Abstract
During development in the bone marrow (BM), NK-cell positioning within specific niches can be influenced by expression of chemokine or adhesion receptors. We previously demonstrated that the maintenance in the BM of selected NK-cell subsets is regulated by the CXCR4/CXCL12 axis. In the present study, we showed that CX3CR1 is prevalently expressed on KLRG1+ NK cells, a subset considered terminally differentiated. Two KLRG1+ NK-cell populations endowed with distinct homing and functional features were defined according to CX3CR1 expression. In the BM, KLRG1+/CX3CR1− NK cells were mainly positioned into parenchyma, while KLRG1+/CX3CR1+ NK cells exhibited reduced CXCR4 expression and were preferentially localized in the sinusoids. We also showed that α4 integrin plays a pivotal role in the maintenance of NK cells in the BM sinusoids and that α4 neutralization leads to strong reduction of BM KLRG1+/CX3CR1+ NK cells. Moreover, we found that KLRG1+/CX3CR1+ cells originate from KLRG1+/CX3CR1− NK-cell population and display impaired capability to produce IFN-γ and to lyse YAC-1 target cells on cytokine stimulation. Altogether, our findings show that CX3CR1 represents a marker of a KLRG1+ NK-cell population with unique properties that can irreversibly differentiate from the KLRG1+/CX3CR1− NK cells during steady state conditions.
Introduction
Natural killer (NK) cells represent a lymphocyte population of the innate immunity distributed in several lymphoid and nonlymphoid organs and involved in host defense against microbial infections and cell transformation.1-4
Bone marrow (BM) is the main site of NK-cell generation, providing both cellular substrates and signals required to sustain cell proliferation and differentiation.5,6 During maturation, mouse NK cells gradually modulate the expression of several surface receptors including cytokine receptors (ie, CD122 or CD127), adhesion molecules (ie, integrins) and receptors with activating or inhibitory functions (ie, CD94, NKG2 and Ly49 receptors).7
The acquisition of CD49b expression (α2 integrin chain, recognized by DX5 monoclonal antibody) is an early event occurring during development and defines a heterogeneous population of mature NK cells.8 According to the expression of the integrin chain CD11b (also known as Mac-1 or αm) 2 major NK-cell subsets have been defined: a less differentiated population expressing low receptor levels (CD11blow) and a more differentiated one expressing higher levels (CD11bhigh) of the integrin.9 In addition, down-modulation of the tumor necrosis factor receptor superfamily member, CD27, by the CD11bhigh subset coincides with the acquisition of the killer lectin-like receptor G1 (KLRG1) expression, an inhibitory receptor that binds to several members of the cadherin family.10-13 KLRG1 is generally considered a marker of terminally differentiated NK and T cells14-17 that can also be acquired after homeostatic proliferation, and defines a subset of NK cells with reduced ability to proliferate and mediate effector functions in response to cytokines.12,18
Developing NK cells differ for proliferation rates and functional skills, as well as for their ability to traffic among several organs and tissues.19 In particular, BM is highly enriched of immature NK cells (DX5− or CD11blow) compared with blood, spleen, or other peripheral organs, where CD11bhigh and KLRG1+ NK-cell subsets are mainly found.
The molecular mediators that contribute to regulate NK-cell homing to and egress from BM during homeostatic conditions are currently poorly understood. Recent findings have highlighted the importance of the chemokine system and bioactive lipids, in particular sphingosine-1-phosphate (S1P), in the regulation of BM NK-cell retention and exit.20-22 In this regard, we have previously demonstrated that distinct NK-cell populations show a specific array of chemokine receptors, including CXCR3, CCR1, and CXCR4, and that the combined action of chemokines can modulate the retention of NK cells in the BM in a CXCR4-dependent fashion.21 Nonetheless, the paucity of available data concerning the spatial expression of chemokines or chemokine-producing stromal cells in the BM as well as the lack of highly specific NK-cell markers, have made difficult to find NK-cell niches in this organ. Sinusoids represent a BM environment lined by endothelial cells and surrounded by adventitial reticular cells, where hematopoietic cells can enter in, exit from circulation, or stop to undergo maturation.23
Recently, it has been shown that an abundant fraction (∼ 14%) of NK cells reside in BM sinusoidal venous blood vessels.22 However, the mechanisms underlying NK-cell retention or release from BM sinusoids have not been characterized.
CX3CL1/CX3CR1 chemokine/chemokine receptor pair is of particular interest in the context of NK cell–mediated responses. Indeed, CX3CL1 binding to its specific receptor, CX3CR1, can promote human NK-cell adhesion and chemotaxis and enhance NK-cell ability to lyse target cells in vitro.24-26 Moreover, this axis has been involved in the regulation of NK-cell recruitment and/or activation in a number of inflammatory responses in vivo.27,28
Despite the numerous evidences on the role of CX3CR1 in NK-cell recruitment during the onset of inflammation, little is known regarding CX3CR1 expression on NK cells from naive mice during maturation.
Here we report that CX3CR1 is mainly expressed by the KLRG1+ NK-cell subset. Moreover, in the BM, KLRG1+/CX3CR1+ NK cells preferentially reside in the sinusoids and this localization is dependent on CXCR4 down-modulation and α4 integrin function. Finally, we show that CX3CR1 expression defines a KLRG1+ NK-cell subset with impaired effector functions.
Methods
Mice
Female wild-type (wt) C57BL/6 and CX3CR1+/GFP mice29 were bred and housed in the animal facility of the Physiology and Pharmacology Department at the “Sapienza” University of Rome, and female C57BL/6 mice on the CD45.1 and CD45.2 background were purchased from Charles River (Calco, Italy). For all experiments, mice between 5 and 10 weeks of age were used. Animals were kept under specific pathogen-free conditions in accordance with institutional guidelines for animal care and use, and animal protocols were approved by the ethical committee of the “Sapienza” University of Rome.
Antibodies, chemokines, and reagents
Several antibodies (Abs) directly conjugated to fluorescein isothiocyanate (FITC), phycoerythrin (PE), peridinin chlorophyll protein (PerCP) 5.5, allophycocyanin (APC), PE- cyanine (cy)7 or biotin, and specific for the following antigens (clone name in parentheses) were used in this study: NK1.1 (PK136), CD3ϵ (145-2C11), CD11b (M1/70), KLRG1 (2F1), CD44 (IM7), CD94 (18D3), DNAM-1 (TX42.1), CD127 (A7R34), c-kit (2B8), CXCR4 (2B11), CD49b (DX5), α4 integrin/CD49d (R1-2), IFN-γ (XMG1.2), CD45.1 (A20) and CD45.2 (104). Abs and PE-Cy7, PE–conjugated streptavidin and PE–conjugated Goat anti–rat IgG and Rat anti–mouse IgG2a/b were purchased from Pharmingen (Becton Dickinson), eBioscience, and Biolegend. PE-conjugated mouse anti–human IgG was purchased from Jackson Immunoresearch Laboratories. Purified rat anti–mouse Ly49G2 (4D11), mouse anti–mouse Ly49C/I (5E6) mAb were from Pharmingen. Alpha4 integrin blocking mAb (PS/II) was kindly provided by Dr E. Butcher (Stanford University, Stanford, CA). Recombinant mouse IL-12, and human CXCL12, IL-15, and IL-2 were from Peprotech EC. BSA, saponin, PKH-26, and AMD-3100 were from Sigma-Aldrich.
Cell preparation
BM cells were isolated by extensive flushing of femurs and tibias with PBS; cells from spleen and liver were obtained by mechanical disruption using a 70-μm cell strainer (Falcon; Becton Dickinson) with a rubber syringe plunger in RPMI 10% FBS. Blood samples were obtained by tail bleeding and collected into heparin containing tubes. For staining experiments, liver cells were separated using Lympholyte (CL5031; Cedarlane Lab), washed, and resuspended in RPMI 10% FBS.
Immunofluorescence and FACS analysis
Cells from the indicated organs were washed and resuspended in staining buffer (PBS without Ca2+/Mg+, BSA 0.5%, EDTA 1 mM, and NaN3 0.05%). The anti-CD16/32 (24G2) mAb was added for 10 minutes on ice to block non specific and Fc-mediated binding. Then, cells were stained with the indicated mAbs diluted in staining buffer for 20 minutes on ice.
For indirect staining, 1.5 × 106 splenocytes were incubated with saturating doses of anti–Ly49G2 mAb or anti-Ly49C/I mAb. Cells were washed and stained with PE–conjugated anti–rat IgG or anti–mouse IgG2a/b. Cells were washed, blocked with 5 μg rat IgG, or mouse serum (1:20 dilution) and incubated with the appropriate cocktail of conjugated Ab to visualize the relevant NK-cell populations. Ly49H+ cells were identified using the fusion protein of MCMV m157 gene product and human-Fc.
Staining for chemokine receptor was performed after cell incubation for 1 hour at 37°C in a CO2 incubator, to allow resensitization of receptors. CX3CR1+ expressing cells were identified as GFP+ cells in CX3CR1+/GFP mice. CX3CR1 surface expression on wt mice was analyzed using the human CX3CL1 fused with the human-Fc (CX3CL1-Fc).
Staining was performed using the culture supernatant of 293-HEK cells transiently transfected with a CX3CL1-Fc (kind gift of Dr Andreas Ludwig, Aachen University, Aachen, Germany), m157-Fc or NKG2D-Fc expression vectors.
Intracellular staining for IFN-γ was performed using freshly isolated cells from BM and spleen of CX3CR1+/GFP mice previously cultured in complete medium in the presence of IL-2 (500U/mL) IL-12 (100ng/mL), for 18 hours. Brefeldin A (10 μg/mL) was added during the last 6 hours. At the end of treatment, cells were stained with anti-NK1.1–, anti-KLRG1–, and anti-CD3ϵ–specific mAbs. After fixation with 1% paraformaldehyde (PFA) in PBS and permeabilization with 0.5% saponin in PBS, cells were stained with anti–IFN-γ–specific mAb, and analyzed by flow cytometry.
Flow cytometric analysis was performed using FACSCanto II (Becton Dickinson) and data were elaborated using Diva Version 6.1.3 or FlowJo Version 8.5.2 software.
Migration assay
Chemotaxis assay of cells isolated from BM of individual animals in response to CXCL12 (200 ng/mL), was performed in 5-μm pore Transwell insert as described.21 After 1 hour of incubation at 37°C 5% CO2, BM cells (1 × 106) were loaded on the upper well. After 90 minutes, the content of the lower chemotaxis well was transferred to a polypropylene tube, and stained with a mixture of the appropriate antibodies. Migrated cells were counted by flow cytometry and expressed as percentage of input cells.
51Cr release cytotoxicity assay
Spleen cells were stained with anti-NK1.1–, anti-KLRG1–, and anti-CD3ϵ–specific mAbs. Successively, KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ NK-cell subsets were purified by sorting using FACSAria (Becton Dickinson). Sorted KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ NK-cell subsets were cultured in the presence of IL-15 (100 ng/mL) and used as effector cells. Mouse lymphoma YAC-1 (H-2a) cells (5 × 105), were labeled by incubation with 51Cr in complete medium for 1 hour, at 37°C, 5% CO2 and used as target cells. Target cells were washed and mixed with effector cells at effector:target ratios ranging from 2.5:1 down to 0.62:1, and incubated at 37°C, 5% CO2. After 4 hours, 30 μL of supernatant was harvested, added to a LUMAPLATE (PerkinElmer), and counted in a TopCount-NXT counter (PerkinElmer).
The percent specific lysis was determined as follows: % specific lysis = 100 × [sample release − spontaneous release]/[total release − spontaneous release].
In vivo adoptive transfer experiments
For homing experiments, spleen cells were labeled with the red fluorescent dye PKH-26 according to the manufacturer's instructions, and injected intravenously. After 18 hours, NK-cell subset distribution in the BM, spleen and liver was evaluated by immunofluorescence and flow cytometry.
Sorted KLRG1+/CX3CR1-GFP+ and KLRG1+/CX3CR1-GFP− (purity > 95%) splenic NK cells from CX3CR1+/GFP CD45.2 mice were adoptively transferred by intravenous tail injection into wt C57BL/6 mice carrying the allelic variant CD45.1, and after 12 days KLRG1 and GFP expression was evaluated on donor NK cells from BM, spleen and liver by immunofluorescence and flow cytometry as described in “Immunofluorescence and FACS analysis.”
Labeling of sinusoidal BM NK-cell populations and in vivo mice treatments
BM sinusoidal NK cells were identified using the procedure described by Pereira and coworkers.30 Briefly, 1 μg of anti–mouse DX5-PE or anti-CD45–PE mAb in 300 μL of PBS was injected intravenously. After 2 minutes, mice were killed, and cells from BM and blood (the latter used as control) were rapidly collected and stained as indicated in Figure 2C. The cells stained in vivo with DX5 or anti-CD45 mAb are referred as sinusoidal, while DX5− and CD45− cells are considered parenchymal.
AMD-3100 (100 μg diluted in PBS in 50 μL) or PS-II mAb (100 μg diluted in PBS in 50 μL) were injected subcutaneously and intravenously, respectively, as previously described.21,30 At the end of treatment, mice were killed, and cells were isolated as described in “Cell preparation.” After removal of contaminating erythrocytes, cells were counted and stained with the Abs indicated in the figure legends and analyzed by flow cytometry.
The number of total CD3−/NK1.1+ NK cells as well as of NK-cell subsets in the organs was obtained taking into account their percentages within the total lymphocytes.
Statistics
The Student t test was used to analyze data. A P value of .05 was chosen as the limit of statistical significance.
Results
CX3CR1 is preferentially expressed on mouse KLRG1+ NK cells
To evaluate CX3CR1 expression during NK-cell maturation, we used a genetically modified mouse model in which the exon encoding Cx3cr1 was replaced by an exon encoding GFP, under the control of Cx3cr1 promoter.29 CX3CR1 expressing cells are identified as GFP+ cells on heterozygous mice (CX3CR1+/GFP), and are referred as CX3CR1-GFP+ cells.
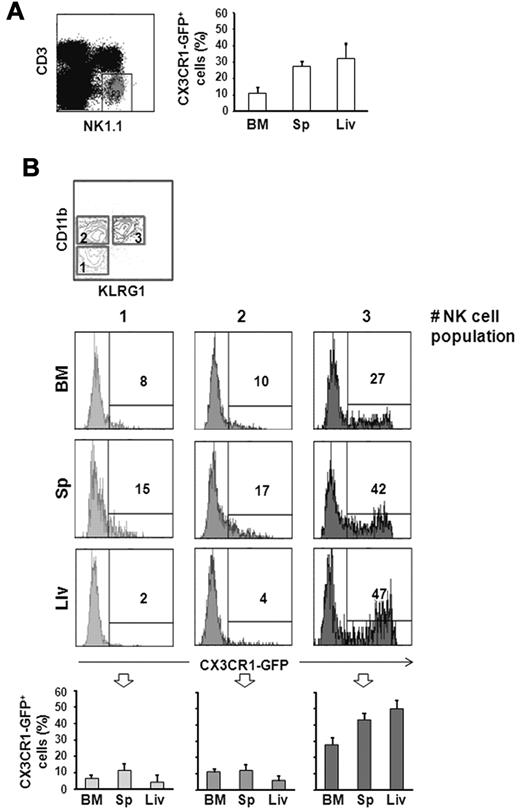
CX3CR1-GFP expression was evaluated on NK cells (defined as CD3−/NK1.1+) from different organs by immunofluorescence and flow cytometric analysis. Among CD3−/NK1.1+ NK cells, only a discrete population expressed CX3CR1-GFP in BM (12% ± 3%), spleen (27% ± 4%) and liver (33% ± 8%; Figure 1A).
CX3CR1-GFP expression on different NK-cell subsets. Cells from BM, spleen (Sp), and liver (Liv) from 6-10 week old CX3CR1+/GFP mice were isolated. Immunofluorescence staining with anti-NK1.1– and anti-CD3ϵ–specific mAbs, in combination with anti-CD11b– and anti-KLRG1–specific mAbs was performed, and expression of CX3CR1-GFP was analyzed on CD3−/NK1.1+ NK cells (A), and on the different NK-cell subsets (B): CD11blow/KLRG1− cells (population 1), CD11bhigh/KLRG1− cells (population 2) and CD11bhigh/KLRG1− cells (population 3). Numbers in the histogram plots indicate the percentage of positive cells of a representative experiment of at least 3 performed. In the bottom panels, the mean values ± SD of the percentage of CX3CR1-GFP+ NK cells in the different populations from the organs of a total of 9 animals analyzed in independent experiments, are represented.
CX3CR1-GFP expression on different NK-cell subsets. Cells from BM, spleen (Sp), and liver (Liv) from 6-10 week old CX3CR1+/GFP mice were isolated. Immunofluorescence staining with anti-NK1.1– and anti-CD3ϵ–specific mAbs, in combination with anti-CD11b– and anti-KLRG1–specific mAbs was performed, and expression of CX3CR1-GFP was analyzed on CD3−/NK1.1+ NK cells (A), and on the different NK-cell subsets (B): CD11blow/KLRG1− cells (population 1), CD11bhigh/KLRG1− cells (population 2) and CD11bhigh/KLRG1− cells (population 3). Numbers in the histogram plots indicate the percentage of positive cells of a representative experiment of at least 3 performed. In the bottom panels, the mean values ± SD of the percentage of CX3CR1-GFP+ NK cells in the different populations from the organs of a total of 9 animals analyzed in independent experiments, are represented.
To better define the subsets expressing CX3CR1-GFP, we dissected the NK-cell population according to CD11b and KLRG1 expression. When these 2 markers are analyzed in combination, 3 NK-cell subsets can be defined: a less mature population characterized as CD11blow/KLRG1− (indicated as population 1, in Figure 1B); an intermediate population defined as CD11bhigh/KLRG1− (population 2); and finally, a terminally differentiated CD11bhigh/KLRG1+ NK-cell population.
As shown in Figure 1B, CX3CR1-GFP is expressed with higher frequency on the more mature KLRG1+ NK-cell subset, while a smaller fraction of CD11blow/KLRG1− and CD11bhigh/KLRG1− NK cells express CX3CR1-GFP at lower levels. Similar results were obtained by analyzing CX3CR1 surface expression on splenic NK cells from wt C57BL/6 mice using a recombinant human-CX3CL1-Fc fusion protein. CX3CR1 was preferentially detected on KLRG1+ NK cells, while only a few KLRG1− NK cells expressed CX3CR1 at lower levels (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Our data demonstrate that CX3CR1 expression on NK cells is developmentally regulated, with a similar pattern observed in all organs analyzed, albeit the frequency of KLRG1+/CX3CR1-GFP+ cells is lower in the BM compared with the periphery.
To understand whether the low frequency of CX3CR1-GFP+ NK cells in the BM could depend on their different homing capacity and/or on the regulation of CX3CR1 expression by BM microenvironment, we performed adoptive transfer experiments, in which splenocytes isolated from CX3CR1+/GFP mice were labeled with the cell tracker PKH-26, and transferred into wt C57BL/6 recipient mice. Eighteen hours after the transfer, mice were killed and the frequency of donor NK cells within recipient lymphocytes (supplemental Figure 2A), the percentage of CX3CR1-GFP+ cells, and the ratio CX3CR1-GFP+/CX3CR1-GFP− within transferred NK cells were calculated (supplemental Figure 2B left and right panels). These experiments evidenced that adoptively transferred CX3CR1-GFP+ NK cells distributed with lower frequency in the BM compared with spleen and liver, and paralleled their tissue distribution in CX3CR1+/GFP mice, thus indicating that tissue localization of CX3CR1+ NK cells is regulated by their homing behavior.
KLRG1+/CX3CR1-GFP+ NK cells show reduced CXCR4 expression and CXCL12 responsiveness and preferentially localize in BM sinusoids
Based on our observation that CX3CR1-GFP is expressed by a fraction of KLRG1+ NK cells, we analyzed whether other chemokine receptors could be differently expressed by KLRG1+/CX3CR1-GFP− and NK-cell populations.
Thus, we focused our attention on CXCR4 expression, because this receptor is involved in NK-cell retention and homing to the BM, and is down-regulated during their differentiation.21
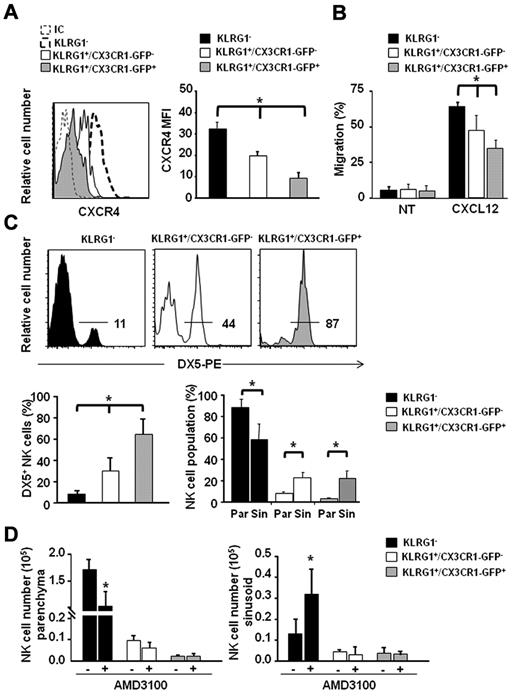
Consistent with their differentiation stage, both KLRG1+/CX3CR1-GFP+ and KLRG1+/CX3CR1-GFP− expressed lower CXCR4 levels compared with the less mature KLRG1− NK-cell population. Interestingly, we found that the KLRG1+/CX3CR1-GFP+ NK-cell subset expressed significantly lower CXCR4 levels compared with KLRG1+/CX3CR1-GFP− cells (Figure 2A).
KLRG1+/CX3CR1-GFP+ NK cells localize in the BM sinusoids and display reduced CXCR4 expression and CXCL12 responsiveness. (A) Immunofluorescence staining of BM cells with anti-NK1.1–, anti-CD3ϵ–, anti-KLRG1–, and anti-CXCR4–specific antibodies was performed, followed by FACS analysis. (Left panel) histogram overlays show CXCR4 geometric mean fluorescence intensity (MFI) of KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ NK cells, of 1 representative experiment of at least 3 performed. (Right panel) histograms show the mean values ± SD of CXCR4 geometric MFI of KLRG1−, KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ BM NK cells of a total of 8 animals analyzed in independent experiments. (B) Ex vivo chemotaxis assays with CXCL12 (200 ng/mL) were performed in 5 μm pore Transwell insert. Migrated cells were stained with anti-NK1.1–, anti-KLRG1–, and anti-CD3ϵ–specific mAbs and counted by FACS analysis. Histograms show migration of the indicated BM NK-cell subsets in response to migration medium (NT) or CXCL12. Data are expressed as percentage of input cells and represent the means ± SD of percentage of migrated cells from a total of 6 animals analyzed in independent experiments. Student t test was performed by comparing CXCR4 expression or CXCL12-supported migration of KLRG1−, vs KLRG1+/CX3CR1-GFP− and vs KLRG1+/CX3CR1-GFP+ NK cells; *P < .05. (C) BM sinusoidal NK cells were labeled by intravenous injection of 1 μg of DX5-PE mAb in 300 μL PBS. The percentage of DX5-PE+ cells within KLRG1−, KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ NK cells was evaluated. Histogram plots (top panel) show a representative experiment of at least 3 performed. Histograms (bottom left panel) show the mean values ± SD of the percentage of DX5+ cells in the different BM NK-cell populations of a total of 9 animals analyzed in independent experiments. Histograms (bottom right panel) show the fraction of parenchymal (Par, DX5-PE−) and sinusoidal (Sin, DX5-PE+) NK cells of the indicated phenotypes within total NK cells of the parenchyma and sinusoids, respectively. NK-cell number (×103) in parenchyma and sinusoids were, respectively, 173 ± 19 and 12.8 ± 7.3 for KLRG1−; 10.1 ± 2.0 and 4.3 ± 1.3 for KLRG1+/CX3CR1-GFP−; 2.5 ± 0.8 and 4.1 ± 2.7 for KLRG1+/CX3CR1-GFP+. (D) CX3CR1+/GFP mice were injected subcutaneously with 50 μL PBS alone (−) or containing 2 mg/mL AMD-3100, and sinusoidal NK cells were stained by intravenous administration of DX5-PE mAb in the last 2 minutes of treatment. Mice were killed and BM cells were counted and stained with anti-CD3ϵ–, anti-NK1.1–, anti-KLRG1–specific mAbs. Histograms represent the mean values ± SD of total cell number of the indicated NK-cell subsets in BM parenchyma (left panel) and sinusoids (right panel) from a total of 6 animals/group analyzed in independent experiments. Student t test was performed to compare the BM distribution of NK-cell subsets in mice treated with vehicle control with that of mice treated with AMD-3100. *P < .05.
KLRG1+/CX3CR1-GFP+ NK cells localize in the BM sinusoids and display reduced CXCR4 expression and CXCL12 responsiveness. (A) Immunofluorescence staining of BM cells with anti-NK1.1–, anti-CD3ϵ–, anti-KLRG1–, and anti-CXCR4–specific antibodies was performed, followed by FACS analysis. (Left panel) histogram overlays show CXCR4 geometric mean fluorescence intensity (MFI) of KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ NK cells, of 1 representative experiment of at least 3 performed. (Right panel) histograms show the mean values ± SD of CXCR4 geometric MFI of KLRG1−, KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ BM NK cells of a total of 8 animals analyzed in independent experiments. (B) Ex vivo chemotaxis assays with CXCL12 (200 ng/mL) were performed in 5 μm pore Transwell insert. Migrated cells were stained with anti-NK1.1–, anti-KLRG1–, and anti-CD3ϵ–specific mAbs and counted by FACS analysis. Histograms show migration of the indicated BM NK-cell subsets in response to migration medium (NT) or CXCL12. Data are expressed as percentage of input cells and represent the means ± SD of percentage of migrated cells from a total of 6 animals analyzed in independent experiments. Student t test was performed by comparing CXCR4 expression or CXCL12-supported migration of KLRG1−, vs KLRG1+/CX3CR1-GFP− and vs KLRG1+/CX3CR1-GFP+ NK cells; *P < .05. (C) BM sinusoidal NK cells were labeled by intravenous injection of 1 μg of DX5-PE mAb in 300 μL PBS. The percentage of DX5-PE+ cells within KLRG1−, KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ NK cells was evaluated. Histogram plots (top panel) show a representative experiment of at least 3 performed. Histograms (bottom left panel) show the mean values ± SD of the percentage of DX5+ cells in the different BM NK-cell populations of a total of 9 animals analyzed in independent experiments. Histograms (bottom right panel) show the fraction of parenchymal (Par, DX5-PE−) and sinusoidal (Sin, DX5-PE+) NK cells of the indicated phenotypes within total NK cells of the parenchyma and sinusoids, respectively. NK-cell number (×103) in parenchyma and sinusoids were, respectively, 173 ± 19 and 12.8 ± 7.3 for KLRG1−; 10.1 ± 2.0 and 4.3 ± 1.3 for KLRG1+/CX3CR1-GFP−; 2.5 ± 0.8 and 4.1 ± 2.7 for KLRG1+/CX3CR1-GFP+. (D) CX3CR1+/GFP mice were injected subcutaneously with 50 μL PBS alone (−) or containing 2 mg/mL AMD-3100, and sinusoidal NK cells were stained by intravenous administration of DX5-PE mAb in the last 2 minutes of treatment. Mice were killed and BM cells were counted and stained with anti-CD3ϵ–, anti-NK1.1–, anti-KLRG1–specific mAbs. Histograms represent the mean values ± SD of total cell number of the indicated NK-cell subsets in BM parenchyma (left panel) and sinusoids (right panel) from a total of 6 animals/group analyzed in independent experiments. Student t test was performed to compare the BM distribution of NK-cell subsets in mice treated with vehicle control with that of mice treated with AMD-3100. *P < .05.
By performing ex vivo chemotaxis assays, we observed a gradual modulation of NK-cell migratory response to CXCL12 during differentiation that paralleled CXCR4 expression levels, being KLRG1+/CX3CR1-GFP+ NK cells less responsive than KLRG1+/CX3CR1-GFP− and KLRG1− NK cells (Figure 2B).
Thus, the acquisition of CX3CR1 expression on KLRG1+ NK cells coincides with decreased CXCR4 expression and function, unraveling the existence of 2 distinct subsets differing for their trafficking abilities within the KLRG1 NK-cell population.
Leukocyte egress from the BM is thought to occur via the sinusoids. To analyze the positioning of NK-cell populations inside the BM, we took advantage of a recent method that allows to selectively label the cells contained in BM sinusoids by short in vivo exposure to intravenously injected mAb conjugated with PE.30 Thus, we evaluated the number of labeled BM sinusoidal NK cells after injection of DX5-PE mAb that recognizes all mature NK cells (> 90% of total BM NK cells). We found that ∼ 13% of BM CD3−/NK1.1+ NK cells were DX5-PE+, and this was consistent with previous observations.22 A similar result was obtained when an anti-CD45–PE was used, indicating that the short in vivo binding of anti-α2β1 integrin DX5-PE mAb on NK cells does not alter their distribution into sinusoids (supplemental Figure 3).
The surface phenotype of sinusoidal NK cells was also analyzed using several maturation markers. As shown in supplemental Figure 4 CD127, CD11b, KLRG1 were similarly expressed by NK cells from sinusoidal and peripheral blood NK cells while c-kit expression levels on sinusoidal NK cells were comparable with those of parenchymal cells. These data indicate that the great majority of NK cells that reside in the sinusoid display a mature phenotype, while only a minor fraction of c-kit+, more immature population is maintained in this compartment.
We then characterized the fraction of DX5-PE+ sinusoidal NK cells within the CX3CR1-GFP+ and CX3CR1-GFP− subsets. We observed that a higher proportion (64%) of KLRG1+/CX3CR1-GFP+ NK cells, was contained in sinusoids compared with KLRG1+/CX3CR1-GFP− (30%) and KLRG1− (8%) NK-cell populations (Figure 2C). This analysis also evidenced that while the 2 KLRG1+ populations are poorly represented among BM parenchymal NK cells (CX3CR1−: 6% ± 1.2, CX3CR1+: 1% ± 0.3) they constitute a significant fraction of sinusoidal NK cells (CX3CR1−: 21% ± 6, CX3CR1+: 19% ± 10; Figure 2C bottom right panel).
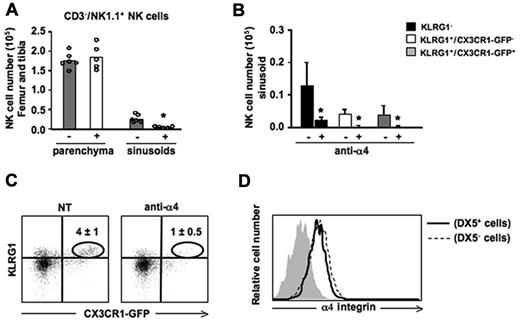
We previously demonstrated that administration of AMD-3100, a pharmacologic antagonist of CXCR4, induces a rapid mobilization of NK cells from BM to blood; in addition, CXCL12 has been shown to be abundantly expressed by BM parenchymal and endothelial cells that line the sinusoids.31 Thus, we investigated how CXCR4/CXCL12 axis could affect NK-cell positioning within these 2 BM compartments. To calculate the number of different NK-cell subsets in the BM parenchyma (DX5− cells after in vivo labeling) and sinusoids (DX5+ cells), mice were treated with AMD-3100 for 1 hour, injected with DX5-PE mAb and then killed. As shown in Figure 2D, NK-cell populations responded differently to CXCR4 blockade. KLRG1− NK cells were mainly affected by AMD-3100 treatment, undergoing a significant reduction in the BM parenchyma (Figure 2D left panel) that was associated with a parallel increase in the BM sinusoids (Figure 2D right panel) and in the blood (data not shown), compared with control mice. On the other hand, we did not observe any significant changes in the number of the 2 KLRG1+ NK-cell populations both in the parenchyma and in the sinusoids.
Collectively these data demonstrate a central role for CXCR4 in NK-cell retention inside the BM parenchyma, and suggest that developmental down-regulation of CXCR4 expression could lead mature NK cells to move from BM parenchyma to sinusoids, where CXCR4-mediated retention seems to be less relevant.
BM localization of KLRG1+/CX3CR1-GFP+ NK cells is dependent on α4 integrin
The interaction between α4(CD49d) β1(CD29) integrin and VCAM-1 is important to retain hematopoietic stem cells and B cells in the BM sinusoids.30,32 On the other hand, the mechanisms underlying NK-cell localization in the sinusoids have not been investigated. In particular, no information are available on the role of α4 integrin on BM NK-cell retention in vivo, although previous observations indicate the VCAM-1/α4β1 interaction mediates NK-cell infiltration into hepatic and pulmonary parenchymas during inflammatory conditions.33 Thus, we treated mice with an α4 blocking antibody for 3 hours, and labeled sinusoidal NK cells by in vivo administration of DX5-PE mAb. Mice were then killed and the number of NK cells in BM parenchyma and sinusoids were counted. Neutralization of α4 integrin selectively reduced NK cells in the sinusoidal compartment, regardless of their maturation stage (Figure 3A-B). Remarkably, α4 blockade determined a drastic reduction of KLRG1+/CX3CR1-GFP+ NK cells inside the BM as this subset is prevalently localized in sinusoids (Figure 3C). On the contrary KLRG1+/CX3CR1-GFP+ NK-cell retention in spleen and liver was not affected by anti-α4 mAb treatment (data not shown).
In vivo administration of α4 integrin neutralizing mAb promotes NK-cell mobilization from the BM sinusoids. Mice were treated with an α4 integrin neutralizing mAb for 3 hours, and BM sinusoidal NK cells were labeled by intravenous injection of 1 μg of DX5-PE mAb. Mice were killed after 2 minutes and BM cells were isolated and stained with anti-NK1.1–, anti-CD3ϵ–, and anti-KLRG1–specific mAbs. (A) Histograms show the numbers of CD3−/NK1.1+ NK cells in the BM parenchyma (DX5− cells) and sinusoids (DX5+ cells) of untreated (−) or treated (+) mice. Each dot represents the number of BM NK cells from individual animals analyzed in independent experiments. (B) Histograms show the number ± SD of sinusoidal (DX5-PE+) cells evaluated within KLRG1−, KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ BM NK-cell subsets. Student t test was performed to compare untreated versus treated mice. *P < .05. (C) Dot plots show the reduction of KLRG1+/CX3CR1-GFP+ NK cells in anti-α4 integrin neutralizing mAb-treated mice compared with control mice. Numbers in dot plots indicate the percentage of the KLRG1+/CX3CR1-GFP+ NK cells (gated on CD3−/NK1.1+ cells). (D) Histogram overlay shows α4 expression on parenchymal (DX5−) and sinusoidal (DX5+) BM NK cells from 1 representative experiment of at least 3 performed.
In vivo administration of α4 integrin neutralizing mAb promotes NK-cell mobilization from the BM sinusoids. Mice were treated with an α4 integrin neutralizing mAb for 3 hours, and BM sinusoidal NK cells were labeled by intravenous injection of 1 μg of DX5-PE mAb. Mice were killed after 2 minutes and BM cells were isolated and stained with anti-NK1.1–, anti-CD3ϵ–, and anti-KLRG1–specific mAbs. (A) Histograms show the numbers of CD3−/NK1.1+ NK cells in the BM parenchyma (DX5− cells) and sinusoids (DX5+ cells) of untreated (−) or treated (+) mice. Each dot represents the number of BM NK cells from individual animals analyzed in independent experiments. (B) Histograms show the number ± SD of sinusoidal (DX5-PE+) cells evaluated within KLRG1−, KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ BM NK-cell subsets. Student t test was performed to compare untreated versus treated mice. *P < .05. (C) Dot plots show the reduction of KLRG1+/CX3CR1-GFP+ NK cells in anti-α4 integrin neutralizing mAb-treated mice compared with control mice. Numbers in dot plots indicate the percentage of the KLRG1+/CX3CR1-GFP+ NK cells (gated on CD3−/NK1.1+ cells). (D) Histogram overlay shows α4 expression on parenchymal (DX5−) and sinusoidal (DX5+) BM NK cells from 1 representative experiment of at least 3 performed.
Neutralization of the integrin alone did not result in NK-cell exit from the BM parenchyma nor it enhanced the mobilizing effect of CXCR4 antagonism as assessed by the in vivo combined administration of α4 blocking mAb and AMD3100 (data not shown). The differential effect of anti-α4 mAb on sinusoidal versus parenchymal NK cells was not attributable to different levels of α4 integrin expressed by the cells in the 2 compartments as assayed by immunofluorescence and cytofluorimetric analysis (Figure 3D).
Collectively, these data demonstrate that α4 integrin is the main player of KLRG1+/CX3CR1-GFP+ NK-cell BM retention in the sinusoid compartment.
KLRG1+/CX3CR1-GFP+ NK-cell population displays impaired effector functions
The evidence that CX3CR1 expression defines 2 distinct populations among KLRG1+ NK cells, prompted us to analyze whether they also differ in their effector functions.
To analyze the ability of KLRG1+ NK-cell subsets to produce cytokines, we treated cells isolated from BM and spleen for 18 hours with IL-2 (500U) and IL-12 (100 ng/mL), and analyzed the percentage of IFN-γ+ cells on KLRG1+/CX3CR1-GFP+ and KLRG1+/CX3CR1-GFP− NK-cell populations by intracellular immunofluorescence staining and FACS analysis. The analysis of the KLRG1+ populations evidenced an impaired ability of CX3CR1-GFP+ NK cells to produce IFN-γ both in BM and spleen, compared with the CX3CR1-GFP− counterpart (Figure 4A).
CX3CR1 expression defines 2 functionally distinct KLRG1+ NK-cell subsets. (A) Freshly isolated cells from BM and spleen of CX3CR1+/GFP mice were incubated for 18 hours, at 37°C, 5% CO2 in the presence of IL-2 (500U) and IL-12 (100 ng/mL). Brefeldin A (10 μg/mL) was added during the last 6 hours. At the end of treatment, cells were stained with anti-NK1.1–, anti-KLRG1–, and anti-CD3ϵ–specific mAbs. Permeabilized cells were stained with anti–IFN-γ–specific mAb, and analyzed by FACS. Dot plots show a representative experiment of 3 performed. Numbers in dot plots indicate the percentage of IFN-γ–producing NK cells relative to CX3CR1-GFP expression on KLRG1+ NK-cell population. Histograms show the mean values ± SD of the percentage of IFN-γ+ NK cells in the 2 different KLRG1+ populations from BM and spleen of a total of 8 animals analyzed in independent experiments. *P < .05. □ indicates KLRG1+CX3CR1-GFP−;  indicates KLRG1+/CX3CR1-GFP+. (B) Sorted KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+; NK-cell subsets were treated as described in panel A and intracellular IFN-γ expression was analyzed by FACS. Numbers in the dot plots indicate the mean percentage ± SD of IFN-γ+ cells within each NK-cell subset from the spleen of a total of 5 animals analyzed in independent expriments. (C) Sorted KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ splenic NK-cell subsets were cultured 24 hours in the presence of IL-15 (100 ng/mL), and used as effector cells in a 4 hour 51Cr release assay against Yac-1 target cells at the indicated effector:target ratios. Data shown are representative of 1 of at least 3 experiments performed.
indicates KLRG1+/CX3CR1-GFP+. (B) Sorted KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+; NK-cell subsets were treated as described in panel A and intracellular IFN-γ expression was analyzed by FACS. Numbers in the dot plots indicate the mean percentage ± SD of IFN-γ+ cells within each NK-cell subset from the spleen of a total of 5 animals analyzed in independent expriments. (C) Sorted KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ splenic NK-cell subsets were cultured 24 hours in the presence of IL-15 (100 ng/mL), and used as effector cells in a 4 hour 51Cr release assay against Yac-1 target cells at the indicated effector:target ratios. Data shown are representative of 1 of at least 3 experiments performed.
CX3CR1 expression defines 2 functionally distinct KLRG1+ NK-cell subsets. (A) Freshly isolated cells from BM and spleen of CX3CR1+/GFP mice were incubated for 18 hours, at 37°C, 5% CO2 in the presence of IL-2 (500U) and IL-12 (100 ng/mL). Brefeldin A (10 μg/mL) was added during the last 6 hours. At the end of treatment, cells were stained with anti-NK1.1–, anti-KLRG1–, and anti-CD3ϵ–specific mAbs. Permeabilized cells were stained with anti–IFN-γ–specific mAb, and analyzed by FACS. Dot plots show a representative experiment of 3 performed. Numbers in dot plots indicate the percentage of IFN-γ–producing NK cells relative to CX3CR1-GFP expression on KLRG1+ NK-cell population. Histograms show the mean values ± SD of the percentage of IFN-γ+ NK cells in the 2 different KLRG1+ populations from BM and spleen of a total of 8 animals analyzed in independent experiments. *P < .05. □ indicates KLRG1+CX3CR1-GFP−;  indicates KLRG1+/CX3CR1-GFP+. (B) Sorted KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+; NK-cell subsets were treated as described in panel A and intracellular IFN-γ expression was analyzed by FACS. Numbers in the dot plots indicate the mean percentage ± SD of IFN-γ+ cells within each NK-cell subset from the spleen of a total of 5 animals analyzed in independent expriments. (C) Sorted KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ splenic NK-cell subsets were cultured 24 hours in the presence of IL-15 (100 ng/mL), and used as effector cells in a 4 hour 51Cr release assay against Yac-1 target cells at the indicated effector:target ratios. Data shown are representative of 1 of at least 3 experiments performed.
indicates KLRG1+/CX3CR1-GFP+. (B) Sorted KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+; NK-cell subsets were treated as described in panel A and intracellular IFN-γ expression was analyzed by FACS. Numbers in the dot plots indicate the mean percentage ± SD of IFN-γ+ cells within each NK-cell subset from the spleen of a total of 5 animals analyzed in independent expriments. (C) Sorted KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ splenic NK-cell subsets were cultured 24 hours in the presence of IL-15 (100 ng/mL), and used as effector cells in a 4 hour 51Cr release assay against Yac-1 target cells at the indicated effector:target ratios. Data shown are representative of 1 of at least 3 experiments performed.
To further support the functional differences observed, purified (FACS-sorted) splenic KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ NK cells were treated with IL-2/IL-12 or with IL-15 (100 ng/mL) to evaluate IFN-γ expression or the capability to lyse YAC-1 target cells, respectively (Figure 4B). The results obtained indicate that among KLRG1+ NK cells, CX3CR1-GFP+ cells are poor IFN-γ producers and display decreased lytic capacity compared with the CX3CR1-GFP− population (Figure 4C).
Surface phenotype of CX3CR1-GFP− and CX3CR1-GFP+ NK-cell subsets
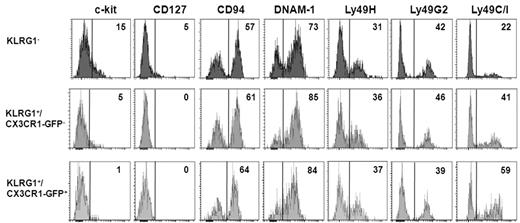
Because during maturation, NK cells down-regulate CD127, c-kit, and CD27 and acquire activating and inhibitory Ly49 MHC class I receptors,7,9 we examined more in depth the surface receptor phenotype of KLRG1− and KLRG1+ (CX3CR1− and CX3CR1+) spleen NK-cell subsets (Figure 5). KLRG1− NK cells specifically expressed IL-7Rα (CD127) and preferentially expressed c-kit. Interestingly, low levels of c-kit were detected on KLRG1+/CX3CR1-GFP− but not on KLRG1+/CX3CR1-GFP+ cells. The proportion of cells expressing Ly-49C/I was lower in KLRG1− NK cells (Figure 5), and it was higher on the CX3CR1-GFP+ cells within KLRG1+ NK cells. Overall, these results suggest that the KLRG1+/CX3CR1-GFP+ subset represents a later stage of NK-cell development.
Characterization of surface receptor expression on KLRG1− and KLRG1+ mouse NK-cell subsets. Spleen cells were isolated from CX3CR1+/GFP mice and stained with anti-NK1.1, anti-CD3, and anti-KLRG1 together with the indicated surface marker. Histograms show expression levels of the indicated molecules on KLRG1−, KLRG1+/CX3CR1-GFP− and KLRG1+CX3CR1+ cells within NK1.1+CD3− NK-cell gate. Control for m157-Fc (Ly49H) staining was NKG2D-Fc followed by PE-conjugated anti–human IgG, while anti-Ly49C/I and anti-Ly49G2 staining controls were PE-conjugated anti–mouse IgG2a/b and anti–rat IgG, respectively (data not shown). Numbers in the histogram plots indicate the percentage of positive cells of a representative analysis of 3 experiments performed.
Characterization of surface receptor expression on KLRG1− and KLRG1+ mouse NK-cell subsets. Spleen cells were isolated from CX3CR1+/GFP mice and stained with anti-NK1.1, anti-CD3, and anti-KLRG1 together with the indicated surface marker. Histograms show expression levels of the indicated molecules on KLRG1−, KLRG1+/CX3CR1-GFP− and KLRG1+CX3CR1+ cells within NK1.1+CD3− NK-cell gate. Control for m157-Fc (Ly49H) staining was NKG2D-Fc followed by PE-conjugated anti–human IgG, while anti-Ly49C/I and anti-Ly49G2 staining controls were PE-conjugated anti–mouse IgG2a/b and anti–rat IgG, respectively (data not shown). Numbers in the histogram plots indicate the percentage of positive cells of a representative analysis of 3 experiments performed.
CX3CR1 expression is acquired by KLRG1+ NK cells during homeostatic conditions
Our data clearly show that CX3CR1 expression is acquired during NK-cell development and defines a KLRG1+ NK-cell subset with distinctive characteristics.
To elucidate the relationships in terms of differentiation occurring between KLRG1+/CX3CR1-GFP+ and KLRG1+/CX3CR1-GFP−, we adoptively transferred these 2 populations isolated from the spleen of heterozygous CX3CR1+/GFP mice (CD45.2) after FACS sorting, by intravenous tail injection into wt C57BL/6 mice carrying the allelic variant CD45.1. Cells from spleen (Figure 6), and BM and liver (data not shown) were harvested after 12 days and stained with specific anti-CD45.1, -CD45.2, -CD3, -NK1.1, and -KLRG1 mAbs. Donor NK cells were identified as CD45.1−/CD45.2+/CD3−/NK1.1+ cells and analyzed for their expression of KLRG1 and GFP. As previously described, KLRG1 expression was maintained by donor NK cells.12 Interestingly, transferred KLRG1+/CX3CR1-GFP− cells acquired CX3CR1-GFP expression, while no changes in CX3CR1-GFP expression were observed on transferred CX3CR1-GFP+ NK cells. These findings indicate that in homeostatic conditions the KLRG1+/CX3CR1-GFP+ NK-cell subset originates from the CX3CR1-GFP− counterpart, and that CX3CR1 is stably expressed by KLRG1+ NK cells once acquired.
KLRG1+/CX3CR1-GFP+ NK cells derive from the KLRG1+/CX3CR1-GFP− counterpart. Sorted KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ NK-cell populations (purity > 95%) from the spleen of heterozygous CX3CR1+/GFP mice (CD45.2) were adoptively transferred into wt C57BL/6 mice (CD45.1). After 12 days, donor NK cells from BM, spleen, and liver were identified as CD45.1−/CD45.2+/CD3−/NK1.1+ cells and analyzed for their expression of KLRG1 and GFP. Numbers in plots indicate the mean percentage ± SD of CX3CR1-GFP+ donor NK cells in the recipient spleen. Data are representative of 1 of at least 3 experiments performed.
KLRG1+/CX3CR1-GFP+ NK cells derive from the KLRG1+/CX3CR1-GFP− counterpart. Sorted KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ NK-cell populations (purity > 95%) from the spleen of heterozygous CX3CR1+/GFP mice (CD45.2) were adoptively transferred into wt C57BL/6 mice (CD45.1). After 12 days, donor NK cells from BM, spleen, and liver were identified as CD45.1−/CD45.2+/CD3−/NK1.1+ cells and analyzed for their expression of KLRG1 and GFP. Numbers in plots indicate the mean percentage ± SD of CX3CR1-GFP+ donor NK cells in the recipient spleen. Data are representative of 1 of at least 3 experiments performed.
Discussion
In this study we show that CX3CR1 expression is developmentally regulated on NK cells, and that only the mature KLRG1+ NK-cell subset acquires high expression levels of the receptor. We also present in vivo evidence shedding light on the positioning of CX3CR1+ NK cells in the BM, as well as on the molecular mechanisms regulating BM localization of developing NK cells. Finally, we demonstrated that KLRG1+ NK cells are heterogeneous as CX3CR1 expression marks a population with impaired effector functions that likely derives from KLRG1+/CX3CR1− cells.
We evidenced that the reduced BM frequency of CX3CR1-GFP+ NK cells inside the KLRG1+ subset is the result of their lower BM homing and retention, and well fits with their poor CXCR4 expression levels and function.
Hematopoietic cells can be maintained into BM sinusoids to be ready to enter into circulation or inside the parenchyma.30,32 Although previous observations evidenced that immature B cells are enriched in BM sinusoids, our evidence show that NK cells that are maintained in this compartment mainly display a mature phenotype. Remarkably, we found that the majority of KLRG1+/CX3CR1-GFP+ BM NK cells reside in sinusoids, while KLRG1+/CX3CR1-GFP− and KLRG1− BM NK cells are mainly located in the parenchyma.
We also showed that developing NK cells can exploit CXCR4 down-modulation to move from the BM parenchyma to sinusoids and enter in blood circulation. This model is supported by the evidence that blockade of CXCR4 in vivo, by means AMD-3100 administration, induces NK-cell mobilization from parenchyma to sinusoids and blood. After AMD-3100 treatment, the different BM NK-cell populations show distinct behaviors that correlate with their positioning and CXCR4 expression levels. Indeed, KLRG1− NK cells are displaced from parenchyma and move to the sinusoids and to the blood. Moreover, although approximately 70% of KLRG1+/CX3CR1-GFP− cells are localized in parenchyma, this population is poorly affected by CXCR4 blockade. Finally, KLRG1+/CX3CR1-GFP+ NK cells that preferentially localize in sinusoids are not affected by CXCR4 inhibition. Indeed, our data demonstrate that CXCR4 is not directly involved in NK-cell retention on BM sinusoids, despite its role in the regulation of hematopoietic cell adhesion properties,34 and the expression of CXCL12 by BM endothelial cells.31
Our results suggest that other molecular mediators are involved in the route of KLRG1+ NK cells from parenchyma to sinusoids. Among these, a positive regulation of S1P has been suggested, as S1P deficient mice display reduced sinusoidal NK-cell numbers and S1P5 mRNA is up-regulated on the more mature NK-cell subsets.20,22 Collectively, these findings suggest that down-modulation of CXCR4 and the activity of S1P is necessary for mature NK-cell population to leave the parenchyma.
It has been demonstrated that the endothelium of BM sinusoids is endowed with a constitutive expression of α4 integrin ligands, such as VCAM-1, osteopontin and fibronectin.35-37 Our results evidenced that once NK cells reach the sinusoids, their retention is mainly regulated by α4 integrin. Indeed, α4 neutralization in vivo leads to a strong decrease of sinusoidal NK cells, regardless of their maturation stage. This blockade has a dramatic effect on the maintenance of KLRG1+/CX3CR1-GFP+ NK cells inside the BM, because these cells are mainly localized on the sinusoids. Whether the different behavior of parenchymal versus sinusoidal NK cells can be attributable to a different usage of α4 integrin is presently unclear. Indeed, we detected similar levels of α4 expression on NK cells from both compartments, and its ligands are functionally expressed also inside the parenchyma.37 On the other hand, the possible contribution of CXCR4 and/or other adhesion molecules in the maintenance of NK cells in the parenchyma cannot be ruled out. In this regard an interesting candidate is the receptor for hyaluronic acid and osteopontin, CD44, a BM homing receptor for CD34+ stem/progenitor cells. Indeed we found a strong CD44 down-modulation on sinusoidal NK cells compared with the parenchymal counterpart (supplemental Figure 5).38 In addition, although we showed that BM NK cells exit into blood through sinusoids, we still cannot exclude that a fraction of NK cells residing in the BM sinusoids derives from blood circulation.
KLRG1+ NK cells were proposed to be a stage of terminal differentiation with reduced ability to produce IFN-γ after MCMV infection or IL-12/IL18 stimulation.15,18 Our data indicate that KLRG1+ cells expressing high levels of CX3CR1 are a subset at later stage of differentiation that arises from KLRG1+/CX3CR1-GFP− cells in naive mice. Indeed, transferred KLRG1+/CX3CR1− NK cells become CX3CR1+, but not vice versa. In addition, by comparing the effector functions of the 2 KLRG1+ NK-cell populations, we observed that CX3CR1+ NK cells have a strong impairment in the ability to produce IFN-γ and a reduced killing capacity compared with CX3CR1−. CX3CR1-GFP+ cells display a lower percentage of IFN-γ–producing cells even in the BM were KLRG1+ NK cells are able to produce much higher levels of IFN-γ following cytokine or PMA plus ionomycin stimulation, compared with their splenic counterpart (Figure 4A and data not shown).
The role played by CX3CR1 in the differentiation of this subset is still unclear. Nonetheless, our preliminary studies evidenced a significant expansion of the NK-cell compartment in the BM of CX3CR1 deficient (CX3CR1GFP/GFP) mice with respect to CX3CR1+/GFP mice that was attributable to a 2 fold increase of BM GFP+ NK cells. This expansion was observed also in the periphery, thus suggesting that CX3CR1 participates to the maintenance of NK-cell homeostasis.
Although our data indicate that KLRG1+/CX3CR1-GFP+ NK cells exhibit decreased effector activity, we would like to envisage that this NK-cell population might also accomplish unique functions during immune responses. In this regard, it has been observed that CX3CR1+ NK cells are critical for neuroprotection during encephalomyelitis,28 likely because brain-recruited NK cells can produce neurotrophic factors.39
Collectively, our work identifies a discrete subset of NK cells that represents a late stage of differentiation showing reduced effector function capacity. In addition, we defined a BM sinusoidal niche in which different NK-cell subsets reside during homeostasis, and the critical factors involved in their maintenance into this site. The characterization of the functional attributes of discrete NK-cell populations and the factors that govern their differential positioning within tissues will be instrumental to understand the plasticity of NK-cell responses during inflammation.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Cristina Limatola (“Sapienza” University of Rome) and Dr Andreas Ludwig (Aachen University, Aachen, Germany) for kindly providing CX3CR1+/GFP mice and the CX3CL1-Fc fusion protein, respectively; and Dr Angela Quinci and Dr Sara Vitale for helpful suggestions.
This work was supported by grants from the Italian Association for Cancer Research (AIRC Investigator Grant and 5 per Mille), MIUR-FIRB “Futuro In Ricerca,” “Sapienza” University, MIUR-L.297 FAR, and the Center of Excellence (BEMM).
Authorship
Contribution: G.S. designed and performed experiments, analyzed and discussed results, and wrote the paper; G.D.A., G.B., and A.P. performed experiments; S.M. contributed to flow cytometric analysis and cell sorting; and A.S. and G.B designed research, discussed results, and contributed to paper writing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Angela Santoni, Department of Molecular Medicine, “Sapienza” University of Rome, Viale Regina Elena 324, Rome, 00161, Italy; e-mail: angela.santoni@uniroma1.it; or Giovanni Bernardini, Department of Molecular Medicine, “Sapienza” University of Rome, Viale Regina Elena 324, Rome, 00161, Italy; e-mail: giovanni.bernardini@uniroma1.it.
References
Author notes
A.S. and G.B. contributed equally to this study.








 indicates KLRG1+/CX3CR1-GFP+. (B) Sorted KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+; NK-cell subsets were treated as described in panel A and intracellular IFN-γ expression was analyzed by FACS. Numbers in the dot plots indicate the mean percentage ± SD of IFN-γ+ cells within each NK-cell subset from the spleen of a total of 5 animals analyzed in independent expriments. (C) Sorted KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ splenic NK-cell subsets were cultured 24 hours in the presence of IL-15 (100 ng/mL), and used as effector cells in a 4 hour 51Cr release assay against Yac-1 target cells at the indicated effector:target ratios. Data shown are representative of 1 of at least 3 experiments performed.
indicates KLRG1+/CX3CR1-GFP+. (B) Sorted KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+; NK-cell subsets were treated as described in panel A and intracellular IFN-γ expression was analyzed by FACS. Numbers in the dot plots indicate the mean percentage ± SD of IFN-γ+ cells within each NK-cell subset from the spleen of a total of 5 animals analyzed in independent expriments. (C) Sorted KLRG1+/CX3CR1-GFP− and KLRG1+/CX3CR1-GFP+ splenic NK-cell subsets were cultured 24 hours in the presence of IL-15 (100 ng/mL), and used as effector cells in a 4 hour 51Cr release assay against Yac-1 target cells at the indicated effector:target ratios. Data shown are representative of 1 of at least 3 experiments performed.