Abstract
Severe congenital neutropenia (SCN) is an inborn disorder of granulopoiesis that in many cases is caused by mutations of the ELANE gene, which encodes neutrophil elastase (NE). Recent data suggest a model in which ELANE mutations result in NE protein misfolding, induction of endoplasmic reticulum (ER) stress, activation of the unfolded protein response (UPR), and ultimately a block in granulocytic differentiation. To test this model, we generated transgenic mice carrying a targeted mutation of Elane (G193X) reproducing a mutation found in SCN. The G193X Elane allele produces a truncated NE protein that is rapidly degraded. Granulocytic precursors from G193X Elane mice, though without significant basal UPR activation, are sensitive to chemical induction of ER stress. Basal and stress granulopoiesis after myeloablative therapy are normal in these mice. Moreover, inaction of protein kinase RNA-like ER kinase (Perk), one of the major sensors of ER stress, either alone or in combination with G193X Elane, had no effect on basal granulopoiesis. However, inhibition of the ER-associated degradation (ERAD) pathway using a proteosome inhibitor resulted in marked neutropenia in G193X Elane. The selective sensitivity of G913X Elane granulocytic cells to ER stress provides new and strong support for the UPR model of disease patho-genesis in SCN.
Introduction
Severe congenital neutropenia (SCN) is a rare bone marrow–failure syndrome characterized by severe chronic neutropenia, an arrest of granulocytic differentiation at the promyelocyte or myelocyte stage, and a marked propensity to develop acute myeloid leukemia and myelodysplasia. SCN demonstrates multiple modes of inheritance, including autosomal recessive, autosomal dominant, X-linked, and sporadic patterns. Accordingly, genetic studies have identified multiple gene mutations in SCN, including ELANE,1 HAX1,2 G6PC3,3 GFI1,4 and WAS.5 Mutations of ELANE encoding neutrophil elastase (NE) are the most common, accounting for approximately 60% of cases (all in autosomal-dominant or sporadic SCN).1,6-10 To date, 73 distinct mutations of ELANE have been identified in patients with SCN.10 Most of the mutations (∼ 80%) are missense mutations, although mutations leading to splicing defects (∼ 10%) and premature stop codons (∼ 10%) have also been observed.
The molecular mechanisms by which ELANE mutations disrupt granulopoiesis are unclear, but genetic studies provide 2 important clues. First, the ELANE mutations in SCN patients are heterozygous, suggesting a dominant mechanism of action. Second, a case report of paternal mosaicism for an ELANE mutation provides evidence that expression of mutant neutrophil elastase inhibits granulopoiesis in a cell-intrinsic fashion, because no toxic paracrine effects of mutant neutrophil elastase on wild-type granulocytic cells in this mosaic individual were observed.11 The diversity of ELANE mutations in SCN and the lack of a consistent effect of these mutations on NE enzyme activity12 suggest that structural rather than functional perturbations in the NE protein might be responsible for the disruption in granulopoiesis. Indeed, there is evidence that a shared feature of the different NE mutants is their propensity to misfold.13,14 These observations suggest a model in which the accumulation of misfolded NE in the endoplasmic reticulum (ER) triggers the unfolded protein response (UPR) and ultimately leads to a block in granulocytic differentiation.15
Protein folding in the specialized environment of the ER is a dynamic and imperfect process that is dependent on several cellular factors, including nutrient availability, oxidation-reduction balance, and energy homeostasis.16 Accumulation of misfolded proteins in the ER triggers the UPR pathway, a coordinated adaptive program evolved to this type of ER stress. The canonical mammalian UPR pathway has 3 main branches that are regulated by the transmembrane ER proteins protein kinase RNA-like ER kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme (IRE1). Activation of these ER sensors reduces misfolded proteins through 3 major mechanisms: (1) by attenuating global protein synthesis; (2) by inducing the transcription of several ER-resident proteins involved in protein folding, including ER chaperon proteins; and (3) by triggering ER-associated protein degradation (ERAD) of misfolded proteins. However, if the ER stress is severe and ER homeostasis cannot be restored, the UPR triggers cell apoptosis.16
We previously reported that transgenic mice carrying a knock-in mutation of Elane, reproducing the V72M ELANE mutation found in some patients with SCN, had normal granulopoiesis.17 Although valine at amino acid 72 is conserved between human and mouse NE, overall amino acid identity is only 76%. Thus, it possible that the normal phenotype of the V72M Elane mice is due to the failure of the V72M mutation to induce the same structural and/or functional changes in murine NE that it does in human NE. To address this possibility, we generated a second transgenic mouse line carrying a targeted mutation of Elane that reproduces the G192pter mutation found in SCN. This mutation truncates the carboxy-terminal 27 amino acids of the mature NE protein, disrupting 1 of 4 disulfide bonds thought to be important for the proper folding of both human and murine NE. We show that the G193X Elane allele produces a truncated NE protein that is rapidly degraded. Although basal granulopoiesis is normal, G193X Elane mice display marked neutropenia after inhibition of the ERAD pathway using a proteosome inhibitor. This study provides new support for the UPR model of disease pathogenesis in SCN associated with ELANE mutations.
Methods
Mice
Perk+/− mice18 on a 129/SvJ background were generously provided by Dr David Ron (University of Cambridge, Cambridge, United Kingdom) and backcrossed 10 generations onto a C57BL/6 background. Genotyping for the Perk wild-type and null alleles was performed by polymerase chain reaction (PCR), as described previously.19 Elane−/− mice have been described previously,20 and were generously provided by Dr Christine Pham (Washington University, St Louis, MO). Congenic C57BL/6 mice carrying the Ly5.1 gene (B6.SJL-Ptprc* Pep3b BoyJ) were obtained from The Jackson Laboratory. Sex- and age-matched mice between 6 and 16 weeks of age were used in accordance with the guidelines of the Washington University Animal Studies Committee.
Generation of G913X Elane mice
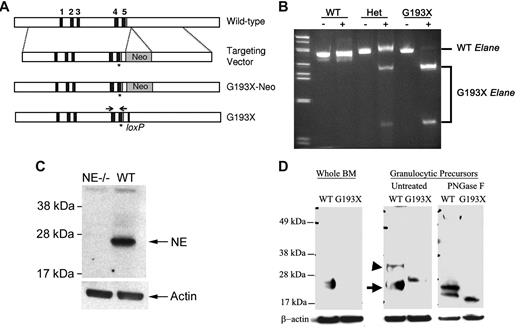
The G193X Elane allele was generated by introducing a G → T substitution in exon 5 of the murine Elane gene (chromosome 10:7350725; NCBIM37) by site-directed mutagenesis. The replacement-type targeting vector contains 4 kb of 5′ targeting sequence, a neomycin phosphotransferase gene driven by the phosphoglycerate kinase I promoter flanked by loxP sites (PGK-neo), and 3 kb of the 3′ targeting sequence (Figure 1A). The targeting vector was digested with NotI- and Xho1 to remove plasmid sequences and then transfected into SCC10 embryonic stem (ES) cells, as described previously.21 Two independent clones that had undergone homologous recombination were identified by Southern blot analysis. The loxP-flanked PGK-neo gene was subsequently removed by cre-recombinase–mediated excision. C57BL/6 blastocysts were microinjected with ES cells from each of these clones and implanted into pseudopregnant Swiss Webster foster females, as described previously.21 Chimeric mice derived from one of the ES clones were capable of germline transmission of the G193X Elane allele. In this study, G193X Elane mice on the following 3 different genetic backgrounds were analyzed: (1) inbred 129/SvJ mice obtained by crossing chimeras with 129/SvJ mice; (2) outbred 129/SvJ × C57Bl/6 mice; and (3) inbred C57Bl/6 background mice obtained by backcrossing 6 generations with C57Bl/6 mice. Mice were genotyped by PCR using the following primers: 5′-tgtgacagtggtgactaacatgtgcc-3′ (Elane exon 4) and 5′-cagcagacatcagacccagtgtacaa-3′ (Elane exon 5). The PCR amplicons were digested with EcoR1 and resolved on a 1.5% agarose gel; the wild-type and G193X Elane alleles produced a band of 500 bp and a doublet at 250 bp, respectively.
RNA-expression analysis
Femurs were each flushed with 1 mL of TRIzol reagent (Invitrogen) and RNA was isolated according to the manufacturer's instructions. For G193X Elane mRNA expression, RNA was reverse transcribed using the Invitrogen Superscript III First-Strand RT-PCR kit according to the manufacturer's instructions. PCR was performed using the oligonucleotides listed above, and the resulting amplicons were digested with EcoR1 and resolved on a 1.5% agarose gel.
Real-time quantitative RT-PCR
Real-time quantitative reverse transcriptase-PCR (RT-PCR) was performed using the TaqMan One-Step RT-PCR Master Mix Reagents Kit (Applied Biosystems) on a GeneAmp 7300 Sequence Detection System (Applied Biosystems). Reactions were repeated in the absence of RT to assess for DNA contamination. RNA content was normalized to murine β-actin. Oligonucleotide primers for each gene are provided in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article); PCR products were detected using Cyber Green or dual-labeled FAM-TAMRA probes, as indicated.
Generation of the rabbit anti–mouse NE antibody
cDNA corresponding to amino acids 1 (Ile) through 191 (Arg) of the mature NE protein was cloned into the pTrcHisA vector and expressed in TOP10 E coli per the manufacturer's instructions (Invitrogen). Cell lysates were fractioned by electrophoresis using a 12% polyacrylamide gel, and the band corresponding to the expected ∼ 20 kDa NE peptide was excised. The peptide in the polyacrylamide gel slice was sent to Covance Research Products to immunize rabbits and generate the antisera.
Western analysis
Bone marrow cells were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (Millipore) and clarified by centrifugation at 16 000g for 30 minutes at 4°C. Selected samples were treated with N-glycosidase F (PNGase F) to remove N-linked oligosaccharides, per the manufacturer's recommendations (New England Biolabs).
Granulocytic cell cultures
Cultures enriched for murine granulocytic precursors were obtained, essentially as described previously.22 In brief, c-Kit+ lineage− cells were isolated using the autoMACS magnetic bead system (Miltenyi Biotec) and cultured for 3 days in α-minimum essential medium containing 15% fetal bovine serum, 100 ng/mL each of granulocyte colony stimulating factor (G-CSF) and murine stem cell factor (PeproTech). For the tunicamycin experiments, tunicamycin (10 mg/mL in 10% dimethylsulfoxide [DMSO]; Sigma) or DMSO alone was added to the cultures on day 3 to a final concentration of 0.5 mg/mL (corresponding to day 0 in Figure 5).
Viable cell quantification
The number of cells in culture was determined using a hemocytometer. The percentage of viable (annexin V− 7-actinomycin D−) cells was determined using the BD Pharmingen Annexin V–Phycoerythrin Apoptosis Detection kit 1 (BD Biosciences), per the manufacturer's instructions. Cells were analyzed using a FACScan 5-color, 2-laser flow cytometer (BD Biosciences and Cytek Development).
Peripheral blood and bone marrow analysis
Blood was obtained by retroorbital venous plexus sampling in polypropylene tubes containing EDTA (ethylenediaminetetraacetic acid). Bone marrow cells were isolated by flushing femurs and tibias with 3-4 mL of ice-cold phosphate-buffered saline. Cell counts were obtained using a Hemavet automated cell counter (CDC Technologies). In some cases, Gr-1+ granulocytic cells were isolated from the bone marrow using the AutoMacs magnetic bead separator (Miltenyi Biotec).
CFU-C assay
Bone marrow was harvested from mice using standard techniques, and 2.0 × 104 nucleated cells suspended in 2.5 mL of methylcellulose medium supplemented with a cocktail of recombinant cytokines (MethoCult 3434; StemCell Technologies) or 10 ng/mL of recombinant human G-CSF (M3231; StemCell Technologies). Cultures were plated in duplicate and placed in a humidified chamber with 6% CO2 at 37°C. Colonies containing at least 50 cells were counted on day 7 of culture.
5-FU and bortezomib administration
5-Fluorouracil (5-FU; Sigma) was dissolved in sterile saline and given as a single intraperitoneal injection at 150 mg/kg. Bortezomib (3 mg/mL; Millenium Pharmaceuticals) or an equal volume of sterile phosphate-buffered saline was injected intravenously at 1 mg/kg. Bone marrow cells were harvested 30 hours after bortezomib administration. Gr-1+ cells were then isolated using the autoMACS magnetic bead system, and RNA was prepared using TRIzol reagent.
Bone marrow transplantation
Bone marrow cells from wild-type or G193X Elane mice (Ly5.2) were mixed at a 1:1 ratio with competitor bone marrow (Ly5.1/5.2), and a total of 5 million bone marrow cells were injected into the tail veins of lethally irradiated wild-type mice (Ly5.2). Recipient mice were conditioned with 1000 cGy from a 137Cs source at a rate of approximately 95 cGy/min 6-24 hours before transplantation. Prophylactic antibiotics (trimethoprim-sulfamethoxazole; Alpharma) were given for the initial 2 weeks after transplantation. A minimum of 2 independent transplantations was performed for each experimental condition.
To obtain fetal liver cells for transplantation, ElaneG193X/+Perk+/− mice (Ly5.2) were intercrossed and 12-14 days-post-conception (dpc) fetuses were harvested. Fetal livers were surgically isolated and cryopreserved in RPMI 1640 medium with 20% fetal bovine serum and 20% DMSO. After thawing, 5 × 106 fetal liver cells were injected into the lateral tail vein of irradiated recipient mice.
Neutrophil elastase activity assay
Cell lysates from unfractionated bone marrow cells harvested from healthy human donors or wild-type mice were assayed for elastase activity using the chromogenic, neutrophil elastase–specific substrate N-methoxysuccinyl Ala-Ala-Pro-Val p-nitroanilide (Sigma-Aldrich), as described previously.17 Elastolytic activity was normalized per microgram of protein.
Statistical analysis
Data are presented as means ± SEM unless otherwise stated. Statistical significance of pairwise sample comparison was performed using a 2-sided Student t test. Datasets with multiple samples were analyzed using 1- or 2-way ANOVA (for time-course experiments) with Bonferroni posttesting.
Results
Generation of G193X Elane mice
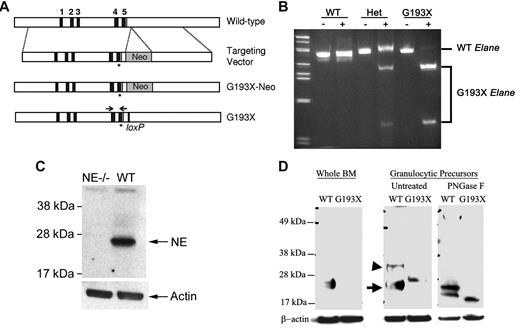
A G → T substitution at nucleotide 684 of the murine Elane cDNA (NM_015779.2) was introduced into the murine Elane gene by homologous recombination in ES cells (Figure 1A). This mutation produces a stop codon at amino acid G193, deleting the carboxyl-terminal 27 amino acids of the mature neutrophil elastase protein. It is orthologous to the G192pter ELANE mutation present in some patients with SCN. Two ES cell clones containing the recombined Elane allele were identified and used to generate chimeric mice. Only one of the chimeras transmitted the G193X Elane allele through the germline. The G193X Elane allele was transmitted in a Mendelian fashion. Animals heterozygous or homozygous for G193X Elane displayed normal growth, development, and fertility, and were grossly indistinguishable from wild-type littermates.
Expression of G193X Elane. (A) The genomic organization of murine Elane is shown in the top panel. The asterisk (*) represents the introduced G193X mutation; an EcoR1 site was just inserted immediately 3′ to this mutation to facilitate genotyping. (B) RT-PCR analysis of bone marrow RNA isolated from wild-type (WT) and mice heterozygous (Het) or homozygous (G193X) for the G193X Elane allele. The genotyping primers are depicted in panel A. After RT-PCR, the amplicons were incubated with EcorR1 (+) or buffer alone (−); EcoR1 cleaves G193X but not wild-type Elane cDNA. (C) Western blot analysis of cell lysates isolated from the bone marrow of wild-type (WT) or Elane−/− (NE−/−) mice. (D) Western blot analysis of cell lysates prepared from unfractionated bone marrow cells (whole BM) or cultured granulocytic precursors. Where indicated, cell lysates were treated with PNGase F before analysis. The positions of mature and precursor wild-type NE are indicated with an arrow or arrowhead, respectively. Data are representative of 3 independent experiments.
Expression of G193X Elane. (A) The genomic organization of murine Elane is shown in the top panel. The asterisk (*) represents the introduced G193X mutation; an EcoR1 site was just inserted immediately 3′ to this mutation to facilitate genotyping. (B) RT-PCR analysis of bone marrow RNA isolated from wild-type (WT) and mice heterozygous (Het) or homozygous (G193X) for the G193X Elane allele. The genotyping primers are depicted in panel A. After RT-PCR, the amplicons were incubated with EcorR1 (+) or buffer alone (−); EcoR1 cleaves G193X but not wild-type Elane cDNA. (C) Western blot analysis of cell lysates isolated from the bone marrow of wild-type (WT) or Elane−/− (NE−/−) mice. (D) Western blot analysis of cell lysates prepared from unfractionated bone marrow cells (whole BM) or cultured granulocytic precursors. Where indicated, cell lysates were treated with PNGase F before analysis. The positions of mature and precursor wild-type NE are indicated with an arrow or arrowhead, respectively. Data are representative of 3 independent experiments.
The G193X Elane allele produces an unstable NE protein
Expression of the G193X Elane mRNA was assessed by RT-PCR assay taking advantage of an EcoR1 site engineered just 3′ to the G913X Elane mutation (Figure 1B). In heterozygous G913X Elane mice, similar expression of wild-type and G193X mRNA was observed, indicating that Elane mRNA expression from the G913X Elane allele is comparable with the wild-type Elane allele. The fidelity of the G193X NE allele was confirmed by direct sequencing of the RT-PCR product obtained from homozygous animals (data not shown).
To assess NE protein expression, we first generated a polyclonal antibody to murine NE by immunizing rabbits with a bacterially expressed peptide containing amino acids 1-191 of mature NE (see “Generation of the rabbit anti–mouse NE antibody”). Relevant to this study, the deleted region of the G193X NE protein was not present in the immunizing peptide. The specificity of this antibody was confirmed using Elane−/− mice (Figure 1C). Whereas a band of the expected size (∼ 28 kDa) was readily observed in cell lysates isolated from the bone marrow of wild-type mice, no band was detected in Elane−/− lysates.
We used this antibody to assess NE protein expression in bone marrow cells from homozygous G193X Elane mice (henceforth simply called G193X Elane mice). Surprisingly, no NE protein was detected (Figure 1D) despite the normal Elane mRNA expression. To determine whether G193X NE protein was being rapidly degraded, we used our previously described myeloid cell-culture system to obtain a population enriched for murine granulocytic precursors.22 In brief, c-Kit+ lineage− hematopoietic progenitors were cultured for 3 days in medium containing Kit ligand and G-CSF. In both wild-type and G193 Elane cultures, approximately 60%-70% of cells had a promyelocyte/myelocyte morphology (data not shown). In wild-type granulocytic precursors, mature NE (28 kDa) was the dominant species detected (Figure 1D). Consistent with a previous report, a precursor form of NE (∼ 37 kDa) containing the 20–amino acid carboxyl-terminal prodomain was also seen.23 In contrast, in G193X Elane granulocytic precursors, only a band of ∼ 29 kDa was observed (G193X NE has a predicted molecular weight of 20.9 kDa). We suspected that this species represented a heavily glycosylated form of the truncated NE. To test this possibility, cell extracts from wild-type or G193X Elane cultures were treated with PNGase F, which removes all N-linked glycosylation. In wild-type cells, PNGase F treatment resulted in the production of bands of ∼ 23 and 27 kDa, corresponding to deglycosylated mature and precursor NE, respectively. In G193X Elane samples, a single band of ∼ 21 kDa was observed, consistent with the predicted size of deglycosylated precursor G193X NE protein. These data suggest that the G193X NE protein is highly unstable and likely rapidly degraded.
Basal granulopoiesis is normal in G193X Elane mice
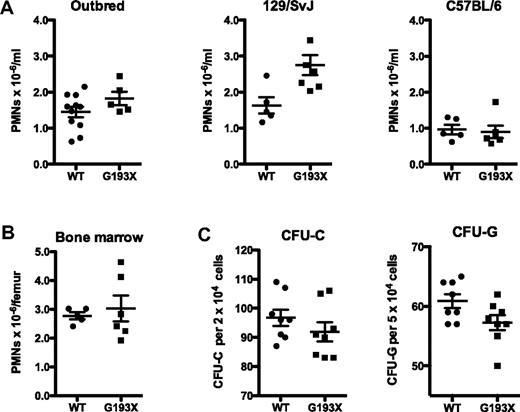
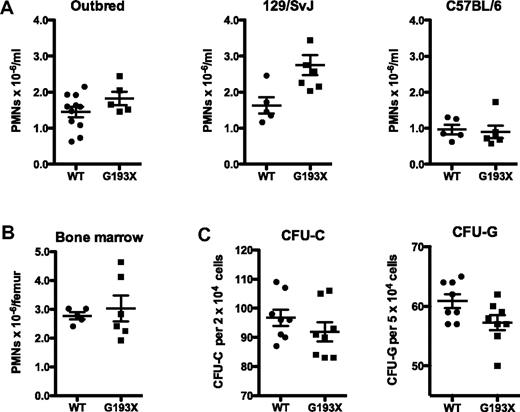
A hallmark of SCN is absolute neutropenia in the peripheral blood and bone marrow. Absolute neutrophil counts in the blood of G193X Elane mice on a SVJ/129, C57BL6, or 129 × B6 outbred background were comparable with strain-matched wild-type mice (Figure 2A). Likewise, the number of mature neutrophils in the bone marrow of G193X Elane mice was similar to that in wild-type mice (Figure 2B 129/SVJ background). Indeed, manual leukocyte differentials showed that the morphology and distribution of granulocytic precursors in the bone marrow of G193X Elane mice were comparable with those in wild-type mice (data not shown). Finally, the number and cytokine responsiveness of myeloid progenitors were assessed using CFU-C assays. Both the number and size of individual colony-forming unit-granulocyte CFU-G and CFU-C were similar in cultures of wild-type and G193X Elane bone marrow cells (Figure 2C). These data show that basal granulopoiesis is normal in G193X Elane mice.
Basal granulopoiesis in G193X Elane mice. (A) Absolute neutrophil count in the peripheral blood of wild-type and G193X Elane mice on the indicated genetic backgrounds. (B) Absolute neutrophil counts in the bone marrow of mice on a SVJ/129 background. (C) Frequency of CFU-C and CFU-G in the bone marrow of mice on a SVJ/129 background (n = 5). All mice were 4-6 weeks old.
Basal granulopoiesis in G193X Elane mice. (A) Absolute neutrophil count in the peripheral blood of wild-type and G193X Elane mice on the indicated genetic backgrounds. (B) Absolute neutrophil counts in the bone marrow of mice on a SVJ/129 background. (C) Frequency of CFU-C and CFU-G in the bone marrow of mice on a SVJ/129 background (n = 5). All mice were 4-6 weeks old.
Granulopoiesis after myeloablative stress is normal in G193X Elane mice
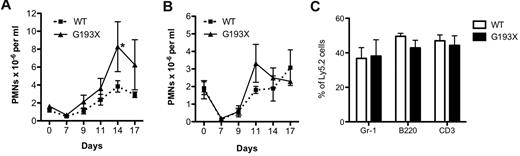
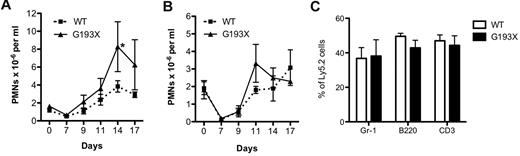
To assess stress granulopoiesis, the neutrophil response to the myelosuppressive agent 5-FU was assessed. In both wild-type and G193X Elane mice on a 129/SvJ background, treatment with 5-FU induced a similar degree of peripheral neutropenia that reached a nadir on day 7 (Figure 3A). Interestingly, a modest but significant increase in the peak neutrophil response after 5-FU administration was observed in the G193X Elane mice. Similar data were observed in mice on a C57Bl/6 background (Figure 3B).
Stress granulopoiesis in G193X Elane mice. (A) Wild-type or G193X Elane mice on a 129/SvJ (A) (n = 9-10) or C57BL/6 (B) (n = 3-4) background were treated with a single intraperitoneal injection of 5-FU (200 mg/kg) on day 0. The absolute neutrophil count was determined at the indicated time points. (C) Wild-type or G193X Elane mice on a C57Bl/6 background (Ly5.2) were mixed with an equal number of congenic wild-type bone marrow cells (Ly5.1/5.2) and injected into irradiated congenic (Ly5.1) mice. The percentage of Ly5.2 cells in the indicated lineage is shown at 16 weeks after transplantation (n = 4-5). Data represent the means ± SEM. *P < .05.
Stress granulopoiesis in G193X Elane mice. (A) Wild-type or G193X Elane mice on a 129/SvJ (A) (n = 9-10) or C57BL/6 (B) (n = 3-4) background were treated with a single intraperitoneal injection of 5-FU (200 mg/kg) on day 0. The absolute neutrophil count was determined at the indicated time points. (C) Wild-type or G193X Elane mice on a C57Bl/6 background (Ly5.2) were mixed with an equal number of congenic wild-type bone marrow cells (Ly5.1/5.2) and injected into irradiated congenic (Ly5.1) mice. The percentage of Ly5.2 cells in the indicated lineage is shown at 16 weeks after transplantation (n = 4-5). Data represent the means ± SEM. *P < .05.
Stress granulopoiesis was further analyzed using a competitive repopulation assay. Bone marrow chimeras were established by transplanting equal numbers of wild-type and G193X Elane bone marrow cells into irradiated congenic recipients. The contribution of G193X Elane cells to the neutrophil, B lymphocyte, and T lymphocyte lineages 16 weeks after transplantation was near input levels. Similar data were observed 6 and 12 weeks after transplantation (data not shown). These data show that expression of G193X Elane has no cell-intrinsic effect on granulopoiesis.
Minimal activation of the UPR is present in G193X Elane granulocytic precursors
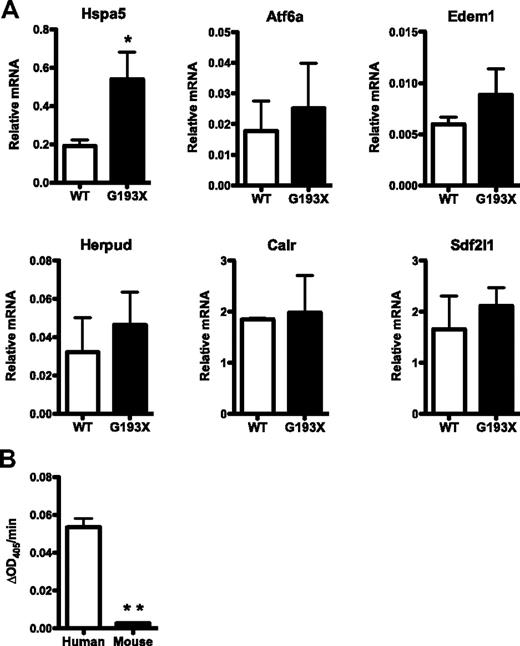
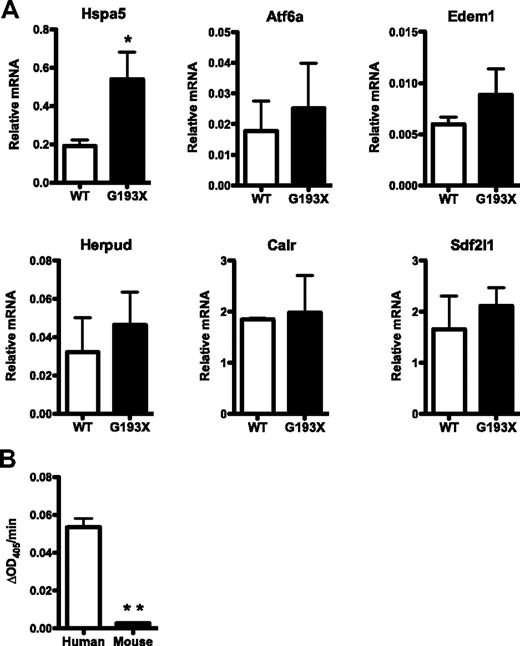
Consistent with a UPR model of disease pathogenesis, we previously reported that the expression of genes associated with UPR activation were increased in granulocytic precursors from patients with SCN and ELANE mutations.13 To assess the basal UPR status in G193X Elane granulocytic precursors, we performed RT-PCR for several genes that are induced with UPR activation (Figure 4A). RNA expression of the ER chaperone Hsap5 (Bip) was induced 2.1-fold in G193X Elane cells compared with wild-type cells. A trend to increased expression of Atf6a, Edem1, Herpud1, and Sdf2l1 also was observed, although none of these differences achieved statistical significance. These data suggest that under basal conditions there is minimal ER stress and UPR activation in G193X Elane granulocytic precursors.
UPR activation in G193X Elane granulocytic precursors. (A) Real-time RT-PCR for the indicated genes was performed on RNA isolated from cultured wild-type (WT) or G193X Elane granulocytic precursors. Shown is the mRNA expression relative to β-actin mRNA. Data represent the means ± SEM of 4-9 samples. (B) Elastolytic activity in human or mouse bone marrow was measured using the chromogenic NE-specific substrate N-methoxysuccinyl Ala-Ala-Pro-Val p-nitroanilide. Shown is the change in optical density at 405nm (OD405) per minute after normalizing for input protein. Data represent the means ± SEM of 2-4 samples. *P < .05; **P < .001.
UPR activation in G193X Elane granulocytic precursors. (A) Real-time RT-PCR for the indicated genes was performed on RNA isolated from cultured wild-type (WT) or G193X Elane granulocytic precursors. Shown is the mRNA expression relative to β-actin mRNA. Data represent the means ± SEM of 4-9 samples. (B) Elastolytic activity in human or mouse bone marrow was measured using the chromogenic NE-specific substrate N-methoxysuccinyl Ala-Ala-Pro-Val p-nitroanilide. Shown is the change in optical density at 405nm (OD405) per minute after normalizing for input protein. Data represent the means ± SEM of 2-4 samples. *P < .05; **P < .001.
The magnitude of UPR activation is critically dependent on the amount of misfolded protein delivered to the ER, which is a product of protein expression and the propensity to misfold. We speculated that that lower expression of NE in murine compared with human granulocytic precursors might account for the reduced UPR activation and lack of impaired granulopoiesis in G193X Elane mice. To test this possibility, we measured the amount of elastolytic activity in human and mouse bone marrow using the NE-specific substrate N-methoxysuccinyl Ala-Ala-Pro-Val p-nitroanilide. A previous study showed that recombinant human and murine NE had similar activity against this synthetic substrate.24 Surprisingly, the elastolytic activity of murine bone marrow was less than 5% of that of human cells (Figure 4B). Even accounting for the decreased percentage of granulocytic cells in murine bone marrow (approximately 50% vs 67% in human bone marrow), these data suggest that NE protein expression in murine granulocytic cells is substantially reduced compared with human cells.
G193X Elane granulocytic precursors display increased sensitivity to tunicamycin
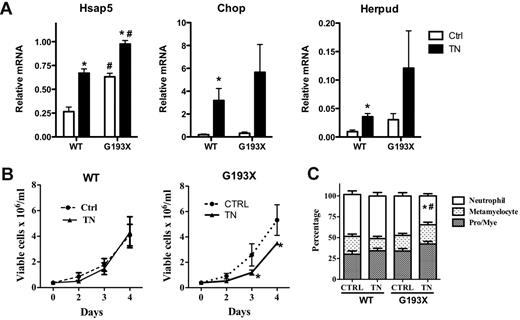
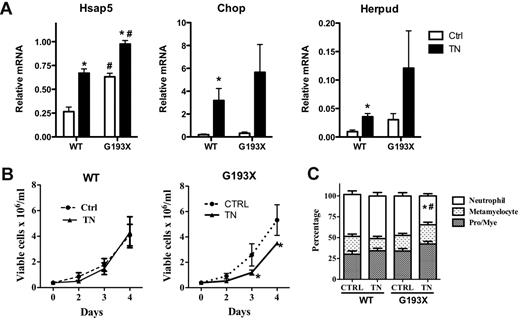
Granulocytic precursors from patients with SCN carrying mutations of ELANE or G6PC3 display increased sensitivity to ER stress induction with tunicamycin, a glycosylation inhibitor.3,13,14 To determine whether G193X Elane cells were similarly sensitive to ER stress induction, we treated granulocytic precursors with tunicamycin and assessed cell growth and terminal granulocytic differentiation. ER stress, as measured by Hsap5, Chop, and Herpud mRNA expression, was induced to a greater degree in G193X Elane cells compared with wild-type cells (Figure 5A). In cultures of wild-type cells, treatment with tunicamycin had no effect on cellular proliferation (Figure 5B). In contrast, in cultures of G193X Elane cells, treatment with tunicamycin was associated with a modest but significant decrease in viable cells on days 3 and 4 of culture. Granulocytic differentiation was assessed on day 4 after tunicamycin treatment by morphology (Figure 5C and supplemental Figure 1). In cultures of G193X Elane cells treated with vehicle alone, the percentage of mature (segmented or band) neutrophils was similar to that seen in wild-type cultures. However, in tunicamycin-treated cultures, a significant decrease in mature neutrophils was observed with G193X Elane but not wild-type cells. These data show that G193X Elane granulocytic precursors are sensitive to ER stress induction with tunicamycin, resulting in reduced cell growth and impaired terminal granulocytic differentiation in vitro.
Tunicamycin-induced alterations in the proliferation and differentiation of G193X Elane granulocytic precursors. Cultured granulocytic precursors were treated with tunicamycin (0.5 μg/mL) or vehicle alone on day 0. (A) Real-time RT-PCR for Hsap5, Chop, and Herpud was performed on cells 24 hours after exposure to tunicamycin (TN) or vehicle alone (Ctrl). RNA expression relative to β-actin is shown. (B) Viable cells were quantified by flow cytometry as annexin V− 7-actinomycin D− cells. (C) Manual leukocyte differentials (200-cell count) were performed on day 4 after treatment with tunicamycin. Neutrophil indicates segmented and band neutrophils; Pro/Mye, promyelocytes and myelocytes. Data represent the means ± SEM of 4-8 samples. *P < .05 compared with saline-treated cells of the same genotype; #P < .05 compared with wild-type mice in the same treatment group.
Tunicamycin-induced alterations in the proliferation and differentiation of G193X Elane granulocytic precursors. Cultured granulocytic precursors were treated with tunicamycin (0.5 μg/mL) or vehicle alone on day 0. (A) Real-time RT-PCR for Hsap5, Chop, and Herpud was performed on cells 24 hours after exposure to tunicamycin (TN) or vehicle alone (Ctrl). RNA expression relative to β-actin is shown. (B) Viable cells were quantified by flow cytometry as annexin V− 7-actinomycin D− cells. (C) Manual leukocyte differentials (200-cell count) were performed on day 4 after treatment with tunicamycin. Neutrophil indicates segmented and band neutrophils; Pro/Mye, promyelocytes and myelocytes. Data represent the means ± SEM of 4-8 samples. *P < .05 compared with saline-treated cells of the same genotype; #P < .05 compared with wild-type mice in the same treatment group.
Loss of Perk does not impair granulopoiesis in G193X mice
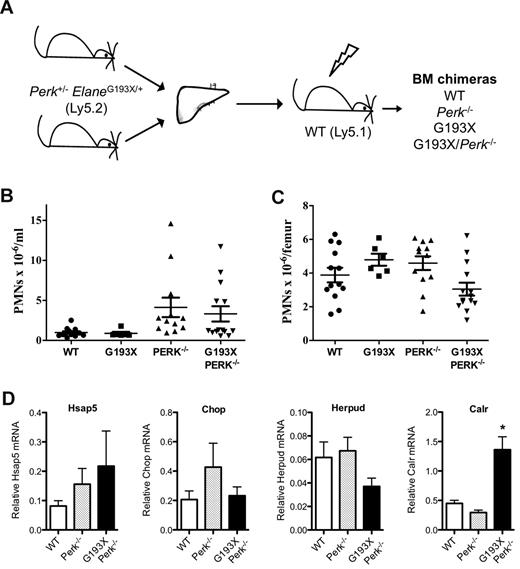
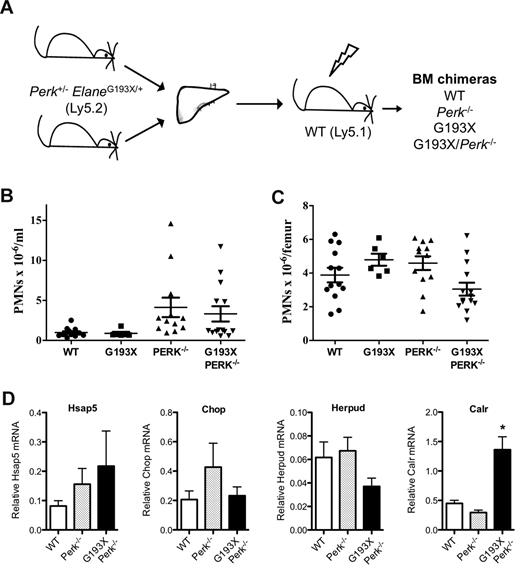
The UPR model of SCN disease pathogenesis predicts that inhibition of adaptive mechanisms to alleviate ER stress would worsen neutropenia. Perk (Eif2ak3) is one of the major proximal sensors of ER stress. Deletion of Perk in humans (Wolcott-Rallison syndrome) or mice is associated with severe ER stress in pancreatic β-islet cells, resulting in early-onset diabetes mellitus.19,25,26 Interestingly, neutropenia is present in approximately 50% of patients with Wolcott-Rallison syndrome.27 To assess the effect of the loss of Perk on granulopoiesis and to determine whether the loss of Perk is able to cooperate with G193X Elane to disrupt granulopoiesis, we generated bone marrow chimeras using fetal liver cells from wild-type, Perk−/−, G193X Elane, or Perk−/− × G193X Elane day 12-15 dpc fetuses (Figure 6A); fetal liver cells were used because Perk deficiency is perinatally lethal in mice.18 Basal granulopoiesis was assessed in the chimeras 12 weeks after transplantation. The absolute number of neutrophils in the blood (Figure 6B) and bone marrow (Figure 6C) was similar in all groups of mice. Surprisingly, UPR activation, as measured by Hsap5, Herpud, Chop, and Calr mRNA expression, was not induced in granulocytic cells by Perk insufficiency, either alone or in combination with G193X Elane, suggesting that Perk provides minor or redundant signals regulating UPR activation in granulocytic cells. In any case, these data show that loss of Perk, either alone or in combination with G193X Elane, is not sufficient to disrupt basal granulopoiesis in mice.
Granulopoiesis in G193X Elane × Perk−/− bone marrow chimeras. (A) Fetal liver cells from 12-15 dpc ElaneG193X/+Perk+/− mice (Ly5.2) intercrosses were transplanted into irradiated wild-type (WT Ly5.1) mice. Neutrophil counts in the blood (B) and bone marrow (C) were measured 12 weeks after transplantation. (D) Gr-1+ myeloid cells were isolated from the bone marrow as described in “Peripheral blood and bone marrow analysis,” RNA was prepared, and real-time RT-PCR for the indicated gene performed. Shown is the mRNA expression relative to β-actin. Data represent the means ± SEM of 6-14 mice. *P < .05 compared with WT cells.
Granulopoiesis in G193X Elane × Perk−/− bone marrow chimeras. (A) Fetal liver cells from 12-15 dpc ElaneG193X/+Perk+/− mice (Ly5.2) intercrosses were transplanted into irradiated wild-type (WT Ly5.1) mice. Neutrophil counts in the blood (B) and bone marrow (C) were measured 12 weeks after transplantation. (D) Gr-1+ myeloid cells were isolated from the bone marrow as described in “Peripheral blood and bone marrow analysis,” RNA was prepared, and real-time RT-PCR for the indicated gene performed. Shown is the mRNA expression relative to β-actin. Data represent the means ± SEM of 6-14 mice. *P < .05 compared with WT cells.
Treatment with bortezomib induces neutropenia in G193X Elane mice
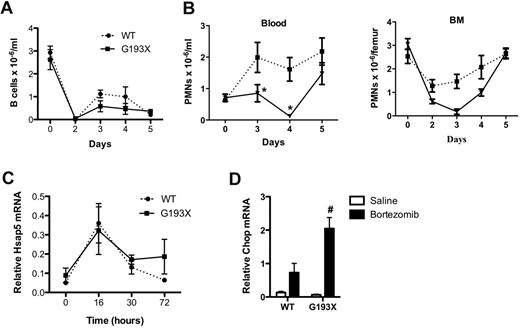
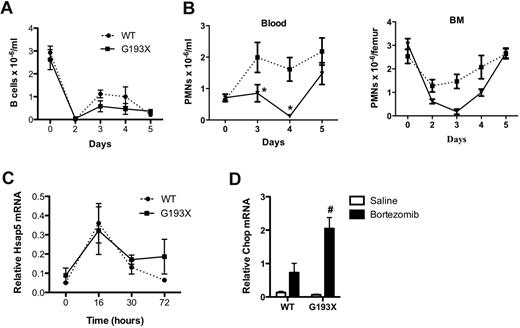
ERAD is the major pathway by which misfolded proteins in the ER are degraded. Treatment with proteosome inhibitors such as bortezomib inhibits the ERAD pathway and has been shown to exacerbate ER stress in several cell types.28,29 Because the G193X NE protein is rapidly degraded (Figure 1C), we next investigated whether treatment with bortezomib disrupted granulopoiesis in G193X Elane mice. Treatment with bortezomib results in B lymphopenia in mice that is thought to be secondary to the induction of ER stress and activation of the UPR.30 Consistent with this finding, a significant decrease in blood B lymphocytes was observed in both wild-type and G193X Elane mice after treatment with a single dose of bortezomib (1 mg/kg on day 0; Figure 7A). In wild-type mice, no significant change in neutrophil number in the blood or bone marrow was observed (Figure 7B). However, in G193X Elane mice, bortezomib treatment resulted in a marked decrease in neutrophils in the bone marrow and blood, with nadirs seen on day 3 and 4 after bortezomib treatment, respectively. By day 5, neutrophil counts in the blood and bone marrow of G193X Elane mice had normalized. To directly assess UPR activation, granulocytic cells were isolated from the bone marrow of mice after bortezomib treatment, and expression of Hsap5 and Chop mRNA was assessed. As expected, Hsap5 expression was induced after bortezomib treatment in both wild-type and G913X Elane cells. However, whereas Hsap5 mRNA expression returned to baseline 72 hours after bortezomib treatment in wild-type granulocytic cells, it remained elevated in G913X cells. Moreover, expression of Chop, a gene associated with UPR-induced apoptosis, was induced to a greater degree in G193X granulocytic cells after bortezomib administration (Figure 7D). These data suggest that G193X Elane granulocytic cells are selectively sensitive to ER stress induction by proteosome inhibition in vivo.
Bortezomib-induced neutropenia in G913X Elane mice. Mice (n = 3-6) were treated with a single subcutaneous injection of bortezomib (1 mg/kg) and at the indicated times, the number of B lymphocytes in the blood (A) and neutrophils in the blood and bone marrow (B) were measured. (C) Granulocytic (Gr-1+) cells were harvested from the bone marrow at the indicated times after treatment with bortezomib, and real-time RT-PCR for Hsap5 performed. RNA expression relative to β-actin is shown. Data represent the means ± SEM of 3-7 samples. (D) Granulocytic (Gr-1+) cells were harvested from the bone marrow 30 hours after treatment with bortezomib or saline alone, and real-time RT-PCR for Chop performed. RNA expression relative to β-actin is shown. Data represent the means ± SEM of 3 samples. *P < .05 compared with wild-type (WT) mice in the same treatment group; #P < .05 compared with saline-treated cells of the same genotype.
Bortezomib-induced neutropenia in G913X Elane mice. Mice (n = 3-6) were treated with a single subcutaneous injection of bortezomib (1 mg/kg) and at the indicated times, the number of B lymphocytes in the blood (A) and neutrophils in the blood and bone marrow (B) were measured. (C) Granulocytic (Gr-1+) cells were harvested from the bone marrow at the indicated times after treatment with bortezomib, and real-time RT-PCR for Hsap5 performed. RNA expression relative to β-actin is shown. Data represent the means ± SEM of 3-7 samples. (D) Granulocytic (Gr-1+) cells were harvested from the bone marrow 30 hours after treatment with bortezomib or saline alone, and real-time RT-PCR for Chop performed. RNA expression relative to β-actin is shown. Data represent the means ± SEM of 3 samples. *P < .05 compared with wild-type (WT) mice in the same treatment group; #P < .05 compared with saline-treated cells of the same genotype.
Discussion
In this study, we report the phenotype of transgenic mice carrying a targeted mutation of Elane (G193X) reproducing the G192X ELANE mutation found in patients with SCN. There are 5 other mutations of ELANE reported in patients with SCN that result in the production of a truncated NE protein, including C179X, G185X, C194X, S196X, and Y199X.1,7,10 Thus, the G193X Elane mutation is representative of a larger group of human ELANE mutations found in SCN. In contrast to patients carrying truncation mutations of ELANE, we show that basal granulopoiesis is normal in G193X Elane mice. Interestingly, modeling in mice of other gene mutations associated with human congenital neutropenia has yielded variable results. Mutations of G6PC3 are associated with severe neutropenia in humans,3 but only mild neutropenia in G6pc3−/− mice.31 Likewise, whereas HAX1 mutations are associated with SCN in humans, neutropenia was not reported in Hax1−/− mice.32 These observations add to the accumulating evidence that the regulation of human and murine granulopoiesis may be distinct. For example, a recent study reported that the transit time for human neutrophils in the bone marrow (5.8 days) was more than twice that observed for murine neutrophils (2.3 days).33
Recent data support a UPR model of disease pathogenesis in which protein misfolding of mutant NE results in ER stress, UPR activation, and ultimately impaired granulocytic differentiation. A bioinformatic analysis of 32 different ELANE mutations suggested that structural changes likely leading to protein misfolding were a common feature.34 The expression of mutant, but not wild-type, NE in myeloid cell lines or primary human granulocytic precursors induces UPR activation and apoptosis.13,14 Finally, myeloid cell lines expressing mutant NE are more sensitive to ER-stress–inducing agents.14 In the present study, we show that the G913X NE protein is unstable and rapidly degraded in granulocytic precursors. Expression of the G193X NE protein in granulocytic precursors is associated with biochemical evidence of mild basal activation of the UPR. We suggest that this low level of UPR activation is not sufficient to disrupt basal granulopoiesis in mice. It is not clear why UPR activation is lower in murine granulocytic precursors, but a comparison of NE activity in human and murine bone marrow suggests one possibility: we observed that the level of NE activity in murine bone marrow cells was less than 5% that of human cells (Figure 4D). Because UPR activation is critically dependent on the amount of misfolded protein delivered to the ER, these observations suggest that the reduced expression of NE in murine versus human granulocytic precursors may account for the reduced UPR activation in granulocytic precursors and normal granulopoiesis in G193X Elane mice.
G193X Elane granulocytic cells are selectively sensitive to ER stress induction. Specifically, treatment with tunicamycin, which induces ER stress by inhibiting N-linked glycosylation, is associated with reduced cell growth and impaired terminal granulocytic differentiation of G193X Elane granulocytic cells compared with wild-type cells. Moreover, treatment with bortezomib, a proteosome inhibitor, is associated with striking neutropenia in G193X Elane mice but not in wild-type mice. Bortezomib is best known for its ability to inhibit the proteosome.35 Proteosome inhibition, through suppression of the ERAD pathway, can increase ER stress and activate the UPR in susceptible cells. For example, recent evidence suggest that UPR activation is a major mechanism by which bortezomib exerts its antitumor effect in multiple myeloma.36 Consistent with these observations, treatment with bortezomib resulted in significantly greater UPR activation in G193X Elane granulocytic cells compared with wild-type cells. G193X Elane cells are not more sensitive to myeloablative stress induced by 5-FU or bone marrow transplantation. This selective sensitivity of G193X Elane granulocytic cells to ER stress provides new and strong support for the UPR model of disease pathogenesis in SCN associated with ELANE mutations. In addition to proteosome inhibition, bortezomib has been shown to inhibit many cellular processes, which raises an important caveat35 : it is possible that “off-target' effects of bortezomib may contribute to neutropenia in G193X Elane mice, but it is not clear why this would be selective for G193X Elane mice.
Wolcott-Rallison syndrome (OMIM 226980) is a rare autosomal recessive syndrome caused by mutations in PERK (EIF2AK3). This syndrome is characterized by neonatal or early-infancy insulin-dependent diabetes, multiple epiphyseal dysplasia, and growth retardation. In the largest case series, neutropenia was reported in approximately 50% of cases.27 Perk insufficiency in mice is associated with marked UPR activation and apoptosis in pancreatic β-islet cells that results in early onset diabetes mellitus and postnatal lethality. We show that in Perk−/− fetal liver chimeras, granulopoiesis is normal. Moreover, Perk insufficiency is unable to cooperate with G193X Elane to disrupt granulopoiesis. At first glance, this result is surprising, because Perk is a key regulator of the UPR in many cell types. However, Perk insufficiency did not result in UPR activation in either wild-type or G913X Elane granulocytic cells. Thus, these data suggest that in granulocytic cells, Perk provides minor or redundant signals regulating UPR activation and argue against a cell-intrinsic defect in granulopoiesis in Wolcott-Rallison syndrome.
In summary, we show that in transgenic mice expressing G193X Elane, granulopoiesis is selectively disrupted by the induction of ER stress. These data support the UPR model of disease pathogenesis for SCN associated with ELANE mutations. Therefore, pharmacologic agents that modulate the UPR may have therapeutic benefit in SCN.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Perk+/− mice were generously provided by Dr David Ron (University of Cambridge, United Kingdom).
This work was supported by National Institutes of Health grant RO1 HL079562 (to D.C.L.).
National Institutes of Health
Authorship
Contribution: S.N. designed and performed research, analyzed data, and wrote the manuscript; M.M., D.S.G., J.W., J.X., and M.S. performed research and collected data; and D.C.L. supervised all of the research and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel C. Link, MD, Division of Oncology, Department of Medicine, 660 South Euclid Ave, Campus Box 8007, St Louis, MO 63110; e-mail: dlink@dom.wustl.edu.