Abstract
CCR6 is a chemokine receptor expressed on Th17 cells and regulatory T cells that is induced by T-cell priming with certain cytokines, but how its expression and stability are regulated at the molecular level is largely unknown. Here, we identified and characterized a noncoding region of the human CCR6 locus that displayed unmethylated CpG motifs (differentially methylated region [DMR]) selectively in CCR6+ lymphocytes. CCR6 expression on circulating CD4+ T cells was stable on cytokine-induced proliferation but partially down-regulated on T-cell receptor stimulation. However, CCR6 down-regulation was mostly transient, and the DMR within the CCR6 locus remained demethylated. Notably, in vitro induction of CCR6 expression with cytokines in T-cell receptor-activated naive CD4+ T cells was not associated with a demethylated DMR and resulted in unstable CCR6 expression. Conversely, treatment with the DNA methylation inhibitor 5′-azacytidine induced demethylation of the DMR and led to increased and stable CCR6 expression. Finally, when cloned into a reporter gene plasmid, the DMR displayed transcriptional activity in memory T cells that was suppressed by DNA methylation. In summary, we have identified a noncoding region of the human CCR6 gene with methylation-sensitive transcriptional activity in CCR6+ T cells that controls stable CCR6 expression via epigenetic mechanisms.
Introduction
Specific patterns of chemokine receptor expression ensure an orchestrated migration of leukocytes throughout the body, where immune cells home to their target tissues under both steady-state and inflammatory conditions. The chemokine receptor CCR6 is widely expressed on human blood and tissue leukocytes, including subsets of dendritic cells, CD45RO+ effector/memory T cells, CD25high regulatory T cells (Tregs), naive and memory B cells, NKT cells, and NK cells.1-6 On memory T cells, CCR6 is expressed on a population of autoreactive interleukin-10 (IL-10)–producing cells7 and subsets of skin- and mucosa-homing cells. Several studies have further shown that CCR6 is consistently expressed on inflammatory IL-17-producing CD4+ T cells,8-10 which are involved in a wide array of adverse inflammatory diseases, such as rheumatoid arthritis, psoriatic disease, inflammatory bowel disease, or encephalopathies in mice and humans.10-14 More recently, CCR6 was identified as a new risk factor for rheumatoid arthritis; moreover, a nucleotide polymorphism within the CCR6 locus was shown to be associated with susceptibility to rheumatoid arthritis, Graves disease, and Crohn disease.15,16
CCR6 was shown to play a pivotal role in directing inflammatory and regulatory cells to inflamed tissues or the gut, where the ligand of CCR6 CCL20 is expressed.13,17,18 Although most chemokine-chemokine receptor interactions are characterized by a high promiscuity, CCR6 binds only CCL20 and β-defensins.1
It is widely assumed that the differentiation of T cells into specialized memory subsets also involves the acquisition and stable expression of homing- and chemokine receptor repertoires, allowing tissue- or inflammation-specific trafficking of these subsets.19 Evidence has been provided in recent years that differentiation of T cells into distinct lineages with stable phenotypes and functions involves epigenetic regulation of critical effector molecules20-22 or lineage-specific transcription factors, such as Foxp3 in Tregs.23-26 Only few studies demonstrated that molecules involved in trafficking are subject to epigenetic regulation in T cells27,28 or cancer cells.29,30 In addition, a considerable plasticity in the expression of homing receptors has also been observed in some cases.31,32 Whether this also applies to the expression of CCR6 is not known. In vitro, CCR6 expression can be induced de novo on T-cell receptor (TCR)-stimulated naive T cells by a cocktail of proinflammatory cytokines in combination with transforming growth factor-β (TGF-β).9
The transcriptional regulation of CCR6 is not well understood; a region with promoter activity has been identified in the mouse CCR6 gene,33 and overexpression of the transcription factor RORγt, the master regulator of Th17 cells, leads to CCR6 expression on human and murine T cells.10,34 However, whether stable CCR6 expression on human T cells is controlled by epigenetic mechanisms has not been studied so far. We therefore investigated whether epigenetic mechanisms, including DNA methylation of the CCR6 locus, contribute to the regulation of stable CCR6 expression in human T cells. We could identify a noncoding region in the CCR6 gene, harboring transcriptional activity in primary T cells and being differentially methylated in human CCR6− and CCR6+ T cells. These observations and the inducing effect of the DNA-methylation inhibitor 5′-azacytidine suggest that epigenetic mechanisms are involved in the regulation of stable CCR6 expression and the imprinting of distinct homing properties in human memory T cells.
Methods
Cells, antibodies, and flow cytometry
Peripheral blood mononuclear cells (PBMCs) from buffy coats (DRK Blutspendedienst) were separated with a Ficoll-Hypaque gradient (Sigma-Aldrich). Cell surface antigens were analyzed by single-parameter or multiparameter fluorescence-activated cell sorter (FACS) analysis using the following monoclonal antibodies: phycoerythrin-anti-CCR6 (11A9) and Alexa700-anti-CD4 (RPA-T4) (both from BD Biosciences), allophycocyanin-anti-CD25 (BC96; eBioscience), phycoerythrin-Cy5-anti-CD8 (B9.11), phycoerythrin-Cy5-anti-CD56 (N901), and fluorescein isothiocyanate-anti-CD25 (B1.49.9, all Beckman Coulter). Antibodies generated in house (DRFZ) are: fluorescein isothiocyanate-anti-CD45RA (4G11), Alexa405-anti-CD4 (TT1), and Alexa405-anti-CD3 (OKT3). In some experiments, CCR6 expression was detected by indirect immunofluorescence using biotinylated CCR6 (11A9, BD Biosciences) followed by staining with allophycocyanin-conjugated streptavidin (Southern Biotechnology). Stains were performed in phosphate-buffered saline containing 0.5% bovine serum albumin to block unspecific binding. A FACSCanto II (BD Biosciences) was used for data acquisition, and analysis was performed using the FlowJo Version 9 software (TreeStar).
T-cell isolation and FACS sorting
Total CD4+ and CD8+ T cells were enriched from PBMCs using anti-CD4 or anti-CD8 magnetic beads, respectively, and the autoMACS separation system according to the manufacturer's instructions (Miltenyi Biotec). After subsequent staining with anti-CD4, anti-CD8, anti-CD25, anti-CD45RA, and anti-CCR6, the following cell populations were sorted on a FACSAria or FACSDiva cell sorter (BD Biosciences): naive CD4+ T cells (CD4+CD25−CD45RA+), CD4+ Tregs (CD4+CD25high), CCR6− naive CD4+ T cells, CCR6− memory CD4+ T cells (CD4+CD25−CD45RA−), CCR6+ memory CD4+ T cells, CCR6− naive CD8+ T cells (CD8+CD25−CD45RA+), CCR6− memory CD8+ T cells (CD8+CD25−CD45RA−), and CCR6+ memory CD8+ T cells. On reanalysis, sorted cells routinely showed more than 95% purity. Ex vivo isolated cells were either used for methylation analyses or for in vitro cell cultures.
T-cell culture of naive and memory CD4+ T cells
Cells were cultured in complete RPMI 1640 Glutamax medium (Invitrogen) containing 5% human serum (complete medium [CM]). For stimulation of naive CD4+ T cells, 1 × 105 cells were cultured in CM supplemented with 20 ng/mL IL-2 (R&D Systems) and 2 μg/mL neutralizing antibodies against IL-4, IL-12, and interferon-γ (neutral conditions, antibodies from BD Biosciences) in flat-bottom microtiter plates and 1 × 105 magnetic beads coated with anti-CD3 and anti-CD28 (Dynabeads, Invitrogen) for 4 days followed by transfer in CM containing 1000 U/mL IL-2 (Proleukin, Chiron). For induction of CCR6 on naive CD4+ cells, the following cytokines were added at the beginning of the culture: 10 ng/mL TGF-β, 10 ng/mL IL-6, 10 ng/mL IL-1, and 10 ng/mL tumor necrosis factor-α (all R&D Systems). In some experiments, stimulation of cells was repeated up to 2 times under the same conditions. For induction of CCR6 on naive CD4+ T cells with the methylation inhibitor 5′-azacytidine (Aza, Sigma-Aldrich), 5μM Aza was added after 48 hours of culture.
Memory CD4+CCR6+ and CD4+CCR6− cells were stained with carboxyfluorescein succinimidyl ester and cultured either in CM containing magnetic beads as described above in this section or 10 ng/mL IL-7 and 10 ng/mL IL-15 (R&D Systems). After 4 days, only the cultures containing Dynabeads were transferred in CM containing 1000 U/mL IL-2 (Proleukin, Chiron). At the end of the cultures, cells were analyzed by flow cytometry and used for methylation analyses (see “Methylation analysis”).
Cloning of luciferase reporter plasmids and in vitro methylation
An extended version of the differentially methylated region of the human CCR6 locus was amplified by polymerase chain reaction (PCR) using genomic DNA as a template and the following primers: (A) 5′-AGTCATGCATCTCTCAGATGGAGGAGGGAC-3′, (B) 5′-TCAGACTAGTAGAGGCCGTGTGAGTTAAAGAC-3′. For generation of a luciferase reporter vector in a CpG-free background, the 1073-bp PCR product was inserted into the pCpGL-cytomegalovirus/EF1 vector35,36 from which the cytomegalovirus promoter was removed. Sequencing of the cloned region revealed a 100% identity with the CCR6 region sequence of the Homo sapiens genome stored at Ensembl. A reporter plasmid without insert served as control.
The luciferase reporter vector pCpGL-CCR6-differentially methylated region (DMR)/EF1 was methylated in vitro using the methylase SssI (New England Biolabs) according to the manufacturer's recommendations, followed by purification using the QIAquick PCR clean-up kit (QIAGEN). In control samples, using pCpGL-EF1 and pCpGL-CCR6-DMR/EF1, the methyl-group donor S-adenosylmethionine was omitted. Successful methylation was verified by restriction enzyme treatment of the reporter plasmid containing the CCR6 DMR with the methylation-sensitive and methylation-insensitive enzymes HpaII and MspI (New England Biolabs), respectively.
Transfection and luciferase reporter assays
Cells from 4 different donors were sorted for CD4+ memory CCR6+ and CCR6− populations and activated using anti-CD3/CD28 Dynabeads (Invitrogen) for 60 hours. Magnetic beads were removed, and 5 × 106 of pooled cells were transfected using 2.5 μg of either methylated or nonmethylated pCpGL-CCR6-DMR/EF1 vector or insert-less control plasmid in triplicates. Synthetic Renilla luciferase reporter vector (pRL-TK; Promega) was cotransfected (1.5 μg) and served as an internal control for transfection efficiency. Nucleofection was performed according to the Amaxa technique using the primary Human T Cell Nucleofector Kit (Lonza Walkersville). Cells were immediately resuspended for culture in prewarmed RPMI 1640 medium containing IL-2 (Proleukin, Chiron; without penicillin/streptomycin). After 48 hours, cells were harvested and luciferase activity was measured on Orion L luminometer using the Dual Luciferase Assay system (Promega). Firefly relative light unit data were normalized to Renilla luciferase activity, and x-fold induction was calculated by comparison with insert-less control vector.
Methylation analysis
Genomic DNA was isolated from sorted T-cell subsets using the DNeasy tissue kit (QIAGEN) following the protocol for cultured animal cells. Bisulfite treatment of genomic DNA was performed as previously described.37 PCR was performed in a final volume of 25 μL containing 1 × PCR buffer, 1 U Taq DNA polymerase (QIAGEN), 200μM deoxynucleoside triphosphate, 12.5 pmol of each primer, and 7 ng bisulfite-treated genomic DNA at 95°C for 15 minutes, and 40 cycles of 95°C for 1 minute, 55°C for 45 seconds, and 72°C for 1 minute, and a final extension step of 10 minutes at 72°C. PCR products were purified using ExoSAP-IT (USB Corp) and sequenced by applying the PCR primers and the ABI Big Dye Terminator, Version 1.1-chemistry (Applied Biosystems) followed by capillary electrophoresis on an ABI 3100 genetic analyzer. AB1 files were interpreted using epigenetic sequencing methylation analysis software (ESME).38
Primers used for the reactions are given in supplemental Figure 2 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
In silico analyses
For prediction of putative transcription factor-binding sites, the tool MatInspector (Genomatix) was used.
Statistics
Data are mean plus or minus SD. Differences between groups were assessed using the Wilcoxon rank test as indicated. P < .05 was considered significant.
Results
A noncoding region in the CCR6 locus displays differential methylation that correlates with CCR6 expression
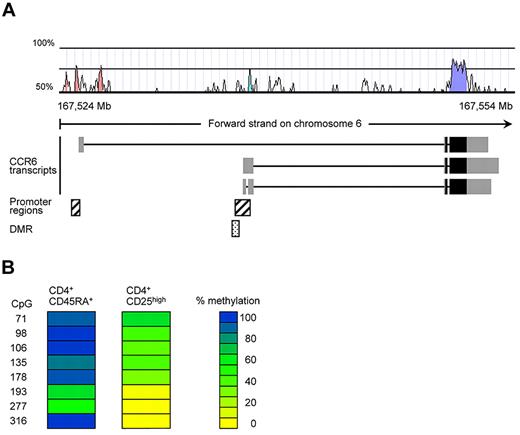
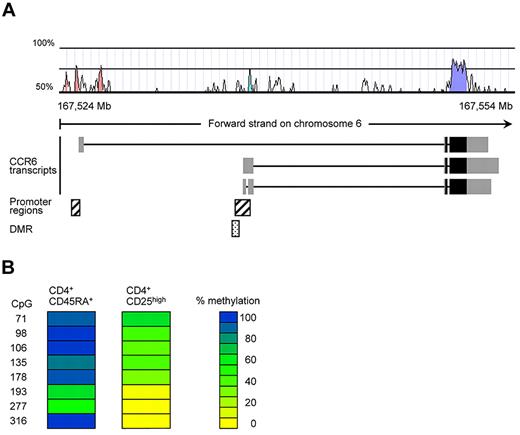
We have previously conducted a screen for epigenetically regulated genes in human Tregs by comparing DNA-methylation patterns of conventional naive CD4+ T cells and CD25highCD4+ Tregs using the differential methylation hybridization (DMH) technique.25 In this DMH screen, a CpG-rich region within the CCR6 locus turned out to be fully methylated in naive CD4+ T cells and demethylated in Tregs (data not shown). The identified DMR is located upstream of 2 reported CCR6 transcripts and overlaps with a putative CCR6 promoter39 in a region displaying substantial (30%-70%) homology between human and mouse (Figure 1A; supplemental Figure 2). To validate the DMH data, we here performed bisulfite sequencing using genomic DNA from conventional naive CD45RA+CD4+ T cells and CD25highCD4+ Tregs isolated from human peripheral blood. In accordance with the DMH data, CD25highCD4+ Tregs showed a largely demethylated DMR (average CpG methylation rate 23.8%), whereas conventional naive T cells were almost fully methylated (average methylation rate 81%, Figure 1B; supplemental Figure 2).
A noncoding region in the CCR6 gene displays differential methylation in human PBMCs. (A) Localization of the CpG-rich region within the human CCR6 locus. The exon structure of the putative CCR6 transcripts, obtained from the Ensembl database (GRCh37), were plotted together with a homology chart (human-mouse; Vista), predicted promoter regions (Genomatix) and the identified DMR in the CCR6 locus. The gray boxes in the CCR6 transcripts represent noncoding exon regions, whereas the black boxes contain coding regions. (B) DNA methylation pattern for CCR6 were analyzed in sorted CD4+CD45RA+ and CD4+CD25high T cells from pooled PBMCs (5 donors). Each column represents one blood cell subset with each row representing a single CpG site. The numbers indicate the position of the CpG motif as indicated in supplemental Figure 2. DNA methylation was measured by bisulfite sequencing. CpG methylation levels are color-coded according to a scale ranging from yellow (0% methylation) to blue (100% methylation).
A noncoding region in the CCR6 gene displays differential methylation in human PBMCs. (A) Localization of the CpG-rich region within the human CCR6 locus. The exon structure of the putative CCR6 transcripts, obtained from the Ensembl database (GRCh37), were plotted together with a homology chart (human-mouse; Vista), predicted promoter regions (Genomatix) and the identified DMR in the CCR6 locus. The gray boxes in the CCR6 transcripts represent noncoding exon regions, whereas the black boxes contain coding regions. (B) DNA methylation pattern for CCR6 were analyzed in sorted CD4+CD45RA+ and CD4+CD25high T cells from pooled PBMCs (5 donors). Each column represents one blood cell subset with each row representing a single CpG site. The numbers indicate the position of the CpG motif as indicated in supplemental Figure 2. DNA methylation was measured by bisulfite sequencing. CpG methylation levels are color-coded according to a scale ranging from yellow (0% methylation) to blue (100% methylation).
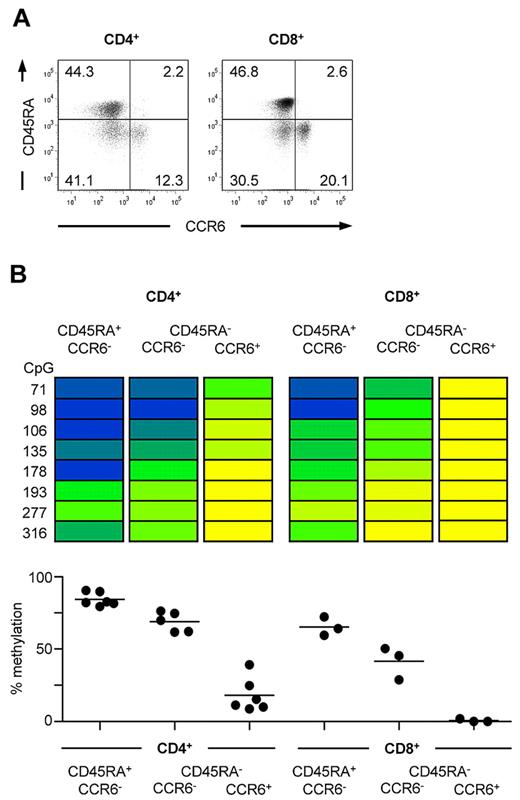
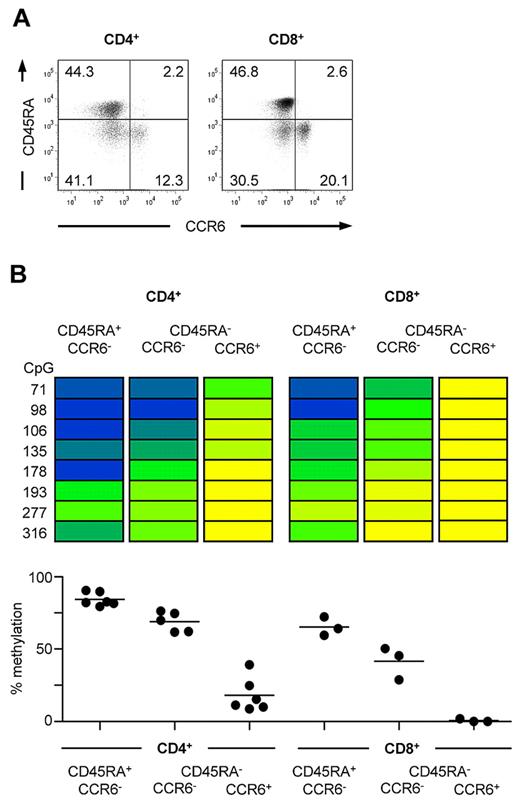
In addition to Tregs,6 expression of CCR6 has been reported for both CD4+ and CD8+ memory T cells.5,7,9 To understand whether the observed demethylation of the CCR6 locus is confined to the fraction of cells that express CCR6, ex vivo CD4+ and CD8+ T-cell subsets were sorted according to CCR6 and CD45RA expression (Figure 2A) and analyzed by bisulfite sequencing of the CCR6 locus. Both naive CD45RA+CD4+ T cells lacking CCR6 expression as well as CCR6−CD45RA−CD4+ memory T cells were strongly methylated, whereas CCR6-expressing CD4+ memory T cells turned out to be almost completely demethylated in the analyzed DMR (Figure 2B). Although CD8+ T cells displayed an overall lower level of DNA methylation, CCR6+CD8+ memory T cells clearly displayed a less methylated DMR compared with CCR6−CD8+ memory T cells (Figure 2B). In accordance with these data, peripheral blood leukocyte subsets lacking CCR6 expression, such as resting CD14+ monocytes and CD15+ granulocytes,1,40,41 displayed an almost completely methylated DMR (average methylation > 84%), whereas CCR6-expressing CD56+ NK and NKT cells2,4 and mature B cells3 were fully demethylated at this site (data not shown). Together, our data suggest that CCR6 expression correlates with demethylation of the DMR in human leukocytes.
Demethylation of the CCR6 region correlates with CCR6 expression in human CD4+ and CD8+ T cells. (A) Representative FACS staining of CCR6 and CD45RA on CD4+CD25− and CD8+CD25− lymphocytes. (B) CD4+ and CD8+ cells were isolated from PBMCs and sorted into CCR6−CD45RA+ naive and CCR6− and CCR6+CD45RA− memory T cells resulting in subsets with a purity more than 95%. Methylation analyses of sorted subsets are shown for one representative donor. The degree of methylation is color-coded according to the scale in Figure 1B. Graphs below show the mean of methylation from all CpG sites from 5 to 6 donors for CD4+ T cells and 3 donors for CD8+ T cells, respectively.
Demethylation of the CCR6 region correlates with CCR6 expression in human CD4+ and CD8+ T cells. (A) Representative FACS staining of CCR6 and CD45RA on CD4+CD25− and CD8+CD25− lymphocytes. (B) CD4+ and CD8+ cells were isolated from PBMCs and sorted into CCR6−CD45RA+ naive and CCR6− and CCR6+CD45RA− memory T cells resulting in subsets with a purity more than 95%. Methylation analyses of sorted subsets are shown for one representative donor. The degree of methylation is color-coded according to the scale in Figure 1B. Graphs below show the mean of methylation from all CpG sites from 5 to 6 donors for CD4+ T cells and 3 donors for CD8+ T cells, respectively.
CCR6+ T cells stably express CCR6 on cell division and maintain a demethylated DMR
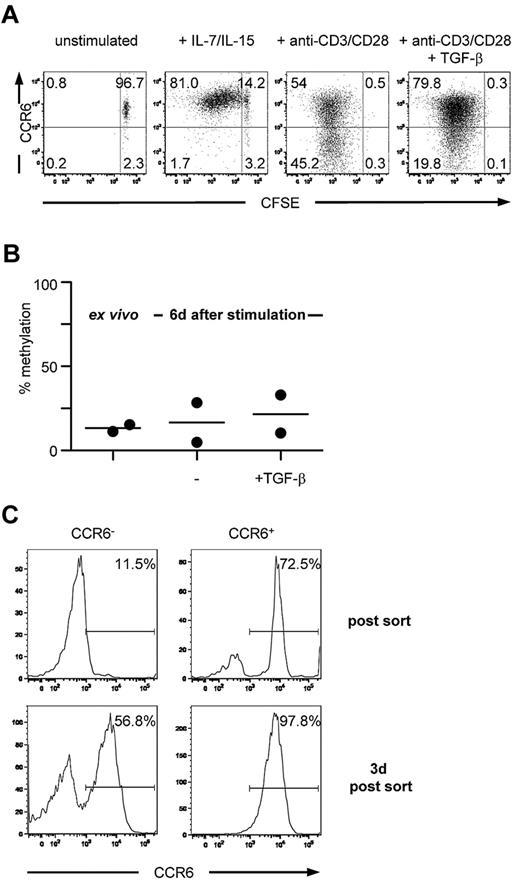
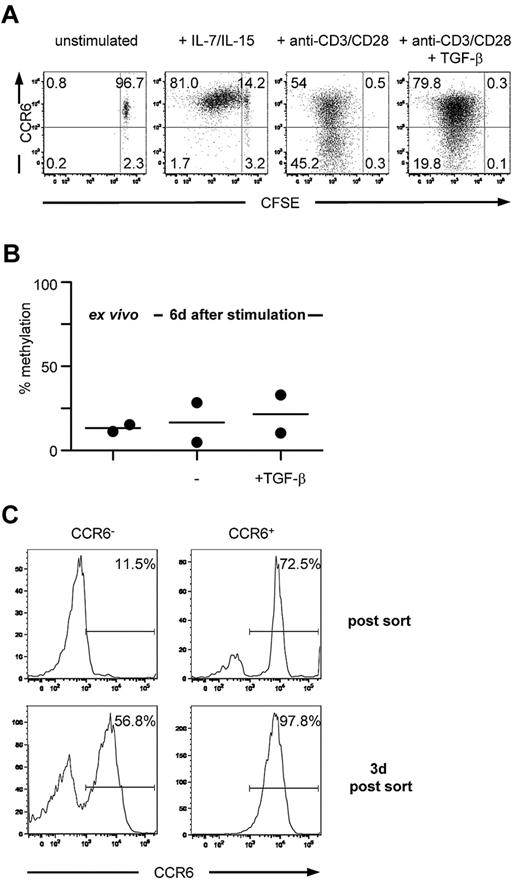
To investigate whether both CCR6 expression on CD4+ memory T cells and the corresponding methylation pattern of the DMR were stable on cell division, sorted CCR6+CD25−CD45RA−CD4+ T cells were labeled with carboxyfluorescein succinimidyl ester and either stimulated with cytokines that mediate homeostatic turnover of memory cells or by triggering via the TCR. Consistent with previous reports,42 memory T cells proliferated in the presence of homeostatic cytokines, such as IL-7 and IL-15, and all proliferating cells maintained high levels of CCR6 expression, even after multiple cell divisions (Figure 3A). TCR stimulation resulted in a down-regulation of CCR6 expression as has been reported,43 which was inhibited by TGF-β (Figure 3A). However, under all stimulation conditions tested, the methylation status of the DMR remained unchanged and showed a comparable low level of methylation (Figure 3B; and data not shown). Importantly, TCR-induced down-regulation of CCR6 expression was mostly transient because a majority (50%–80% among 4 donors) of FACS-purified CCR6−/low cells that had lost surface CCR6 expression on TCR stimulation rapidly reexpressed CCR6 after culture in the absence of TCR stimulation (Figure 3C). Thus, demethylation of the DMR is linked to long-term stability of CCR6 expression.
Circulating CCR6+ T cells stably express CCR6 on in vitro expansion and maintain a demethylated DMR. (A) Carboxyfluorescein succinimidyl ester-labeled CCR6+CD4+ memory T cells were cultured for 6 days in medium containing recombinant human IL-7 and IL-15 or anti-CD3/anti-CD28 Dynabeads with or without TGF-β and reanalyzed for CCR6 expression. One representative donor of 4 is shown. (B) Ex vivo isolated CCR6+CD4+ memory T cells were stimulated with anti-CD3/anti-CD28 Dynabeads in the absence or presence of TGF-β. After 6 days, methylation analysis of the DMR was assessed. (C) Ex vivo isolated CCR6+CD4+ memory T cells were stimulated with anti-CD3/anti-CD28 Dynabeads, sorted on day 6 in CCR6− and CCR6+ cells, and cultured for an additional 3 days in medium containing IL-2. Data are representative for 2 donors.
Circulating CCR6+ T cells stably express CCR6 on in vitro expansion and maintain a demethylated DMR. (A) Carboxyfluorescein succinimidyl ester-labeled CCR6+CD4+ memory T cells were cultured for 6 days in medium containing recombinant human IL-7 and IL-15 or anti-CD3/anti-CD28 Dynabeads with or without TGF-β and reanalyzed for CCR6 expression. One representative donor of 4 is shown. (B) Ex vivo isolated CCR6+CD4+ memory T cells were stimulated with anti-CD3/anti-CD28 Dynabeads in the absence or presence of TGF-β. After 6 days, methylation analysis of the DMR was assessed. (C) Ex vivo isolated CCR6+CD4+ memory T cells were stimulated with anti-CD3/anti-CD28 Dynabeads, sorted on day 6 in CCR6− and CCR6+ cells, and cultured for an additional 3 days in medium containing IL-2. Data are representative for 2 donors.
In vitro induction of CCR6 expression is unstable and does not lead to demethylation of the DMR
CCR6 expression can be induced in vitro on a fraction of naive CD4+ T cells on activation in the presence of a cocktail of cytokines, including tumor necrosis factor-α, IL-6, IL-1, and TGF-β.9 We asked whether this de novo induction of CCR6 expression is associated with a demethylation of the DMR.
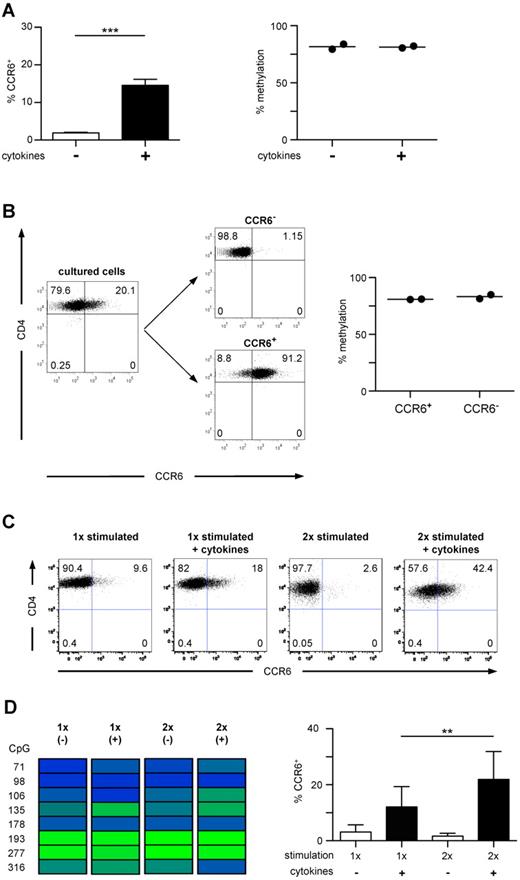
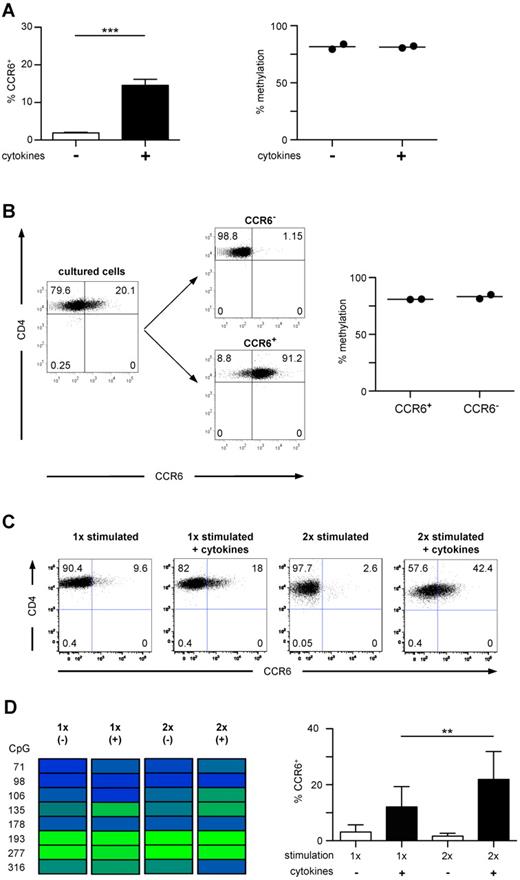
Naive CD45RA+CCR6−CD4+ T cells were stimulated with anti-CD3/anti-CD28 Dynabeads in the presence of the inflammatory cytokine-cocktail and TGF-β. At day 6 of in vitro culture, a significantly higher fraction of T cells expressed CCR6 compared with T cells stimulated in the absence of exogenous cytokines (Figure 4A), whereas IL-17-producing capacities were not acquired under these conditions (data not shown). However, both cell populations were almost completely methylated within the DMR, showing no difference to unstimulated naive T cells (Figures 4A, 2B). Even after sorting into CCR6− and CCR6+ cells, no selective demethylation of the DMR could be observed within de novo induced CCR6+ cells (Figure 4B). Although repetitive stimulation in the presence of the cytokine cocktail further increased the frequency of CCR6+ cells, demethylation of the DMR was still not detectable (Figure 4C-D), suggesting that inflammatory cytokines raise the expression rate but do not induce a demethylation of the CCR6 locus. In contrast to ex vivo isolated CD4+ memory T cells, CCR6 de novo induction in naive T cells by TCR stimulation in the presence of inflammatory cytokines led only to transient CCR6 expression, and virtually all cells lost CCR6 expression on prolonged in vitro culture in the absence of the inducing cytokines (supplemental Figure 1). Thus, the failure of cytokines to induce demethylation of the DMR in vitro is associated with unstable CCR6 expression.
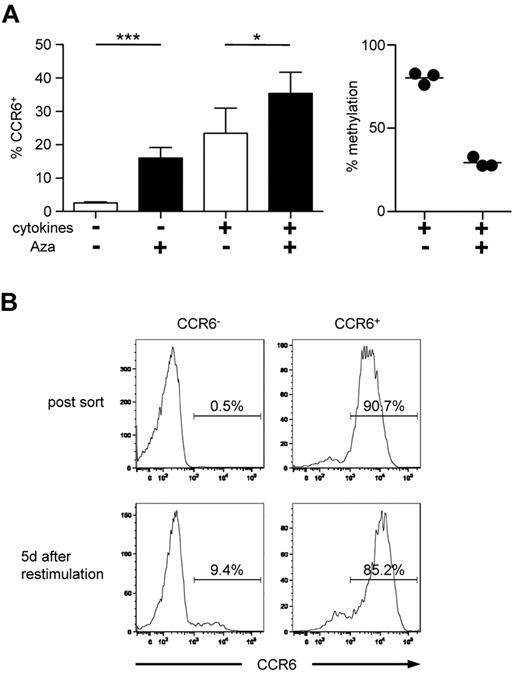
In vitro induced CCR6 expression is unstable and not associated with a demethylated DMR. (A) Naive CD4+ T cells were stimulated with anti-CD3/anti-CD28 Dynabeads without (white bar) or with proinflammatory cytokines and TGF-β (black bar) for 4 days, followed by culture in IL-2. After 6 to 7 days, cells were analyzed for CCR6 expression (n = 21, left graph). The methylation profile of CD4+ T cells cultured with (+) and without (−) cytokines is shown in the right graph (n = 2). ***P < .001. (B) Cells stimulated with cytokines as in panel A were sorted into CCR6+ and CCR6− cells (shown for 1 representative donor) and analyzed for methylation of the DMR. (C) Naive CD4+ T cells were stimulated and cultured as in panel A. At day 6, a fraction of the cultured cells were stimulated again under the same conditions. Cells were analyzed for CCR6 expression (one representative donor). (D) Methylation analysis of the CCR6 DMR (one representative donor, left panel) and FACS analysis of CCR6 expression in cells cultured as described in C (n = 11, right panel). **P < .01.
In vitro induced CCR6 expression is unstable and not associated with a demethylated DMR. (A) Naive CD4+ T cells were stimulated with anti-CD3/anti-CD28 Dynabeads without (white bar) or with proinflammatory cytokines and TGF-β (black bar) for 4 days, followed by culture in IL-2. After 6 to 7 days, cells were analyzed for CCR6 expression (n = 21, left graph). The methylation profile of CD4+ T cells cultured with (+) and without (−) cytokines is shown in the right graph (n = 2). ***P < .001. (B) Cells stimulated with cytokines as in panel A were sorted into CCR6+ and CCR6− cells (shown for 1 representative donor) and analyzed for methylation of the DMR. (C) Naive CD4+ T cells were stimulated and cultured as in panel A. At day 6, a fraction of the cultured cells were stimulated again under the same conditions. Cells were analyzed for CCR6 expression (one representative donor). (D) Methylation analysis of the CCR6 DMR (one representative donor, left panel) and FACS analysis of CCR6 expression in cells cultured as described in C (n = 11, right panel). **P < .01.
Inhibition of DNA methylation leads to stable CCR6 expression in TCR-stimulated naive T cells
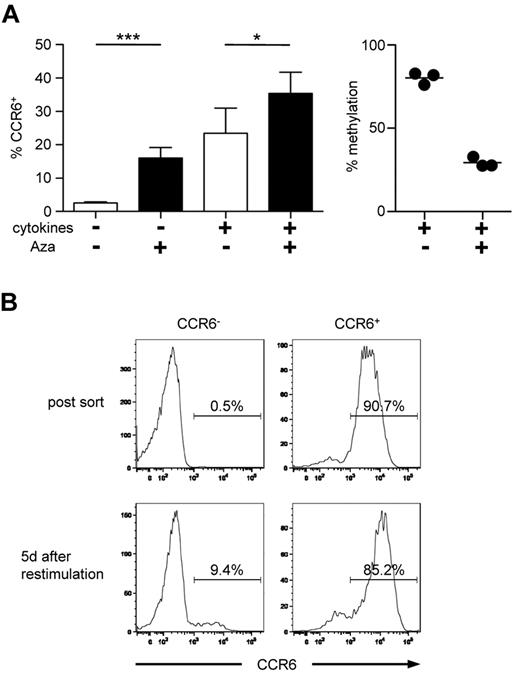
The DNA methylation status can be pharmacologically manipulated by induction of DNA replication in the presence of DNA-methyltransferase inhibitors, such as Aza. To further investigate whether demethylation of the DMR is involved in the stabilization of CCR6 expression, we activated naive CD4+ T cells with anti-CD3 and anti-CD28 Dynabeads in the presence of Aza. Strikingly, Aza treatment induced CCR6 expression in a significant fraction of cells even in the absence of exogenous cytokines (Figure 5A left panel). Combined treatment with cytokines and Aza resulted in an increase in CCR6 induction compared with cells treated with cytokines alone. In addition, in these Aza plus cytokine-treated cells, a clear demethylation of the DMR could be detected (Figure 5A right panel). Interestingly, when Aza-induced CCR6+ cells were sorted and restimulated under neutral conditions in the absence of Aza, a remarkable stability of CCR6 expression was observed in these cells, suggesting that demethylation of the CCR6 locus controls stable chemokine receptor expression in T cells (Figure 5B).
Inhibition of DNA methylation leads to stable CCR6 expression. (A) Naive CD4+ T cells were stimulated with or without proinflammatory cytokines and TGF-β. After 48 hours, Aza was added to the media for another 48 hours. Expression of CCR6 (n > 18, left panel) and methylation of the DMR (n = 3, right panel) were analyzed on day 6. *P < .05. ***P < .001. (B) Naive CD4+ T cells were stimulated with Aza and without cytokines as in panel A. After 6 days, cells were sorted according to their CCR6 expression and restimulated under neutral conditions in the absence of Aza for additional 5 days followed by reanalysis of CCR6 expression. Data shown are representative for 2 independently performed experiments.
Inhibition of DNA methylation leads to stable CCR6 expression. (A) Naive CD4+ T cells were stimulated with or without proinflammatory cytokines and TGF-β. After 48 hours, Aza was added to the media for another 48 hours. Expression of CCR6 (n > 18, left panel) and methylation of the DMR (n = 3, right panel) were analyzed on day 6. *P < .05. ***P < .001. (B) Naive CD4+ T cells were stimulated with Aza and without cytokines as in panel A. After 6 days, cells were sorted according to their CCR6 expression and restimulated under neutral conditions in the absence of Aza for additional 5 days followed by reanalysis of CCR6 expression. Data shown are representative for 2 independently performed experiments.
The differentially methylated element of the CCR6 locus mediates transcriptional activity
The differential methylated region of the CCR6 locus starts 444 of 446 bases upstream of 2 predicted CCR6 transcripts (Figure 1A) and partially overlaps with an in silico predicted promoter region. However, prediction strength was weak, and a promoter-typical transcriptional start site was absent in the analyzed region. Several putative binding sites for transcriptional regulators, such as ETS1, GATA, NFAT, CREB, and others, could be identified (Genomatix; applying a matrix similarity > 0.9; supplemental Figure 2).
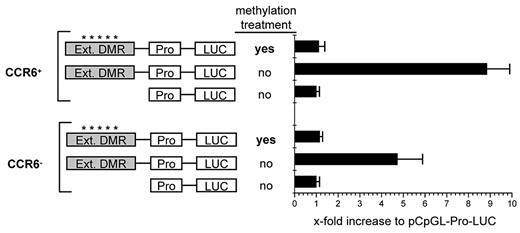
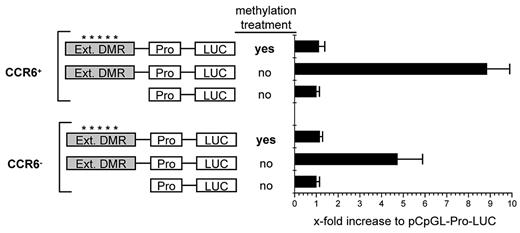
To analyze the role of the DMR for the transcriptional regulation of CCR6 expression, we cloned this element into a CpG-free luciferase reporter vector (kindly supplied by M. Rehli, Regensburg) which contains a human EF1 promoter and allows the detection of transcriptionally active enhancer elements as well as its dependency on DNA methylation.35 To include further possibly important sequences, we extended the region into both directions to cover a total of 1072 bp (supplemental Figure 2); The CpGs in the extended region displayed the same differential methylation pattern (supplemental Figure 2) as found for the DMR (Figure 2). On transfection of either CCR6+ or CCR6− memory CD4+ T cells isolated from peripheral blood, transcriptional activity of the luciferase reporter construct containing the extended DMR was increased 6- to 21-fold compared with the empty control vector (pCpGL-EF1) in CCR6+ cells and 5- to 10-fold in CCR6− cells (Figure 6). The notion of an enhancer function of the extended DMR is furthermore supported by the presence of an activating histone modification mark (H3K4monomethyl) typical for enhancers in this region, as to be found in the database of the “ENCODE” project for a lymphoblastoid cell line.44
The CCR6 DMR mediates transcriptional activity. Activated CCR6+ and CCR6− CD45RA−CD4+ memory T cells were transfected with control plasmid (pCpGL-EF1 promoter) or pCpGL-EF1-CCR6-DMR, where the extended DMR was cloned into the pCpGL-EF1 promoter plasmid. The relative luciferase light units were normalized to Renilla luciferase activity. Values for the pCpGL-EF1-promoter plasmid were set to 1 to obtain the x-fold activation. Shown is the mean luciferase activity of one representative of 3 experiments with cells pooled from 4 donors each. *A methylated slate of CpGs.
The CCR6 DMR mediates transcriptional activity. Activated CCR6+ and CCR6− CD45RA−CD4+ memory T cells were transfected with control plasmid (pCpGL-EF1 promoter) or pCpGL-EF1-CCR6-DMR, where the extended DMR was cloned into the pCpGL-EF1 promoter plasmid. The relative luciferase light units were normalized to Renilla luciferase activity. Values for the pCpGL-EF1-promoter plasmid were set to 1 to obtain the x-fold activation. Shown is the mean luciferase activity of one representative of 3 experiments with cells pooled from 4 donors each. *A methylated slate of CpGs.
Strikingly, transcriptional activation was completely lost after in vitro methylation of the CpGs in the extended DMR, demonstrating that the methylation status critically affects its enhancer function. These findings demonstrate that the extended DMR of the CCR6 locus displays methylation-sensitive enhancer activity independently of TCR-mediated signals, suggesting that this element and its demethylation is functionally involved in the maintenance of stable CCR6 expression in memory T cells.
Discussion
Differential expression of distinct chemokine receptors on immune cells ensures a coordinated temporal-spatial distribution of various functional subsets and differentiation stages of effector and regulatory cells to and within lymphoid and nonlymphoid tissues. Some of the migratory phenotypes become apparently permanently imprinted on differentiation28,45 and allow a selective delivery of memory populations to specific compartments of the body (“homing”). How stable homing phenotypes are acquired and maintained in the progeny of T and B cells is not well understood. Epigenetic mechanisms, notably the methylation/demethylation of CpG-rich DNA regions involved in transcriptional regulation, are ideally suited to provide a long-term memory of phenotypic changes because of the imprinting of “heritable” methylation signatures.22,46 We here investigated whether differential DNA methylation might be involved in the acquisition of stable CCR6 expression in primary human T cells.
A region within the CCR6 locus that was differentially methylated between Tregs and naive CD4+ T cells was initially identified by a DMH approach.25 This DMR was now verified by bisulfite sequencing of sorted human blood leukocyte subsets. The observed methylation patterns matched largely the expression of CCR6 in various leukocyte subsets, where CCR6-expressing cells, such as CD56+ NK and NKT cells,2,4 certain CD4+ and CD8+ T-cell subsets,47,48 mature B cells,3 and CD4+ Tregs,6 displayed a partially or even completely demethylated DMR, whereas the locus was completely methylated in resting CD14+ monocytes and CD15+ granulocytes lacking CCR6 expression.1,40,41 Among CD4+ and CD8+ memory T cells, demethylation of the DMR was confined to those subsets expressing CCR6 on their surface, suggesting a role for this genetic element and its methylation status in CCR6 regulation.
Demethylation of essential regulatory DNA regions is usually regarded as a signature for a stable, heritable expression of the regulated gene, as we could recently exemplify for the Treg-specific transcription factor Foxp3.24 As CCR6 expression can be either transient or permanent, we investigated in the present study whether such an epigenetic imprinting also occurs at the DMR in the CCR6 locus.
Like many other receptors for inflammatory chemokines, CCR6 is expressed on a large proportion of circulating memory T cells but is absent from naive T cells,5,47 suggesting that CCR6 expression is acquired during T-cell priming. Along this line, CCR6 expression can be induced on priming of naive CCR6−CD4+ T cells using a cytokine cocktail containing IL-1, IL-6, TGF-β, and tumor necrosis factor-α.9,47 Importantly, T cells that had acquired CCR6 expression in vivo displayed a demethylated DMR and showed stable CCR6 expression in dividing cells in the absence of TCR stimulation. The methylation status in memory cells was independent from the cytokine profile, as we detected demethylation of the DMR in both IL-17-secreting and nonsecreting CCR6+ T cells (S.S. and S.F., unpublished data, 2010). Although CCR6 was down-regulated on memory cells on TCR triggering, this down-regulation was mostly transient and did not lead to remethylation of the DMR. The observed CCR6 down-regulation under these conditions might even be the result of receptor recycling rather than transcriptional down-regulation. In contrast, in vitro induction of CCR6 on naive T cells using cytokines was neither stable nor associated with demethylation of the DMR. Even though the percentage of CCR6+ cells increased with repetitive stimulations, CCR6 expression remained unstable and the DMR fully methylated. A similar observation was recently made for the Treg-specific transcription factor Foxp3, where the Treg-specific demethylated enhancer region in the Foxp3 locus was only found to be demethylated in ex vivo isolated stable Tregs, but not in in vitro induced Tregs, which only transiently expressed Foxp3.49 Even repetitive stimulations with the inducing cytokine TGF-β were neither sufficient to convey stability of Foxp3 expression nor demethylation of the enhancer region. In other cases, however, repetitive stimulations under permissive conditions have been shown to achieve stability of certain phenotypes, such as selectin-ligand expression in CD4+ T cells45 and cytokine production in polarized Th1 and Th2 cells.50 Together, our present data suggest that specific, so far unknown signals are required to imprint a stable CCR6 expression profile in CD4+ T cells by demethylation of the DMR in the CCR6 locus.
Our hypothesis that the methylation status of a regulatory gene element in the CCR6 locus dictates long-term stability of CCR6 expression was further corroborated by experiments involving pharmacologic DNA hypomethylation. Application of the DNA methyltransferase inhibitor 5′-azacytidine during T-cell stimulation not only increased CCR6 expression, even in the absence of exogenous CCR6-inducing cytokines, but also resulted in CCR6+ cells that showed stable CCR6 expression on in vitro expansion, a finding that was previously also observed for Foxp3 expression in in vitro induced Tregs.49
To understand the function of the DMR in transcriptional regulation of CCR6, we cloned an extended version of this element in a human EF1 promoter/luciferase reporter gene construct. To cover a larger part of the conserved region, we extended the initially identified region downstream up to the beginning of the predicted CCR6 transcripts. This resulted in a strong increase in transcriptional activity of the reporter, suggesting that the element is contributing to transcriptional regulation (eg, by acting as an enhancer). Several putative transcription factor-binding sites were predicted for this region, although only one was conserved between mouse and humans. Some of them, such as ETS1 or CREB, have been reported to bind in a methylation-dependent manner to their target sequences.36,51 Strikingly, the only transcription factor-binding site conserved between mouse and human in the extended DMR under the criteria used here predicts binding of ETS1.
As to be expected, transcriptional activity was highest in CCR6+ memory cells providing a permissive milieu for gene activity. Use of a reporter vector devoid of CpG motifs35 allowed us to test the consequence of DNA methylation for the activity of the extended CCR6 DMR. Consistent with the hypothesis that differential methylation of this region is decisive for the capacity to express stably CCR6, the methylated form of the extended DMR was not able to mediate transcriptional activity, even in the permissive CCR6+ cells, similar to what has been found in case of Foxp3.36
In conclusion, we here provide, for the first time, experimental evidence that epigenetic mechanisms are not only regulating the transcriptional activity of a chemokine receptor but moreover might play a key role in the imprinting of a permanent expression pattern, thus endowing differentiated memory T cells with topographic memory and shaping their long-term migratory behavior.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the members of the FACS facility, DRFZ for cell sorting, Dr Julia Polansky for carefully reading the manuscript and helpful suggestions, and Michael Rehli, Regensburg, for kindly providing the CpG-free plasmid.
This work was supported by the Collaborative Research Center (program SFB650 and SFB633), the German Research Foundation (KFO250), and the Technologiestiftung Berlin.
Authorship
Contribution: S.S. and S.F. performed research, analyzed data, and wrote the paper; D.E. and B.H. performed research and analyzed data; U.B., L.R., and B.S. performed research; A.G. designed and performed research; S.O. designed research; and J.G., J.H., and A.H. designed research and wrote the paper.
Conflict-of-interest disclosure: U.B. and S.O. are employees and owners of Epiontis GmbH, who owns CCR-6 methylation patent applications. The remaining authors declare no competing financial interests.
The current affiliation of L.R. is Immunology Program, National University of Singapore, Centre for Life Sciences, Singapore.
Correspondence: Alf Hamann, Experimental Rheumatology, Charité University Medicine Berlin, c/o DRFZ, Charitéplatz 1, 10117 Berlin, Germany; e-mail: hamann@drfz.de.
References
Author notes
S.S. and S.F. contributed equally to this study.
J.G., J.H., and A.H. contributed equally to this study.