Abstract
Sézary syndrome (SS) is an aggressive subtype of cutaneous T-cell lymphoma that is characterized by circulating leukemic Sézary cells. The accumulation of these malignant cells has been shown to be the result of the resistance to apoptosis, in particular, activation-induced cell death. However, the mechanism of apoptosis resistance remains unknown. By characterizing the gene transcription profiles of purified CD4+CD7− Sézary cells from patients with SS and cultured Sézary cells, it was found that Sézary cells are deficient in the expression of special AT-rich region binding protein 1 (SATB1), a key regulator of T-cell development and maturation. Retrovirus-mediated gene transduction revealed that SATB1 restoration in cultured Sézary cells (Hut78) triggered spontaneous cell death and sensitized Hut78 cells to activation-induced cell death, with associated activation of caspase 8 and caspase 3. Furthermore, endogenous expression of FasL in Sézary cells was increased in transcriptional and translational levels on restoration of SATB1 expression in cultured Sézary cells. These results suggest that deficiency in SATB1 expression in Sézary cells plays an important role in SS pathogenesis by causing apoptosis resistance. Thus, restoration of SATB1 expression may represent a potential molecular targeted therapy for SS, which does not have a cure at present.
Introduction
Cutaneous T-cell lymphoma (CTCL) is composed of a heterogeneous group of non-Hodgkin lymphomas characterized by clonal accumulation of mature, postthymic T cells originating in the skin.1 Sézary syndrome (SS) is an aggressive leukemic variant of CTCL that typically presents with erythroderma, peripheral lymphoadenopathy, severe pruritus, and characteristic circulating Sézary cells.1 With an estimated 5-year survival of 15% to 24%, Sézary patients have poor prognosis, and so far this disease does not have a cure.1,2 The pathogenesis of SS remains largely unclear. Immunophenotypically, Sézary cells are mature memory CD4+CD45RO+ T cells with atypical cerebriform nuclei.2 They mainly display a T-helper 2 (Th2) cytokine profile and often lose expression of CD73 and CD26 antigens.4 The mechanism of Sézary cell accumulation in SS was found, not because of increased proliferation, but because of resistance to apoptosis.5 In particular, the resistance to activation-induced cell death (AICD) is characteristic in Sézary cells.6 During the course of an immune response, antigen-reactive T cells clonally expand and then are removed by apoptosis to maintain immune homeostasis. The majority of effector T cells are eliminated by AICD.7 The induction of AICD depends on stimulation of the T-cell receptor (TCR) complex on activated T cells and the consequent up-regulation of death receptor ligands, including the Fas ligand (FasL/CD95L).8 After ligation of Fas (APO-1/CD95) on the cell surface with FasL, the death-inducing signaling complex is formed. Within the complex, caspase 8 is cleaved and activates effector caspases, such as caspase 3, leading to apoptosis.8 However, the molecular basis for the apoptosis resistance of Sézary cells has not been fully elucidated.
Recent genome-wide studies have identified a series of gene aberrations in Sézary cells. These include overexpression of MUM1,9 AP-1,10 KIR3CL2, CDO1, and DNM3,11 S100P, LIR9, IL7R, and CD52,12 EPHA4 and TWIST,11 JUNB,13 GATA3 and RhoB,2 AHI-1 and PLS3,14-16 and down-regulation of CD7 and CD26,4 TGFBR2, BCL11A, STAT4, CREBBP, MXI1, and RIZ1,11 and TBX21, NKG7, and SCYA5.12 Other studies have pointed to the presence of chromosomal alterations, indicating genomic instability in Sézary cells.13,17 However, the contribution of these gene changes to Sézary cell pathogenesis is largely unknown. In this study, a microarray transcriptome analysis was performed to identify differentially expressed genes directly in primary CD4+CD7− Sézary cells from SS patients. We report that one of these candidates, the global transcriptional regulator, special AT-rich region binding protein 1 (SATB1), is aberrantly and specifically down-regulated in both primary CD4+CD7− leukemic Sézary cells and in cultured Sézary cell lines. Retrovirus-mediated SATB1-transduction in Sézary cells revealed that this specific down-regulation contributes to the resistance to AICD by regulating transcription of the FasL/CD95L.
Methods
Preparation of primary Sézary cells and primary CD4+ T cells from healthy donors
SS patients and healthy control subjects were recruited from the Skin Lymphoma Clinic of the British Columbia Cancer Agency and the outpatient dermatology clinics of the Vancouver General Hospital, with approval by the Clinical Ethics Board of University of British Columbia and British Columbia Cancer Agency, in accordance with the Declaration of Helsinki principles. All the SS patients met the diagnostic criteria proposed by the International Society for Cutaneous Lymphomas in 200218 : erythroderma, lymphadenopathy, increased CD4+/CD8+ ratio in peripheral blood, and Sézary cell number more than 5% in the peripheral blood. A total of 9 patient samples were studied. Six patients were diagnosed with typical SS with the presence of TCR clonality, and 3 patients with negative TCR clonality were diagnosed with atypical SS. The 6 patients involved in gene expression profiling study include 4 typical SS patients (with TCR clonality) and 2 atypical SS patients (with no demonstrable TCR clonality). All of these 6 patients have deficient CD7 expression (CD4+CD7− cells ≥ 40%) on T cells and typical clinical presentation of SS (Table 1). Leukemic Sézary cells were purified by negative selection with monoclonal antibodies directed against granulocytes, B cells, and CD8+ and CD7+ T cells using a Rosette Sep kit (StemCell Technologies). The purity of cells was verified by flow cytometry using phycoerythrin-conjugated anti-CD7 antibody and fluorescein isothiocyanate-conjugated anti-CD4 antibody (BD Biosciences). More than 90% purity by immune phenotyping (CD4+CD7−) was obtained as described previously.15 For AICD, isolated primary CD4+ cells from healthy volunteers and primary Sézary cells purified from patients were stimulated with 1μg/mL phytohemagglutinin for 16 hours, followed by culturing for 6 days in the presence of 25 U/mL recombinant human interleukin-2 (IL-2), added every 2 days of culture, as described previously.6
Cell lines and cell culture
Human CTCL cell lines Hut78 and HH (ATCC no. TIB-161 and CRL-2105) and T-cell leukemia cell lines, Jurkat and CCRF-CEM (ATCC no. TIB-152 and CCL-119) were cultured in RPMI 1640 medium with 10% fetal bovine serum, 100 U/mL penicillin, 0.1 mg/mL streptomycin, and 10−4M β-mercaptoethanol (StemCell Technologies).
Gene expression profile analysis
Total RNA was extracted and purified by QIAGEN RNAeasy kit (QIAGEN). A total of 500 ng RNA was reverse-transcribed into cDNA and linearly amplified by in vitro transcription in the presence of fluorescent-labeled CTP using the Low RNA Input Linear Amplification Kit, PLUS, Two-Color from Agilent following the manufacturer's instructions. Each microarray was hybridized with 825 ng of each amplified cDNA labeled with Cy5 or Cy3 at a specific activity between 8 and 15 pmol/μg. Hybridizations were performed on Whole Human Genome Oligo microarrays (G4112F; Agilent Technologies), composed of 41 059 60-nt oligonucleotide probes, most represented as single spots. Agilent microarray slides were scanned using the Agilent DNA Microarray Scanner and quantified using Agilent Feature Extraction software Version 10.5.1.1. Red and green processed signals were then normalized and analyzed with GeneSpring software Version 7.3 to identify significantly differentially expressed genes (data accessible at NCBI GEO database, accession GSE23113 and GSE24951; supplemental File 2, Gene transcriptome raw data table, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Quantitative real-time RT-PCR
Quantification of gene expression on both mRNA level and protein level was performed as previously described.15,16 RNA was reverse transcribed using random primers and SuperScript III reverse transcriptase (Invitrogen). Real-time polymerase chain reaction (PCR) was performed and analyzed, with GAPDH and 18S genes as the internal controls. Primers and gene designations used for PCR amplification are listed in supplemental Table 1.
Western blot analysis
Cell lysates were prepared, and the concentrations were quantified by Bradford assay. Protein expression was assessed by Western blotting with the NuPAGE Novex Bis-Tris Gel Electrophoresis System (Invitrogen), and the filters were probed with specific antibodies. Relative protein expression was determined by densitometry using the image analysis program ImageQuant Version 5.2 (Molecular Dynamics). Antibodies used to detect specific proteins are listed in supplemental Table 2.
Immunofluorescence analysis
Cell suspension (1 × 105 cells per slide) was plated onto poly-L-lysine–coated slides and fixed with 4% ice-cold paraformaldehyde. After permeabilization and blocking, cells were incubated with mouse anti-SATB1 monocloncal antibody (BD Biosciences). This was followed by incubation with Alexa-594-conjugated secondary antibody (Invitrogen). Cell nuclei were counterstained with DAPI before being mounted with Fluorescence Mounting Medium (Dako Canada). Images were collected and processed by fluorescence microscope.
Construction of SATB1 retroviral vector and production of stably transduced cells
A MSCV-SATB1-IRES-GFP retroviral vector was constructed by ligating a 3.0-kb fragment containing full-length SATB1 cDNA (pCMV6-XL5-SATB1; Origene) to the upstream IRES of the MSCV-IRES-GFP (MIG) vector as described.19,20 Constructs were verified by DNA sequencing. Helper-free virus was obtained by transfecting amphotropic Phoenix packaging cells cultured in Dulbecco modified Eagle medium plus 10% fetal bovine serum (StemCell Technologies) using calcium-phosphate precipitation method. The virus-containing medium was harvested and filtered 32 to 72 hours later as described previously.19,20 Hut78 cells were resuspended at 2 × 105 cells/mL in the virus-containing medium diluted 1:2 in Dulbecco modified Eagle medium containing 15% fetal bovine serum and protamine sulfate (5 mg/mL) for 4 hours. Then the cells were incubated at 37°C for a further 72 hours. Fluorescein-activated cell sorting (FACS) was then performed on FACStar Plus (BD Biosciences) to isolate GFP+ cell populations from transduced cells. Single cells were also sorted to establish stably transduced clonal cell lines. MIG empty vector transduced clones (Hut78-MIG) were also generated. In the following cell assays, both the vector-transduced control cells (Hut78-MIG) and the parental untransduced cells (Hut78) served as controls.
Cell viability assays
Cell viability was measured with MTS-based cell viability assay using Cell viability Colorimetric Assay Kit (Promega) according to the manufacturer's instructions. Briefly, aliquots of transduced and control cells cultured in growth factor-free RPMI 1640 medium with 10% fetal bovine serum were distributed in 6-well plates (5 × 105 cells per well) in 3 mL of medium and incubated at 37°C in 5% CO2 for 24, 48, 72, and 96 hours. At each time point, 100-μL cell aliquots were incubated with 20 μL MTS for an additional 4 hours. The viable cell numbers were then determined at 490 nm using 96-well plate spectrometer. Each population was performed in triplicate wells, and biologic replicates were repeated twice.
Apoptosis assays
For the induction of AICD, cells were seeded at 2 × 105/mL in 2 mL RPMI 1640 media with 25 U/mL IL-2 for 48 hours; then the apoptosis was induced by either 10 μg/mL plate-bound anti-CD3 antibody or addition of 25 ng/mL phorbol 12-myristate 13-acetate (PMA) and 50 ng/mL ionomycin (Sigma-Aldrich) for 24 hours as described.6,21 For FasL/CD95L-induced apoptosis, cells were seeded at 5 × 105/mL in 1 mL RPMI 1640 culture media. Super FasL (40 ng/mL; Alexis Biochemicals) was added into the cell culture and incubated for 16 hours. Cell apoptosis was stained with 7-aminoactinomycin D and phycoerythrin-conjugated annexin V (Annexin V-PE Detection Kit I; BD Pharmingen) and then quantified by flow cytometry analysis using a FACSCanto II or a FACScan (BD Biosciences). Data were analyzed with FACS Diva 6.0 and CellQuest Pro software. Specific apoptosis was calculated as (percentage of induced apoptosis − percentage of spontaneous apoptosis)/(100 − percentage of spontaneous apoptosis).6
Cell-cycle analysis
Cell-cycle distribution was determined by PI-mediated flow cytometric analysis. Briefly, 5 × 105 cells of transduced and control cells were collected, fixed, and permeablized with cold 100% ethanol. After treatment with DNAse-free RNAse, the cells were stained with 50 μg/mL propidium iodide. Distribution of the cell-cycle phase with different DNA contents was determined with the sub-G1 phase representing the apoptotic cells.22
Statistical analysis
Statistical analyses were performed with SPSS Version 10.0 software. Nonparametric Mann-Whitney U tests were used to compare the difference between SATB1-transduced clonal cells and control cells.
Results
Differentially expressed genes in primary CD4+CD7− leukemic Sézary cells in SS
To identify critical genes involved in pathogenesis of SS, Sézary cell–enriched clinical samples (CD4+CD7− cells) from peripheral blood of 6 Sézary patients and CD4+ T cells from peripheral blood of 9 healthy donors were subjected to transcriptome analysis. Agilent Whole Human Genome Oligo microarrays (G4112F) were applied, and in total 41 059 transcripts were examined. Extensive gene expression alterations in Sézary cells against normal CD4+ cells were observed. A total of 153 transcripts in Sézary cells showed more than 4-fold increase or reduction compared with the control CD4+ T cells (P < .01; Figure 1A). Hierarchichal unsupervised clustering with these genes readily separated Sézary cells from normal CD4+ T cells (Figure 1B). Several previously reported up-regulated genes in Sézary cells, such as DNM3,11 PLS3,15 NEDD4L,11 and TWIST1,11,23 and down-regulated genes (STAT4 and BCL11A23 ) were also found in this gene list (supplemental Table 3). The genes differentially up-regulated in Sézary cells include genes regulating cell proliferation, cell adhesion, cytoskeleton organization, and signal transduction, as well as several oncogenes identified in other malignancies, such as ACVR2A (breast cancer24 ), CDH1 (hepatocellular cancer25 ), KLF8 (ovarian cancer5 ), and PDGFA (papillary thyroid carcinoma26 ). Genes differentially down-regulated in Sézary cells are involved in regulation of apoptosis, cell proliferation, and T-cell differentiation. In addition, several tumor suppressors found in other cancer types were also found to be down-regulated in Sézary cells, including CBFA2T3 (breast cancer27 ), FCRL2 (chronic lymphocytic leukemia28 ), and SERPINB6 (colon cancer29 ).
Transcriptome analysis of primary Sézary cells. (A) Volcano plot of the 153 differentially expressed genes identified from RNA prepared from CD4+CD7+ Sézary cell from 6 SS patients compared with similarly purified cells from 9 normal volunteers that were used to perform transcriptome analysis on 41 059 gene transcripts. Red dots represent differently expressed genes with fold changes more than 4. P < .01. Genes on the right arm are overexpressed in Sézary cells, whereas those on the left arm are underexpressed in Sézary cells. (B) Hierarchical unsupervised clustering of the 153 genes, which clearly demonstrates a separation between Sézary cells and the normal T lymphocyte controls.
Transcriptome analysis of primary Sézary cells. (A) Volcano plot of the 153 differentially expressed genes identified from RNA prepared from CD4+CD7+ Sézary cell from 6 SS patients compared with similarly purified cells from 9 normal volunteers that were used to perform transcriptome analysis on 41 059 gene transcripts. Red dots represent differently expressed genes with fold changes more than 4. P < .01. Genes on the right arm are overexpressed in Sézary cells, whereas those on the left arm are underexpressed in Sézary cells. (B) Hierarchical unsupervised clustering of the 153 genes, which clearly demonstrates a separation between Sézary cells and the normal T lymphocyte controls.
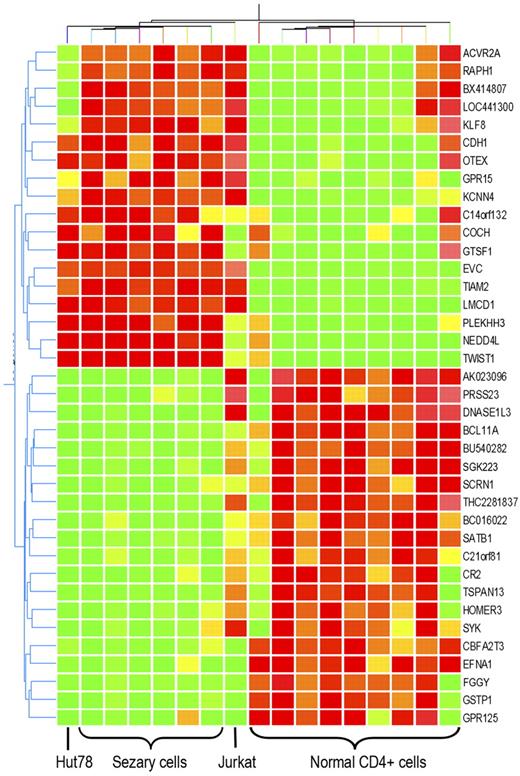
Global gene regulator SATB1 was specifically down-regulated in primary and cultured Sézary cells
To further identify the gene alterations that may be responsible for the unique biologic features of Sézary cells, Sézary cell line Hut78 and non-Sézary T-cell leukemia cell line Jurkat were added into microarray analysis. Not surprisingly, hierarchichal unsupervised clustering with these 153 transcripts classified Hut78 cells in the group of Sézary cells and classified Jurkat cell as a single cluster. However, not all of the Sézary cell gene alterations were preserved in Hut78 cells, whereas some of them were also shared by Jurkat cells. Differential expression in 32 genes was shared by both primary CD4+CD7− Sézary cells and Sézary cell line Hut78, but not present in Jurkat cells. Among those genes, 16 were up-regulated in Sézary cells, whereas 16 were down-regulated (P < .01, Figure 2; Table 2). After quantitative reverse transcription (RT)–PCR and Western blot confirmation with 3 additional Sézary cell samples, CTCL cell line HH, and non-Sézary T-cell leukemia cell line CCL119 for Sézary differentially expressed genes, SATB1, a T-cell lineage–specific global gene regulator,30 demonstrated the most striking expression changes in primary Sézary cells and Sézary cell lines (Hut78 and HH). As shown in Figure 3A-B, SATB1 RNA and protein expression was found to be specifically and consistently down-regulated in patient-derived Sézary cells (typical and atypical) and in Sézary cell lines (Hut78 and HH), although its expression was highly expressed in normal CD4+ T cells as well as in other non-Sézary T-cell leukemia cell lines (Jurkat and CCL119) by quantitative RT-PCR and Western blot analysis. Further investigation by immunofluorescence staining of SATB1 with a specific monoclonal antibody demonstrated the nuclear localization of SATB1 protein in normal CD4+ T cells and T-cell leukemia cell lines, and confirmed the specific SATB1 expression deficiency in primary Sézary cells and Sézary cell lines (Figure 3C).
Genes with expression aberration specific to Sézary cells. Expression change of 32 genes are specific to primary Sézary cells and Sézary cell line Hut78, compared with normal CD4+ T cells and non-SS T-cell leukemia cell line Jurkat cells. Hierarchichal unsupervised clustering of those genes classify primary Sézary cells and Hut78 cells from normal controls, and from Jurkat cells. Among those genes, 16 are overexpressed in Sézary cells, whereas 16 are underexpressed in Sézary cells.
Genes with expression aberration specific to Sézary cells. Expression change of 32 genes are specific to primary Sézary cells and Sézary cell line Hut78, compared with normal CD4+ T cells and non-SS T-cell leukemia cell line Jurkat cells. Hierarchichal unsupervised clustering of those genes classify primary Sézary cells and Hut78 cells from normal controls, and from Jurkat cells. Among those genes, 16 are overexpressed in Sézary cells, whereas 16 are underexpressed in Sézary cells.
Specific suppression of SATB1 expression in Sézary cells. (A) SATB1 RNA expression levels are significantly down-regulated in CD4+CD7− primary Sézary cells from 6 SS patients with TCR clonality [SS(T)] and 3 SS patients without TCR clonality [SS(A)], as well as cultured Sézary cell lines (Hut78 and HH), compared with similarly purified T lymphocytes from normal volunteers (NC), and 2 cultured non-Sézary leukemic cell lines (Jurkat and CCL119). **P = .004. (B) SATB1 protein expressions are absent from patient-derived Sézary cells (SS1 and SS2) and Sézary cell lines, whereas the normal cells (NC1 and NC2) and 2 non-Sézary leukemia cell lines show abundant expression of SATB1 protein. GAPDH serves as the internal reference. (C) Immunofluorescence staining of SATB1 on CD4+ T cells in vitro. Cells were stained with mouse SATB1 monoclonal antibody (1:100) and Alexa-594-conjugated anti–mouse secondary antibody (1:200). DAPI was used to counterstain cell nucleus. Non-Sézary cells (NC, Jurkat and CCL119) showed strong SATB1 staining on the nucleus, whereas patient-derived SS and Sézary cell line (Hut78 and HH) cells showed nearly no staining on the nucleus. Original magnification ×630. Scale bar represents 10 μm.
Specific suppression of SATB1 expression in Sézary cells. (A) SATB1 RNA expression levels are significantly down-regulated in CD4+CD7− primary Sézary cells from 6 SS patients with TCR clonality [SS(T)] and 3 SS patients without TCR clonality [SS(A)], as well as cultured Sézary cell lines (Hut78 and HH), compared with similarly purified T lymphocytes from normal volunteers (NC), and 2 cultured non-Sézary leukemic cell lines (Jurkat and CCL119). **P = .004. (B) SATB1 protein expressions are absent from patient-derived Sézary cells (SS1 and SS2) and Sézary cell lines, whereas the normal cells (NC1 and NC2) and 2 non-Sézary leukemia cell lines show abundant expression of SATB1 protein. GAPDH serves as the internal reference. (C) Immunofluorescence staining of SATB1 on CD4+ T cells in vitro. Cells were stained with mouse SATB1 monoclonal antibody (1:100) and Alexa-594-conjugated anti–mouse secondary antibody (1:200). DAPI was used to counterstain cell nucleus. Non-Sézary cells (NC, Jurkat and CCL119) showed strong SATB1 staining on the nucleus, whereas patient-derived SS and Sézary cell line (Hut78 and HH) cells showed nearly no staining on the nucleus. Original magnification ×630. Scale bar represents 10 μm.
Restored SATB1 expression confers growth disadvantage on Sézary cells and increases spontaneous apoptosis
SATB1 has been defined as an important regulator in the development of mature T cells and the activation of Th2 cells.31,32 Thus, we were interested in determining whether its suppression regulates malignant biologic behaviors in Sézary cells. To further define the role of SATB1 suppression in Sézary cells, we performed retrovirus-mediated gene transduction to stably restore SATB1 gene expression in Hut78 Sézary cells. All transduced clonal cells achieved increased SATB1 protein expression compared with Hut78 cells transduced with a control vector (MIG, Figure 4A). MTS-based cell viability assay demonstrated that decreased rate of cell expansion in SATB1 expression restored cells in liquid culture, compared with Hut78 parental cells and vector-transduced control cells. On day 4, the viable cell numbers of SATB1-transduced cells were significantly lower than those of parental Hut78 cells and vector control cells (Figure 4B, SATB1 cells vs MIG vector control cells, P = .0247).
Retrovirus-mediated SATB1 gene transduction confers growth disadvantage to Hut78 cells. (A) Immunofluorescence analysis show that Hut78 cells transduced with SATB1 gene in retrovirus vector (Hut78-SATB1) stably express SATB1 protein in cell nucleus, whereas control cells transduced with empty vector (Hut78-MIG) do not express SATB1. Original magnification ×630. Scale bar represents 10 μm. (B) Parental Hut78 cells (black) and Hut78-MIG cell clones (navy) present growth advantage over SATB1-transduced cell clones (light blue). On day 4, the viable cell numbers of Hut78-SATB1 cell clones were significantly lower than those of parental Hut78 cells and Hut78-MIG cells. Data were generated from the individual cell clones from each group, and all experiments were triplicated. *P < .05 by Mann-Whitney U test.
Retrovirus-mediated SATB1 gene transduction confers growth disadvantage to Hut78 cells. (A) Immunofluorescence analysis show that Hut78 cells transduced with SATB1 gene in retrovirus vector (Hut78-SATB1) stably express SATB1 protein in cell nucleus, whereas control cells transduced with empty vector (Hut78-MIG) do not express SATB1. Original magnification ×630. Scale bar represents 10 μm. (B) Parental Hut78 cells (black) and Hut78-MIG cell clones (navy) present growth advantage over SATB1-transduced cell clones (light blue). On day 4, the viable cell numbers of Hut78-SATB1 cell clones were significantly lower than those of parental Hut78 cells and Hut78-MIG cells. Data were generated from the individual cell clones from each group, and all experiments were triplicated. *P < .05 by Mann-Whitney U test.
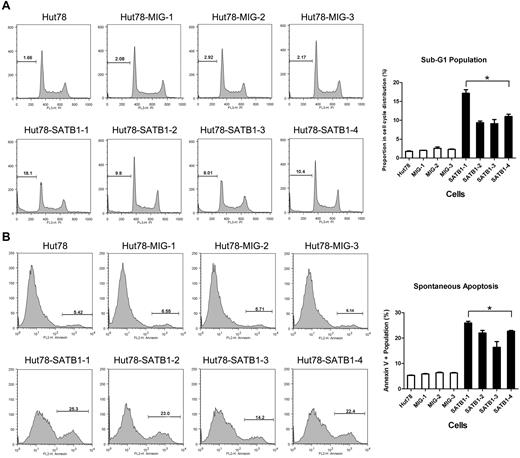
The proliferation and apoptosis features of SATB1-restored Sézary cells were further evaluated by cell-cycle analysis and apoptosis assay, respectively. In cell-cycle analysis, the SATB1-restored cells had increased cell population specifically in the apoptotic sub-G1 phase, (2.17% vs 11.65%, P = .021; Figure 5A) compared with the control cells, whereas there were no significant differences in G1, M, or G2 phases. Annexin V–based apoptosis also revealed increased apoptosis in SATB1-restored cells compared with the control Hut78 cells (5.95% vs 21.76%, P = .021; Figure 5B). These results suggest that there was a significantly increased apoptosis with fewer changes in cell proliferating rate and cell-cycle distribution in SATB1-restored Hut78 cells.
Cell-cycle and apoptosis analysis by flow cytometry. SATB1-transduced and control cell clones were analyzed in propidium iodide cell-cycle analysis (A) and annexin V binding-based apoptosis assay (B). (A) Each cell cycle is composed of G1, S, G2 stages, as well as sub-G1 stage, which represents the apoptotic or dead population of cells. Proportions of sub-G1 stage are indicated as no. (%). Chart on right: SATB1-transduced cells revealed increased sub-G1 population in contrast to the control cells. (B) Proportions of annexin V+ population, which represents apoptotic and dead cells are indicated as no. (%). Chart on right: SATB1-transduced cells revealed increased annexin V population in contrast to the control cells. Hut78 indicates parental untransduced Hut78 cells; MIG-1 to MIG-3, Hut78 cell clones transduced with empty MIG vector; and SATB1-1 to SATB1 4, Hut78 cell clones transduced with MIG vector containing SATB1. *P < .05 by Mann-Whitney U test.
Cell-cycle and apoptosis analysis by flow cytometry. SATB1-transduced and control cell clones were analyzed in propidium iodide cell-cycle analysis (A) and annexin V binding-based apoptosis assay (B). (A) Each cell cycle is composed of G1, S, G2 stages, as well as sub-G1 stage, which represents the apoptotic or dead population of cells. Proportions of sub-G1 stage are indicated as no. (%). Chart on right: SATB1-transduced cells revealed increased sub-G1 population in contrast to the control cells. (B) Proportions of annexin V+ population, which represents apoptotic and dead cells are indicated as no. (%). Chart on right: SATB1-transduced cells revealed increased annexin V population in contrast to the control cells. Hut78 indicates parental untransduced Hut78 cells; MIG-1 to MIG-3, Hut78 cell clones transduced with empty MIG vector; and SATB1-1 to SATB1 4, Hut78 cell clones transduced with MIG vector containing SATB1. *P < .05 by Mann-Whitney U test.
SATB1 restoration sensitizes Sézary cells to AICD
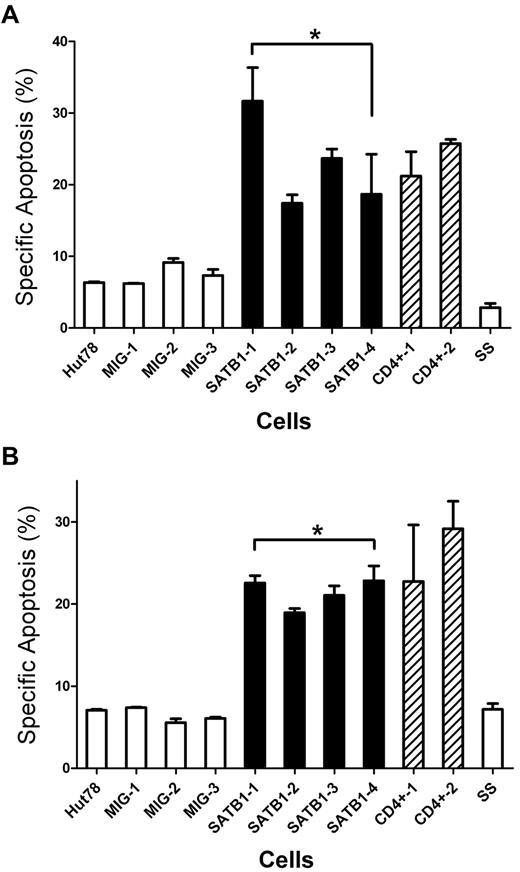
TCR restimulation on activated T cells leads to AICD.8 In vitro, this can be simulated by a model system using cultured IL-2 activated T cells that are restimulated by a TCR agonist, or by stimulation with PMA and ionomycin.21 For primary T cells, with activation and subsequent culture of T cells in IL-2-containing medium, cells switch from an AICD-resistant (day 1) to an AICD-sensitive phenotype (day 6).21 We used this ex vivo model to investigate the effects of SATB1 restoration in the AICD resistance of Sézary cells. On TCR stimulation with anti-CD3 antibody or PMA/ionomycin, in contrast to control Hut78 cells and primary Sézary cells, which had low specific apoptosis, the SATB1-restored Hut78 cells demonstrated significantly higher levels of specific apoptosis. Similar levels of specific apoptosis were observed in primary CD4+ T cells from healty donors, which express high levels of SATB1 (Figure 6). This observation indicates that SATB1 restoration sensitized Hut78 cells to AICD.
SATB1 restoration sensitizes Sézary cells to AICD. SATB1-transduced and control clonal Hut78 cells were cultured with IL-2. Primary peripheral CD4+ T cells and CD4+CD7− Sézary cells were stimulated by phytohemagglutinin and cultured in the presence of IL-2 for 6 days. All cells were restimulated with either plate-bound anti-CD3 antibody or PMA/ionomycin to trigger AICD. Specific apoptosis, which represents the increased apoptosis population after stimulation, was analyzed with annexin V binding-based apoptosis assay and compared. Proportions of annexin V+ population are indicated as no. (%). (A) On anti-CD3 antibody restimulation, SATB1-transduced Hut78 cells revealed increased specific apoptosis in contrast to the control Hut78 cells. The level of specific apoptosis in SATB1-transduced Hut78 cells was comparable with that in primary normal CD4+ T cells, whereas the level in control Hut78 cells was comparable with primary Sézary cells, which presented SATB1 expression defect. *P < .05 by Mann-Whitney U test. (B) On PMA/ionomycin restimulation, SATB1-transduced Hut78 cells and primary CD4+ T cells also revealed increased specific apoptosis in contrast to the control Hut78 cells, or primary Sézary cells. *P < .05 by Mann-Whitney U test. Hut78 indicates parental untransduced Hut78 cells; MIG-1 to MIG-3, Hut78 cell clones transduced with empty MIG vector; SATB1-1 to SATB1-4, Hut78 cell clones transduced with MIG vector containing SATB1; CD4+-1 and CD4+-2, primary peripheral CD4+ T cells; and SS, primary CD4+CD7− T cells purified from Sézary patient.
SATB1 restoration sensitizes Sézary cells to AICD. SATB1-transduced and control clonal Hut78 cells were cultured with IL-2. Primary peripheral CD4+ T cells and CD4+CD7− Sézary cells were stimulated by phytohemagglutinin and cultured in the presence of IL-2 for 6 days. All cells were restimulated with either plate-bound anti-CD3 antibody or PMA/ionomycin to trigger AICD. Specific apoptosis, which represents the increased apoptosis population after stimulation, was analyzed with annexin V binding-based apoptosis assay and compared. Proportions of annexin V+ population are indicated as no. (%). (A) On anti-CD3 antibody restimulation, SATB1-transduced Hut78 cells revealed increased specific apoptosis in contrast to the control Hut78 cells. The level of specific apoptosis in SATB1-transduced Hut78 cells was comparable with that in primary normal CD4+ T cells, whereas the level in control Hut78 cells was comparable with primary Sézary cells, which presented SATB1 expression defect. *P < .05 by Mann-Whitney U test. (B) On PMA/ionomycin restimulation, SATB1-transduced Hut78 cells and primary CD4+ T cells also revealed increased specific apoptosis in contrast to the control Hut78 cells, or primary Sézary cells. *P < .05 by Mann-Whitney U test. Hut78 indicates parental untransduced Hut78 cells; MIG-1 to MIG-3, Hut78 cell clones transduced with empty MIG vector; SATB1-1 to SATB1-4, Hut78 cell clones transduced with MIG vector containing SATB1; CD4+-1 and CD4+-2, primary peripheral CD4+ T cells; and SS, primary CD4+CD7− T cells purified from Sézary patient.
SATB1 restoration causes up-regulation of FasL/CD95L and Sézary cell transcriptome change
To investigate the possible mechanism by which SATB1 sensitizes Sézary cells to spontaneous apoptosis and AICD, we subjected SATB1-restored Sézary cells (Hut78-SATB1 cell clones) to transcriptome analysis, using empty-vector–transduced Sézary cells (Hut78-MIG cell clones) as the controls with methods previously described. As expected, transcription levels of a number of genes were dramatically altered by SATB1 restoration in Hut78 cells. Among them, FasL/CD95L was one of the most remarkably changed genes. FasL/CD95L expression, which is normally down-regulated in primary Sézary cells (Figure 7A), was significantly up-regulated in SATB1-restored Sézary cells, as confirmed by quantitative RT-PCR (Figure 7B) and Western blot (Figure 7C), whereas expression of other apoptosis pathway related genes, including cFLIP, Bcl-2, Bax, NF-κB (p65), TNFR1, STAT3, MYC, Bcl-2L1, and CCND1, remained unchanged (data not shown). Fas/CD95, which is the cell surface receptor to FasL/CD95L and had similar expression level between primary Sézary cells and normal CD4+ T cells (supplemental Figure 1), was also unchanged after SATB1 restoration. Moreover, as shown in supplemental Table 3, SATB1 restoration normalized 18 of 153 (12%) genes abnormally transcribed in the Sézary cells. The expression fold changes of these 18 genes from normal CD4+ T cells versus primary CD4+CD7− Sézary cells and from SATB1-transduced Hut78 cells versus control Hut78 cells are listed in supplemental Figure 2.
Up-regulation of FasL expression and activation of FasL-induced apoptosis pathway in SATB1-transduced Hut78 cells. (A) RNA expression level of FasL in primary CD4+CD7− Sézary cells and Sézary cell line Hut78 are significantly lower than that in normal CD4+ T cells. *P < .05 by t test. (B) RNA expression level of FasL was significantly up-regulated in SATB1-transduced Hut78 cells (Hut78-SATB1), compared with empty MIG vector-transduced cells (Hut78-MIG). **P < .01 by t test. (C) The protein expressions of SATB1, FasL, Fas, cleaved-caspase 8, and cleaved caspase 3 were analyzed by Western blot with specific antibodies. All SATB1-transduced cell clones revealed high expression level of SATB1, increased expression of FasL, cleaved caspase 8, and cleaved caspase 3, whereas the expression of Fas remained unchanged. GAPDH served as an internal reference. (D) SATB1-transduced and control clonal cells were incubated with exogenous super FasL to trigger Fas-mediated cell death. SATB1-transduced cells revealed comparable specific apoptosis rate with the control cells, which showed unaltered Fas function after SATB1 transduction. Hut78 indicates parental Hut78 cells; MIG-1 to MIG-3, clonal cells from Hut78 cells transduced with control MIG vector; and SATB1-1 to SATB1-4, cell clones from Hut78 cells transduced with vector containing SATB1.
Up-regulation of FasL expression and activation of FasL-induced apoptosis pathway in SATB1-transduced Hut78 cells. (A) RNA expression level of FasL in primary CD4+CD7− Sézary cells and Sézary cell line Hut78 are significantly lower than that in normal CD4+ T cells. *P < .05 by t test. (B) RNA expression level of FasL was significantly up-regulated in SATB1-transduced Hut78 cells (Hut78-SATB1), compared with empty MIG vector-transduced cells (Hut78-MIG). **P < .01 by t test. (C) The protein expressions of SATB1, FasL, Fas, cleaved-caspase 8, and cleaved caspase 3 were analyzed by Western blot with specific antibodies. All SATB1-transduced cell clones revealed high expression level of SATB1, increased expression of FasL, cleaved caspase 8, and cleaved caspase 3, whereas the expression of Fas remained unchanged. GAPDH served as an internal reference. (D) SATB1-transduced and control clonal cells were incubated with exogenous super FasL to trigger Fas-mediated cell death. SATB1-transduced cells revealed comparable specific apoptosis rate with the control cells, which showed unaltered Fas function after SATB1 transduction. Hut78 indicates parental Hut78 cells; MIG-1 to MIG-3, clonal cells from Hut78 cells transduced with control MIG vector; and SATB1-1 to SATB1-4, cell clones from Hut78 cells transduced with vector containing SATB1.
SATB1 restoration induces FasL-induced apoptosis via caspase 8-dependent pathway without affecting Fas function
The induction of AICD on TCR stimulation depends on the up-regulation of death receptor ligands, including the FasL/CD95L, and the consequent activation of FasL-Fas-caspase 8-caspase 3 apoptosis pathway.8 To examine whether this apoptosis pathway was indeed activated, cleaved caspase 8 and cleaved caspase 3, which are downstream proteins activated in FasL-induced apoptosis pathway, were detected using Western blot analysis. As predicted, they were increased in SATB1-transduced Hut78 cells compared with the control cells (Figure 7C). This indicates that SATB1 restoration induces apoptosis by up-regulating the expression of FasL leading to the downstream activation of the FasL-Fas-caspase 8 apoptosis pathway. To test whether SATB1 restoration had additional effects on the sensitivity of Fas/CD95 to FasL/CD95L-induced apoptosis, we triggered Fas/CD95 signaling cascade in all cell clones by addition of exogenous FasL (recombinant FasL, 40 ng/mL). As shown in Figure 7D, SATB1-restored Hut78 cells showed similar levels of specific apoptosis as the control cells on FasL induction, suggesting that SATB1 restoration does not have effect on this apoptosis pathway downstream of Fas/CD95 step.
Discussion
Using primary CD4+CD7− leukemic Sézary cells, our gene expression profiling study confirmed several genes previously reported to be altered in Sézary cells, including TWIST1, DNM3, NEDD4L, STAT4, BCL11A, and PLS3, and uncovered a series of gene changes previously unknown in Sézary cells. The dysregulation of oncogenes and tumor suppressors is consistent with the malignant feature of Sézary cells. In addition, it was found that the majority of gene changes in clinically derived Sézary cells were either not preserved in cultured Sézary cell lines or shared by other T-cell malignancies, with the exception of 32 genes. The potential biologic significance of these genes in the pathogenesis of SS warrants further functional analysis. Among these gene alterations, specific down-regulation of SATB1 was identified in both primary Sézary cells from patients and in established Sézary cell lines, such as Hut78 and HH cells, although it was not altered in non-Sézary T-cell malignancies, such as Jurkat and CCL119 cell lines.
SATB1 was originally identified as a T-cell lineage–specific nuclear protein that binds to the matrix attachment region of thymocyte DNA sequences.33 In thymocytes, SATB1 shows a “cage-like” nuclear distribution, surrounds heterochromatin, and participates in the maintenance and compaction of chromatin architecture by tethering loops of chromatin to its network structure.30,34 Furthermore, SATB1 acts as a “docking site” for several chromatin-remodeling complexes, including ACF, ISWI, and HDAC1.30 By dynamically altering the organization and epigenetic status of the chromatin, SATB1 was shown to be a potent and highly pleiotropic regulator of gene expression.35 SATB1 has been shown to be essential in T-cell development because thymocyte development is arrested at the CD4, CD8 double-positive stage in SATB1 knockout mice.32 In the peripheral blood, SATB1 was shown to be highly expressed in naive and activated CD4+ T cells.36 It is also highly expressed in Th2 cells and presents as a key factor in Th2 cell activation by regulating the coordinated expression of Th2 cytokines, including IL-4, IL-5, and IL-13.32 Moreover, up-regulation of SATB1 was observed in patients with atopic dermatitis in which Th2 inflammatory cells are predominant.37 However, in our study, specific down-regulation of SATB1 was identified in Sézary cells. Sézary cells were thought to be derived from persistently activated skin-homing T cells and display a Th2 immunophenotype.38 The down-regulation of SATB1 suggests that Sézary cells are in an erroneous activation state, although this state can be partially compensated because our previous study showed that Hut78 cells could produce high levels of cytokines, including IL-4.14 Of special interest was that down-regulation of SATB1 was also observed in anergic CD4+ T cells, compared with naive and activated T cells.36 This may indicate that Sézary cells possess some features of anergic T cells, which may explain the compromised immune response observed in SS patients, although some of the other anergic cell fingerprints were not revealed, such as overexpression of CD7, PLC-γ, and LCK.36
In addition, in transcriptome analysis on SATB1-transduced Hut78 cells, approximately one-eighth of the genes aberrantly expressed in Sézary cells demonstrated either expression restoration or normalization after SATB1 transduction. This indicates that SATB1 deficiency might be responsible for part of the gene expression aberrations in Sézary cells. Thus, we postulate that the loss of SATB1 in Sézary cells serves as a “switch” from benign inflammation to malignant proliferation. SATB1 aberration has not been reported in T-cell malignancy previously. Recently, ectopic SATB1 expression was found on metastatic breast cancer, and overexpression of SATB1 was strongly correlated with tumorigenesis, metastatic potential, and poor clinical prognosis.35 These seemingly conflicting observations in 2 tumors suggest the likely presence of tissue specificity of SATB1's biologic effects.
In this study, we demonstrated that SATB1 expression restoration in Hut78 cells triggered spontaneous apoptosis and sensitized Hut78 cells to AICD. Appropriate apoptosis and cell death are essential in the control of lymphocyte homeostasis. Several lines of evidence suggest that the pathogenesis of CTCL depends initially on resistance to apoptosis rather than enhanced proliferation to account for the accumulation of tumor cells over time. CTCL usually begins as flat patches and thin plaques rather than bulky tumors. The clinical pace of the early disease often remains indolent until patients develop tumors or visceral involvement.39 Early CTCL tumor cells typically exhibit a low proliferation rate, as assessed by mitotic index and Ki-67 staining.40 In addition to their lack of proliferation, CTCL tumor cells in skin lesion rarely demonstrate apoptosis as detected by terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling assay.41 Consistent with these observations, CTCL is resistant to chemotherapy that targets rapidly dividing tumor cells.42 Recently, several reports consistently demonstrated that CTCL cells, including Hut78 cells, are highly resistant to apoptosis, in particular AICD, which has been postulated to be responsible for the development and progression of CTCLs.6,43,44 The SATB1 expression-restored Hut78 cells in our study demonstrated increased spontaneous apoptosis, compared with control Hut78 cells. When stimulated with anti-CD3 antibody or PMA/ionomycin in the presence of IL-2, which simulates the AICD process in vitro, the SATB1-restored cells showed higher rates of apoptosis. These indicate that Sézary cells with ectopic SATB1 expression are more sensitive to death induction, in particular AICD. Therefore, our results suggest that SATB1 deficiency in Sézary cells plays an important role in the pathogenesis of SS through regulating the apoptosis resistance in tumor cells.
The mechanism leading to apoptosis resistance in Sézary cells has been the subject of much debate, especially with regard to the expression aberrancies of key players of the FasL-Fas apoptosis pathway and their contributions to apoptosis resistance. First, concerning cFLIP, a potent inhibitor of the Fas signaling pathway, although Braun et al45 and Contassot et al46 demonstrated increased cFLIP expression in CTCL/Sézary cells, Klemke et al6 demonstrated that this increase did not cause AICD resistance of CTCL/Sézary cells. Second, regarding Fas protein, several studies reported disparity in Fas expression on Sézary cells,6,44,45 although its role in apoptosis resistance was widely discussed.39,45 However, in our data, no significant differences were observed in Fas or cFLIP expression comparing primary Sézary cells with normal CD4+ cells.
In normal T cells, Baumann et al demonstrated that regulation of FasL expression plays important roles in the induction of, and resistance to, AICD.47 It is therefore intriguing that, in primary and cultured Sézary cells, FasL expression was significantly decreased compared with normal control CD4+ T cells (Figure 7A). This was also observed by several other groups studying primary Sézary cells and Sézary cell lines.6,44 We showed here that SATB1 restoration up-regulated FasL transcription and activated the downstream caspase 8-mediated apoptosis pathway. This leads to the increased spontaneous apoptosis as well as to the increased susceptibility to AICD. This is also consistent with our observation that the Fas-mediated apoptosis induced with exogenous FasL was not affected in SATB1-restored Hut78 cells. Therefore, our results demonstrated that SATB1 regulates Sézary cell apoptosis through the regulation of FasL.
Inducing FasL independent of TCR ligation has been proposed as a potential future therapeutic option to break AICD resistance in SS.6 Extracorporeal photopheresis, one of the few effective therapies for late-stage CTCL and SS, executes its therapeutic effect by inducing malignant lymphocyte apoptosis via up-regulating FasL expression.48 Further, in a population of activated T cells, those expressing FasL were predominantly dead.49 Our study demonstrated up-regulation of FasL, as a result of SATB1 restoration, rendered Sézary cells sensitive to spontaneous apoptosis and susceptible to AICD. It strengthens the notions that FasL expression contributes to the killing of malignant Sézary cells and that FasL defects are at least partially responsible for the apoptosis resistance of Sézary cells. Therefore, restoring SATB1 expression may serve as a promising therapeutic strategy for the treatment of SS.
It is unknown at present how SATB1 regulates FasL expression. FasL is regulated predominantly at the transcriptional level. The FasL promoter is fairly well characterized, and the interplay among a variety of transcription factors has been defined. Among them, c-Myc, NFAT, Erg family proteins, NF-κB, and AP-1 were proposed to be key mediators in FasL expression in T cells.8 In our transcriptome analysis on SATB1-transduced Hut78 cells, none of the previously identified transcription factors for FasL was affected in the transcriptional level (data not shown). Therefore, we speculate that SATB1, as a global chromatin organizer, may enhance FasL expression by packaging transcriptionally active chromatin structure to enhance the accessibility of transcription factors to FasL promoter, as described by Cai et al in Th2 cell activation.31 Alternatively, SATB1 may bind to FasL gene loci directly and assemble histone-modifying factors or other chromatin-remodeling factors to this loci to alter the epigenetic status of this gene, as described by Han et al previously.35 More studies are needed in the future to determine how SATB1 regulates FasL expression in Sézary cells.
In conclusion, our results showed that down-regulation of SATB1 gene is one of the most consistent and specific gene expression changes in Sézary cells, and that through regulating FasL expression, it plays important roles in the development of Sézary cells' resistance to apoptosis. Restoring SATB1 expression in Sézary cells may represent an effective strategy for breaking apoptosis resistance of Sézary cells, which may be useful for the development of SS therapies in the future.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Laboratory for Advanced Genome Analysis, Vancouver Prostate Centre, for their help with the gene transcriptional profiling study.
This work was supported by grants from the Canadian Dermatology Foundation (Y.Z.), and in part by the Canadian Cancer Society (grant 2010-700289), the Cancer Research Society, and the Leukemia & Lymphoma Society of Canada (X.J.). P.T. is a recipient of National Nature Science Foundation of China (grant 81072233). Y.W. is a Canadian Institutes of Health Research-Skin Research Training Scholar. X.J. is Michael Smith Foundation for Health Research Scholar.
Authorship
Contribution: Y.W., M.S., and L.L.Z. designed and performed the experiments and analyzed the data; M.S., P.T., X.Z., and Y.Z. provided clinical data and RNA samples from patients with SS and performed microarray analyses; X.J. and Y.Z. developed the concept of the study and designed and supervised the experiments; Y.W., X.J., and Y.Z. wrote the manuscript; and all authors commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Youwen Zhou, Department of Dermatology and Skin Science, University of British Columbia, 835 West 10th Ave, Vancouver, BC, V5Z 4E8, Canada; e-mail: ywzhou@interchange.ubc.ca; and Xiaoyan Jiang, Terry Fox Laboratory, British Columbia Cancer Agency, 675 West 10th Ave, Vancouver, BC, V5Z 1L3, Canada; e-mail: xjiang@bccrc.ca.


![Figure 3. Specific suppression of SATB1 expression in Sézary cells. (A) SATB1 RNA expression levels are significantly down-regulated in CD4+CD7− primary Sézary cells from 6 SS patients with TCR clonality [SS(T)] and 3 SS patients without TCR clonality [SS(A)], as well as cultured Sézary cell lines (Hut78 and HH), compared with similarly purified T lymphocytes from normal volunteers (NC), and 2 cultured non-Sézary leukemic cell lines (Jurkat and CCL119). **P = .004. (B) SATB1 protein expressions are absent from patient-derived Sézary cells (SS1 and SS2) and Sézary cell lines, whereas the normal cells (NC1 and NC2) and 2 non-Sézary leukemia cell lines show abundant expression of SATB1 protein. GAPDH serves as the internal reference. (C) Immunofluorescence staining of SATB1 on CD4+ T cells in vitro. Cells were stained with mouse SATB1 monoclonal antibody (1:100) and Alexa-594-conjugated anti–mouse secondary antibody (1:200). DAPI was used to counterstain cell nucleus. Non-Sézary cells (NC, Jurkat and CCL119) showed strong SATB1 staining on the nucleus, whereas patient-derived SS and Sézary cell line (Hut78 and HH) cells showed nearly no staining on the nucleus. Original magnification ×630. Scale bar represents 10 μm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/14/10.1182_blood-2010-07-294819/4/m_zh89991168150003.jpeg?Expires=1765952966&Signature=TxN4TbIQCUE7nQpb0smKhnGQ7zdTR2Eb1MHP1g5N1yVgPTzXh8dj~idEehdterLf5p7DkaA-Nvt7Pfg8tnDS6JrPTi4vQuy9bMfRGYoXqvUW~2OVtYJe-ZH2zN1zOVK5mDbnEzlHMQMhbsCrb0bfXNM6dcRg8vqjGMGK6hiqCVCh3~vCpCCZ73AuihYvcCEVmRM7Td1-k1L2ckwIAAhQikFFWe8pVx~9xa-ehFL63tXep5hQ0f3AHFUxco0jnc8YkTJQZD~jU5RJ3PPSjAz0lMzKYGJjZdTcM1oMNRkuAWhSqaFAlNsnrlPNAgiQluT6KBPvmCvZp2-ZEFZEKUXkzg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)