (CB) and bone marrow (BM). (B) H4K20me3 at the γ-promoter was measured by ChIP in chromatin fractions from erythroid progenitors from CB and BM. (C) Localization of H4K20me3 across the β-globin locus measured by ChIP in chromatin fractions from erythroid progenitors from BM. The precipitated DNA was amplified with primers specific for the indicated regions of the β-globin locus.8 HS, hypersensitive site; pro, promoter; G/Aγ, intergenic region between Gγ- and Aγ-globin genes; β-ex3, exon 3 of the β-gene. The error bars correspond to the SD. Each experiment was performed 3 times independently. Normal rabbit IgG served as the control. (D) Western blot analysis of proteins immunoprecipitated from erythroid progenitors from BM with α-PRMT5 Ab (left panel) or α-DNMT3A Ab (right panel). A mock IP with normal rabbit IgG was used as a negative control. Abs used are indicated on the right. (E) Expression of SUV4-20h1 and (F) PRMT5 was analyzed in erythroid progenitors from BM expressing specific shRNAs or scrambled control sequences (scr). (G) Analysis of γ-, β-, and α-globin gene expression in SUV4-20h1-kd, PRMT5-kd and scr control BM cells. The error bars correspond to the SD. Each experiment was performed 4 times independently. (H) Comparison of γ-globin gene expression levels in CB and SUV4-20h1-kd, PRMT5-kd and scr control BM cells by Q-RT-PCR.
SUV4-20h1 induced epigenetic modification of the γ-globin genes is developmentally specific. (A) Quantitative reverse-transcriptase (Q-RT)–PCR analysis of γ-, β-, and α-globin gene expression in cord blood (CB) and bone marrow (BM). (B) H4K20me3 at the γ-promoter was measured by ChIP in chromatin fractions from erythroid progenitors from CB and BM. (C) Localization of H4K20me3 across the β-globin locus measured by ChIP in chromatin fractions from erythroid progenitors from BM. The precipitated DNA was amplified with primers specific for the indicated regions of the β-globin locus.8 HS, hypersensitive site; pro, promoter; G/Aγ, intergenic region between Gγ- and Aγ-globin genes; β-ex3, exon 3 of the β-gene. The error bars correspond to the SD. Each experiment was performed 3 times independently. Normal rabbit IgG served as the control. (D) Western blot analysis of proteins immunoprecipitated from erythroid progenitors from BM with α-PRMT5 Ab (left panel) or α-DNMT3A Ab (right panel). A mock IP with normal rabbit IgG was used as a negative control. Abs used are indicated on the right. (E) Expression of SUV4-20h1 and (F) PRMT5 was analyzed in erythroid progenitors from BM expressing specific shRNAs or scrambled control sequences (scr). (G) Analysis of γ-, β-, and α-globin gene expression in SUV4-20h1-kd, PRMT5-kd and scr control BM cells. The error bars correspond to the SD. Each experiment was performed 4 times independently. (H) Comparison of γ-globin gene expression levels in CB and SUV4-20h1-kd, PRMT5-kd and scr control BM cells by Q-RT-PCR.
To examine the functional role of SUV4-20h1 and PRMT5 in adult BM erythroid progenitors, we employed our specific shRNAs in lentiviral vectors to target the individual enzymes. Levels of SUV4-20h1 were reduced to 30% of a scrambled control (Figure 5E), and PRMT5 to 25% of control (Figure 5F). Up-regulation of γ-gene expression was observed in both knockdowns, whereas the expression of the β- and α-globin genes was not increased (Figure 5G). The level of γ-gene expression in the SUV4-20h1-kd BM was 40% and the PRMT5-kd was 20% of the levels observed in unmanipulated CB (Figure 5H), suggesting that these interventions could have therapeutic relevance.
Discussion
DNA methylation and repressive histone modifications play essential and coordinated roles in gene silencing. Direct links between these processes have been emerging, with the identification of interactions between lysine methyltransferases, and DNA methyltransferases,18,24,–26 methyl-CpG binding proteins27 and methyl-CpG enriched regions.28 We have recently shown that histone methylation and DNA methylation are directly linked in the context of human γ-globin gene silencing by the protein arginine methyltransferase PRMT5, which induces the repressive histone H4R3me2s mark that acts as a template for direct binding of DNMT3A.8 We now report that PRMT5-mediated γ-gene silencing is also dependent on the assembly of a multiple repressor complex containing previously unrecognised partners of PRMT5, including CK2α, and SUV4-20h1, in addition to DNMT3A, and components of the NuRD and Sin3 complexes. The enzymatic activity of PRMT5 is essential for assembly of this complex at the γ-promoter, as it is not observed in cells expressing a PRMT5 mutant lacking methyltransferase activity, despite the mutant retaining the ability to localize to the γ-promoter.
The idea that chromatin inactivation lies upstream of DNA methylation has been proposed previously, although the molecular mechanism by which these events are directly linked has remained elusive. Studies of the epigenetic inactivation of several tumor suppressor genes have demonstrated that methylation of histone H3K9 corresponds to gene inactivation, and precedes DNA methylation.29,30 Similarly, silencing of the human fetal globin genes has been shown to precede DNA methylation in somatic cell hybrids.31 Our studies with PRMT5-kd, PRMT5Δ-f and SUV4-20h1-kd cells suggest that establishment of repressive epigenetic modifications are coordinated, with the H4R3me2s and H4K20me3 marks functioning as necessary triggers for subsequent DNA methylation. This is supported by our finding of the H4K20me3 mark at the β-promoter in adult BM in levels above background, but with no enrichment for H4R3me2s,8 and no gene silencing. Conceivably, SUV4-20h1 could play a dual role, analogous to the methyltransferase G9a, involved in repression of the γ-globin gene with the PRMT5 complex, and activation of the β-globin gene with alternate partners.32 γ-globin gene expression is not detectable in a large proportion of uninduced K562 cells, indicating that the γ-genes are significantly repressed in these cells. This repression is due, in part, to DNA methylation as γ-gene induction is observed with 5-azacytidine, coincident with DNA hypomethylation. It is also due to other repressive epigenetic marks (H4R3me2s, H4K20me3, H3K9me3, H3K27me3) that can be erased with knockdown of components of the PRMT5 complex. Our findings suggest that γ-gene repression (like gene repression at most loci) is multilayered, requiring coordinated histone and DNA modifications.
Methylation of the CpG dinucleotides flanking the CAP site is essential for γ-gene silencing,6 and 2 of these CpGs reside within the region of the γ-promoter where the repressor complex assembles.33 Although the changes in methylation that we observed in our mutant K562 lines were modest compared with the increase in γ-gene expression, this is consistent with in vivo data in the baboon in which relatively modest changes in methylation induced substantial changes in γ-gene expression.14 DNMT3A-induced methylation of the γ-gene CpGs could serve as a template for the subsequent recruitment of MBD2 and MBD3, and this could be facilitated further by a direct interaction between the RG-rich N-terminus of MBD2 and PRMT5.23 MBD2 has recently been implicated in silencing of the human γ-genes in transgenic mice, although in this setting it was not localized to the γ-promoter.34 MBD2 has also been linked to silencing of the chicken embryonic genes, where it binds in a repressor complex containing NuRD components.35 The identification of both MBD2 and MBD3 in the γ-globin gene PRMT5-dependent repressor complex is in contrast to the PRMT5-containing complex that assembles on the P14ARF and P16INK4a CpG islands, which contains only MBD2.23 Unlike our findings, recruitment of PRMT5 in that context is dependent on DNA methylation, although a direct link between histone and DNA methyltransferases was not established.
The identification of the epigenetic markers H4S1ph and H4K20me3s at the γ-genes in the context of PRMT5-mediated transcriptional silencing expands the known roles of these histone modifications. To date, phosphorylation of H4S1 has been most widely studied in yeast, where it has been shown to regulate chromatin compaction during sporulation, a function that may be conserved in spermatogenesis in higher organisms.36 Further yeast studies have demonstrated that CK2α can induce this modification in the setting of transcriptional repression, and DNA repair.15,37 H4S1ph has been shown to oppose acetylation of histone H4 at lysines 5, 8, 12, and 16 by the NuA4 acetyltransferase complex, an effect that was abrogated by the methylation of H4R3.15 We noted a reduction in acetylation of H4K12 and K8 (but not K5 or K16) in the context of γ-gene silencing that was not blocked by the H4R3me2s mark, suggesting that H4S1ph at the γ-globin promoter may impede the function of another histone acetyltransferase complex. Yeast proteomic studies have found CK2 associated with Sin338,39 mirroring their association with PRMT5 in our studies.
Tri-methylation of H4K20 has been studied in Drosophila and mammalian systems, where it has emerged as a hallmark of pericentric heterochromatin in concert with tri-methylation of H3K9 and H3K27.15 Induction of H4K20me3 in mammals is dependent on SUV4-20h isoforms, 2 SET domain HMTases that are recruited to the nucleosome through a direct interaction with HP1 isoforms.16 More recently, these 3 markers have been linked to transcriptional silencing of Hox genes, being enriched in the promoter and coding regions of the Ultrabithorax gene in the “off” state.40 The presence of SUV4-20h1 in the PRMT5 complex provides a direct mechanism by which this mark could be established at the γ-promoter. Interestingly, we did not identify SUV39h1 or HP1 in association with PRMT5, suggesting that recruitment of SUV4-20h1 in this setting may occur through a different mechanism to that observed at pericentric heterochromatin.
In addition to increased levels of the H4K20me3 mark at the γ-promoter, we also observed substantial enrichment of this mark at several of the LCR HSs, raising the possibility that coordinated repressive marks established synchronously at the promoters, and HSs could inhibit the LCR/globin gene interactions.41,42 Alternatively, the complex could assemble on the LCR and spread across the locus, similar to epigenetic modifiers acting at a distance in the β-globin cluster in the setting of activation of β-gene expression.43 Although the peak enrichment at the LCR occurred at HS3, the site at which the H4R3me2s modification also peaks in BM, high levels of the H4K20me3 mark were also observed at HS4 and HS2, which exhibit no enrichment of H4R3me2s.8 Similarly, higher levels of the H4K20me3 mark were detected elsewhere in the locus compared with the IgG control, and the significance of this is currently unclear.
The increased γ-gene expression in both cell lines and erythroid progenitors from BM suggests that the effects of the SUV4-20h1-kd are specific and direct. In keeping with this, we observed reduced PRMT5 localization, reduced levels of the H4R3me2s mark and loss of DNA methylation at the γ-gene in these cells. Activation of γ-gene expression in patients with β-thalassemia and sickle cell disease has been a therapeutic goal for many years. The identification of both PRMT5 and SUV4-20h1 as key regulators of γ-gene silencing provides novel targets for small molecule inhibitors, a strategy that we are actively pursuing.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Sidney Pestka for the PRMT5Δ plasmid, Richard Gaynor for the PRMT5 plasmid, and members of the Jane laboratory for helpful discussions. Human BM and CB were collected after approval by the Melbourne Health, and Royal Women's Hospital Human Research Ethics Committees.
This work was supported by The National Health and Medical Research Council of Australia, National Institutes of Health grants PO1 HL53749-03 and RO1 HL69232-01 (S.M.J.), The Cooley's Anemia Foundation (Q.Z.), and The Natural Science Foundation of China no. BK2007521 and New Century Excellent Talents NCET-07-0433 (Q.Z.).
National Institutes of Health
Authorship
Contribution: G.R., S.M.J., and Q.Z. designed the experiments, analyzed the data, and wrote the manuscript; L.C. performed experiments; and R.J.S. and R.L.M. performed proteomics experiments and analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stephen M. Jane or Quan Zhao, Rotary Bone Marrow Research Laboratory, c/o Royal Melbourne Hospital Post Office, Grattan St, Parkville 3050, Australia; e-mail: jane@wehi.edu.au or qzhao@nju.edu.cn.