Abstract
Fanconi anemia (FA) is a rare genetic disorder characterized by bone marrow failure, congenital abnormalities, and an increased risk for cancer and leukemia. Components of the FA-BRCA pathway are thought to function in the repair of DNA interstrand cross-links. Central to this pathway is the monoubiquitylation and chromatin localization of 2 FA proteins, FA complementation group D2 (FANCD2) and FANCI. In the present study, we show that RAD18 binds FANCD2 and is required for efficient monoubiquitylation and chromatin localization of both FANCD2 and FANCI. Human RAD18-knockout cells display increased sensitivity to mitomycin C and a delay in FANCD2 foci formation compared with their wild-type counterparts. In addition, RAD18-knockout cells display a unique lack of FANCD2 and FANCI localization to chromatin in exponentially growing cells. FANCD2 ubiquitylation is normal in cells containing a ubiquitylation-resistant form of proliferating cell nuclear antigen, and chromatin loading of FA core complex proteins appears normal in RAD18-knockout cells. Mutation of the RING domain of RAD18 ablates the interaction with and chromatin loading of FANCD2. These data suggest a key role for the E3 ligase activity of RAD18 in the recruitment of FANCD2 and FANCI to chromatin and the events leading to their ubiquitylation during S phase.
Introduction
Fanconi anemia (FA) is an autosomal or X-linked recessive disorder characterized by genomic instability, congenital abnormalities, and a predisposition to cancer and leukemia. To date, 15 genes have been identified that, when mutated, result in FA or an FA-like syndrome. On a cellular level, these mutations can be characterized by hypersensitivity to DNA cross-linking agents such as diepoxybutane or mitomycin C (MMC). Consequently, the proteins encoded by these genes are thought to function in a common pathway responsible for the repair of interstrand cross-links (ICLs).1-3 ICLs are complex lesions that covalently link double-stranded DNA, preventing replication and ultimately resulting in a double-strand break during repair. For this reason, several DNA repair factors are thought to function alongside FA proteins, including those involved in homologous recombination, nucleotide excision repair, and translesion synthesis (TLS).4
Eight of the 15 FA proteins (FA complementation group A [FANCA], FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, and FANCM) comprise what is known as the FA core complex, and a complete and functional core complex is required for the monoubiquitylation of FANCD2 and FANCI after DNA damage or during the S phase.5 FANCL, along with the E2 protein UBE2T, functions as the E3 ubiquitin ligase component of the core complex responsible for the monoubiquitylation of FANCD2 and FANCI.6-8 Monoubiquitylated FANCD2 and FANCI are readily loaded onto chromatin,9 where they colocalize in nuclear repair foci with FANCD1, FANCJ, and FANCN, as well as other DNA repair factors such as BRCA1 and RAD51 and the DNA replication processivity factor proliferating cell nuclear antigen (PCNA).10-13
RAD18 is an E3 ubiquitin ligase best known for its role in the monoubiquitylation of PCNA in response to stalled replication forks.14,15 Monoubiquitylation of PCNA on lysine-164 by RAD18 and its partner E2 enzyme, RAD6, triggers a mechanism known as polymerase switching.16 The sliding clamp, PCNA, normally carries a replicative polymerase such as polδ along the DNA strand during replication. When PCNA encounters a lesion caused by various DNA-damaging agents, the replication fork stalls. PCNA is then monoubiquitylated and the replicative polymerase is replaced by a TLS polymerase such as polη, which allows for bypass of the lesion because of its larger active site.17
In addition to its role in the polymerase switch mechanism, RAD18 has been reported to perform the monoubiquitylation reaction for other DNA repair factors such as 53BP118 and has been shown to physically interact with the DNA repair proteins WRNIP119 and RAD51C.20 Recent reports have also suggested a role for RAD18 in the coordination of homologous recombination repair in a manner that is independent of its ubiquitylation activity and solely dependent on its recruitment to sites of DNA damage.20 Given the results of recent studies indicating a role for RAD6 in the ubiquitylation of FANCD2,21 we sought to determine whether RAD18 plays a role in the FA pathway and repair of ICLs.
In this study, we describe the interaction between RAD18 and FANCD2. We show by immunoprecipitation that RAD18-FANCD2 binding occurs both in the presence and absence of DNA cross-linking damage and in a core complex–independent manner. RAD18 is required for efficient monoubiquitylation of FANCD2 and FANCI after treatment with various DNA cross-linking agents, and this effect is not dependent on the modification of PCNA. Depletion of RAD18 results in hypersensitivity to MMC and cisplatin and a significant delay in FANCD2 foci formation. FANCD2 and FANCI are not present on chromatin in untreated, exponentially growing RAD18–deficient cells, whereas core complex proteins appear normal in this regard. Chromatin loading of FANCD2 in untreated RAD18–deficient cells can be corrected by expression of wild-type, zinc-finger (ZNF) domain deletion mutant, and SAF-A/B, Acinus, and PIAS (SAP) domain deletion mutant forms of RAD18. However, mutation of the RING domain of RAD18 (C28F) fails to correct the chromatin loading of FANCD2. In addition, the RING domain point mutant fails to interact with FANCD2 in these cells. These data suggest a key role for the E3 ubiquitin ligase activity of RAD18 in the recruitment of FANCD2 and FANCI to chromatin and the events leading to their ubiquitylation during S phase.
Methods
Antibodies
RAD18 polyclonal antibody was obtained from Bethyl Laboratories. FANCD2 polyclonal and monoclonal antibodies were obtained from Abcam and Santa Cruz Biotechnology, respectively. PCNA, FANCL, hemagglutinin (HA), Ku86, Topo II, and α-tubulin antibodies were obtained from Novus Biologicals, Covance, Santa Cruz Biotechnology, and Calbiochem, respectively. Anti-FANCI antibody was made in rabbits immunized with full-length human FANCI (purified from insect cells with an N-terminal 6xHis tag; the 6xHis tag was removed via TEV cleavage before immunization; see Longerich et al6 for purification details). Serum from the terminal bleed was affinity purified using full-length human FANCI immobilized on a CnBr-Sepharose 4B column according to the manufacturer's instructions (GE Healthcare). Secondary antibodies (HRP-linked ECL anti–rabbit IgG, HRP-linked ECL anti–mouse IgG, and HRP-linked protein A) were obtained from Amersham Health.
Cell culture
HeLa, H1299, FA-A mutant cells GM6914 and GM6914+Flag−FANCA, HCT116, and HCT116 RAD18−/− cells (kindly provided by J. Chen, M. D. Anderson Cancer Center, Houston, TX) were cultured in DMEM (Invitrogen) containing 10% fetal bovine serum (Biowest) and 1% Pen-Strep (Invitrogen). MRC5+His−PCNA−K164R and MRC5+His−PCNA−WT cells (kindly provided by A. Lehmann, University of Sussex, United Kingdom) were cultured in DMEM containing 10% fetal bovine serum, 1% Pen-Strep, and 1.2 mg/mL of G418 (Sigma-Aldrich).
Adenoviral vectors for overexpression of HA-tagged RAD18 or green fluorescent protein as a control were made as described previously.22 H1299 or HeLa cells were infected at approximately 50% confluence with 5 × 109 PFU/mL adenovirus and incubated for 24 hours. Cells were then treated with 500nM MMC for 24 hours.
Immunoprecipitation
Cells were cultured on 15-cm plates and treated with 500nM MMC for 24 hours before harvest by scraping. Cells were lysed in nondenaturing lysis buffer (1% Triton X-100, 50mM Tris-Cl pH7.4, 300mM NaCl, 5mM EDTA, leupeptin 1 μg/mL, pepstatin 1 μg/mL, aprotinin 1 μg/mL, sodium pyrophosphate 2mM, and sodium orthovanadate 2mM) and sonicated for 10 seconds. Extracts were cleared by centrifugation at 17 000g for 10 minutes at 4°C, the supernatant removed, and equal amounts of protein used for each immunoprecipitation. 1 μg of FANCD2 antibody or normal rabbit IgG (Santa Cruz Biotechnology) was added to samples and incubated overnight at 4°C on a rotator. Protein A Sepharose beads (GE Healthcare) were washed in nondenaturing lysis buffer, added to each sample, and then left to rotate at 4°C for 1 hour. Beads were then washed 5 times in wash buffer (0.1% Triton X-100, 50mM Tris-Cl, pH 7.4, 300mM NaCl, and 5mM EDTA) and resuspended in SDS loading buffer.
Western blotting
Protein samples were suspended in SDS loading buffer, boiled for 5 minutes, and briefly centrifuged. Samples were subjected to SDS-PAGE and transferred to nitrocellulose (Trans-Blot; Bio-Rad). Membranes were blocked in 5% milk in PBS-T (PBS + 0.1% Tween 20) overnight at 4°C, and then incubated at room temperature in primary antibody diluted in PBS-T. After 3 5-minute washes in PBS-T, membranes were incubated for 1 hour at room temperature in secondary antibody at 1:5000 in PBS-T with 0.5% milk. After 5 5-minute washes in PBS-T, blots were developed by chemiluminescence (Supersignal West Pico Kit; Pierce).
Immunofluorescence staining
Cells were plated in 8-chamber slides, grown to 40% confluence, and then treated with 500nM MMC for the durations indicated. Slides were rinsed in PBS, fixed in 4% paraformaldehyde for 5 minutes at room temperature, rinsed in PBS again, and permeabilized in 0.3% Triton X-100 for 5 minutes at room temperature. After rinsing in PBS, slides were blocked overnight at 4°C in PBS with 0.1% NP40 and 10% normal goat serum (Amersham Biosciences). Primary antibodies FANCD2 Fl17 mAb and normal mouse IgG (Santa Cruz Biotechnology) diluted 1:200 in PBS with 0.1% NP40 and 0.5% BSA were added and slides were incubated for 1 hour at room temperature or overnight at 4°C. Slides were washed 3 times for 5 minutes in PBS-T at room temperature, and then Cy3-conjugated goat anti–mouse IgG (Jackson ImmunoResearch Laboratories) secondary antibody was diluted 1:5000 in PBS with 0.1% NP40 and 0.5% BSA and added to the slides for 1 hour at room temperature. After 3 5-minute washes in PBS-T at room temperature and two 3-minute washes in PBS at room temperature, slides were mounted using DAPI Vectashield Hard-Set (Vector Laboratories) and analyzed on a Nikon TE2000-E Eclipse inverted fluorescent microscope.
Cellular fractionation
The cellular fractionation protocol was adapted from Montes de Oca et al.9 Cells were grown in 10-cm dishes, treated with 500nM MMC for 24 hours, and collected by scraping. Soluble fractions were extracted by resuspending the pelleted cells in buffer A with 0.5% Triton X-100 (10mM piperazine diethanesulfonic acid, pH 7, 100mM NaCl, 3mM MgCl2, 1mM EGTA, 300mM sucrose, 0.5mM Na3VO4, 50mM NaPO3, 1 μg/mL of aprotinin, 1 μg/mL of leupeptin, 1 μg/mL of pepstatin A, and 1mM PMSF) and incubating at room temperature for 3 minutes. Nuclei were pelleted by centrifugation for 3 minutes at 300g and were then washed 3 times in buffer A, followed by incubation in buffer A with DNase 1 for 30 minutes at room temperature. The extract was centrifuged for 3 minutes at 300g, and the supernatant was set aside. The pellet was extracted again with nondenaturing lysis buffer, and the resulting supernatant was pooled with the buffer A with the DNase 1 supernatant to form the chromatin fraction.
siRNA transfection
All small interfering RNA (siRNA) duplex pools were obtained from Dharmacon Research and transfected using X-tremeGENE siRNA transfection reagent (Roche) and Opti-MEM (Invitrogen) according to the manufacturers' protocols. A nontargeting siRNA for luciferase was used as a control. Cells were plated in 6-well plates and transfected at 50% confluence. Cells transfected with siRNA directed toward RAD18 were treated with MMC for the time points indicated and collected for Western blotting 48 hours after transfection. PCNA knockdown was performed using a single siRNA duplex with the sequence 5′-GCCGAGAUCUCAGCCAUAU-3′, and collected for Western blotting 72 hours after transfection.
RAD18 mutant constructs
RAD18 point and deletion mutant constructs were kindly provided by J. Chen (The University of Texas M. D. Anderson, Houston, TX). Cells were transfected using Lipofectamine 2000 and Opti-MEM according to the manufacturer's protocol (Invitrogen).
Cell survival assays
Cells were seeded into 6-well plates at 10 000 cells per well and treated with MMC or cisplatin 24 hours later. After incubation for 6 days, the medium was aspirated and wells were rinsed once with PBS. Cells were then fixed in 10% methanol/10% acetic acid for 5 minutes at room temperature on a rocker. The fixing solution was aspirated and crystal violet solution (1% in methanol) was then added to cells and rocked for 5 minutes at room temperature. Plates were rinsed in tap water to remove excess crystal violet and allowed to dry. Extraction of crystal violet dye was performed using extraction solution (methanol/SDS), allowing plates to rock for 2 hours. Absorbance was then measured at 595 nm. For colony survival assays using RAD18 mutants, cells were transfected as above, then replated in 6-well plates at 500 cells per well 24 hours after transfection. Twenty-four hours after plating, cells were treated with increasing concentrations of MMC for 1 hour, rinsed with PBS, and then fresh medium was added and cells were incubated for 10 days. Colonies were fixed in 10% methanol/10% acetic acid, stained with crystal violet, and counted.
Cell-cycle analysis
HCT116 cell lines were harvested, washed, and fixed in 70% ethanol overnight at 4°C. Cells were then incubated in RNAse A solution (10 μg/mL) for 5 minutes at room temperature and stained with propidium iodide (Sigma-Aldrich). Flow cytometric analysis was then performed by the Yale FACS Core Facility using a FACSCalibur flow cytometer (BD Biosciences).
Results
RAD18 binds to FANCD2 in vivo
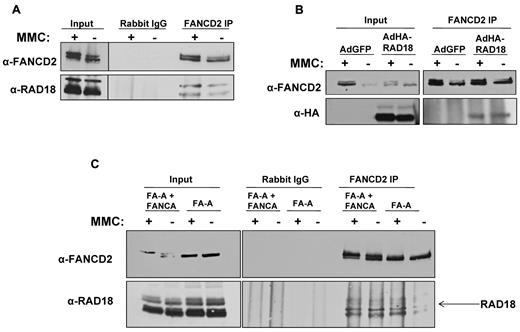
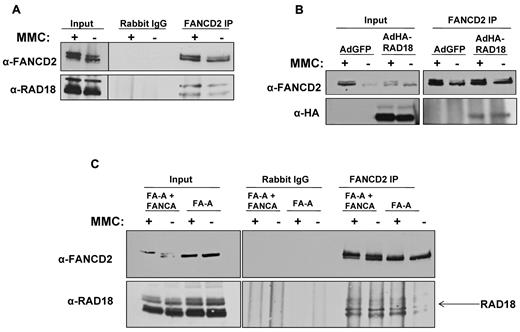
Previous reports have shown that RAD18 directly interacts with proteins involved in homologous recombination and other forms of DNA repair,18-20 suggesting that RAD18 may function as an important upstream signaling protein in several DNA repair pathways. Because it has also been reported that RAD6 regulates FANCD2 monoubiquitylation,21 we examined the potential interaction between RAD18 and FANCD2. FANCD2 was immunoprecipitated from MMC-treated or untreated H1299 whole-cell extracts, followed by immunoblotting for FANCD2 and RAD18. As shown in Figure 1A, endogenous RAD18 coprecipitated with FANCD2 in both MMC-treated and untreated extracts. To demonstrate the interaction of FANCD2 with exogenously expressed RAD18, we introduced HA-RAD18 using transduction of adenoviral vectors into H1299 cells treated or not with MMC. Immunoprecipitation of endogenous FANCD2 from whole-cell extracts, followed by immunoblotting using anti-FANCD2 or anti-HA antibody, revealed that HA-tagged RAD18 coprecipitated with endogenous FANCD2 in both treated and untreated cells (Figure 1B).
RAD18 binds to FANCD2 in vivo. (A) Immunoprecipitation of endogenous FANCD2 from H1299 whole-cell extract followed by immunoblot for endogenous RAD18 shows that RAD18 binds to FANCD2. This interaction is present in untreated cell extracts; however, treatment with MMC slightly increases RAD18 and FANCD2 interaction. (B) HA-tagged RAD18 was expressed in H1299 cells using adenoviral vectors. Immunoprecipitation of endogenous FANCD2 followed by immunoblot for HA showed that exogenously expressed RAD18 interacts with FANCD2. (C) Immunoprecipitation of endogenous FANCD2 from FANCA-deficient and corrected whole-cell extracts followed by immunoblot for endogenous RAD18 showed that RAD18 and FANCD2 interact in both FA-A cells and corrected cells. This interaction is therefore core complex–independent, and RAD18 likely interacts with nonubiquitylated FANCD2.
RAD18 binds to FANCD2 in vivo. (A) Immunoprecipitation of endogenous FANCD2 from H1299 whole-cell extract followed by immunoblot for endogenous RAD18 shows that RAD18 binds to FANCD2. This interaction is present in untreated cell extracts; however, treatment with MMC slightly increases RAD18 and FANCD2 interaction. (B) HA-tagged RAD18 was expressed in H1299 cells using adenoviral vectors. Immunoprecipitation of endogenous FANCD2 followed by immunoblot for HA showed that exogenously expressed RAD18 interacts with FANCD2. (C) Immunoprecipitation of endogenous FANCD2 from FANCA-deficient and corrected whole-cell extracts followed by immunoblot for endogenous RAD18 showed that RAD18 and FANCD2 interact in both FA-A cells and corrected cells. This interaction is therefore core complex–independent, and RAD18 likely interacts with nonubiquitylated FANCD2.
Because RAD18 and FANCD2 coprecipitate in both MMC-treated and untreated cell extracts, we next sought to determine whether RAD18 interacts with FANCD2-S or FANCD2-L. Using FANCA-deficient cells and their corrected counterparts, we again immunoprecipitated endogenous FANCD2 from treated and untreated whole-cell extracts, followed by immunoblotting for endogenous FANCD2 and RAD18. Figure 1C shows that RAD18 coprecipitated with FANCD2 in both MMC-treated and untreated FANCA-deficient and corrected cells. These data show that RAD18 interacts with FANCD2-S, because FANCD2 cannot be monoubiquitylated in FANCA-deficient cells. In addition, these data show that RAD18 interacts with FANCD2 in a core complex– and monoubiquitylation-independent manner. Immunoprecipitation of FANCD2 in FA-D2 cell lines containing no FANCD2, the ubiquitylation-resistant form of FANCD2 (K561R), or corrected with wild-type FANCD2 confirmed that RAD18 interacts with FANCD2-S (data not shown).
RAD18 regulates FANCD2 and FANCI monoubiquitylation
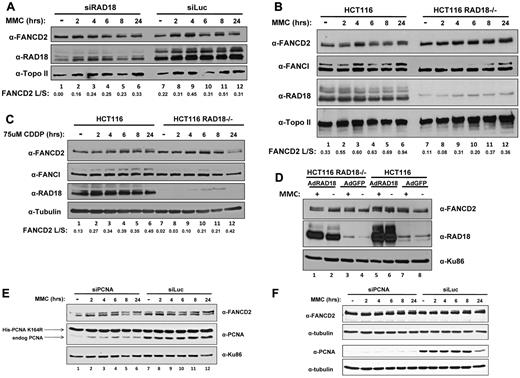
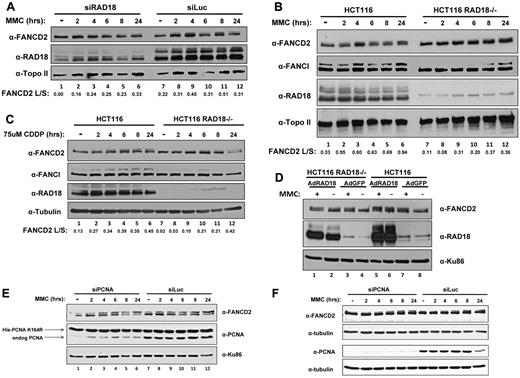
Because previous reports have shown a defect in the monoubiquitylation of FANCD2 after depletion of RAD6,21 we next sought to determine whether depletion of RAD18 would affect levels of monoubiquitylated FANCD2 after treatment with DNA cross-linking agents. Using H1299 cells, we depleted RAD18 by siRNA transfection and subsequently treated those cells with MMC at several time points over 24 hours. As shown in Figure 2A, FANCD2 monoubiquitylation was significantly delayed in cells depleted of RAD18 compared with the control siRNA transfection (luciferase), as determined by immunoblotting. Although cells transfected with nontargeting siRNA directed toward luciferase displayed a small amount of monoubiquitylated FANCD2 even when left untreated, RAD18-depleted cells showed no significant amount of ubiquitylated FANCD2 until approximately 6 hours after treatment (compare lanes 1-4 and 7-10 in Figure 2A). In addition, FANCI monoubiquitylation was also delayed until approximately 8 hours after treatment with MMC in RAD18-knockout cells.
RAD18 regulates FANCD2 and FANCI monoubiquitylation. (A) H1299 cells were depleted of RAD18 by siRNA transfection and then treated with 500mM MMC over several time points for 24 hours. Immunoblotting for FANCD2 shows a 6- to 8-hour delay in the monoubiquitylation of FANCD2 compared with control knockdown. (B) Rad18-deficient and wild-type cells were treated with 500mM MMC over several time points for 24 hours and assessed for FANCD2 and FANCI monoubiquitylation by Western blot. FANCD2 and FANCI monoubiquitylation is delayed by 6-8 hours in RAD18-deficient cells compared with wild-type cells. (C) RAD18-deficient and wild-type cells were treated with 75uM cisplatin over several time points for 24 hours. Immunoblotting reveals that FANCD2 and FANCI monoubiquitylation was again delayed by nearly 6-8 hours in RAD18-deficient cells. (D) HA-tagged RAD18 was expressed in Rad18-deficient and wild-type cells, which were then treated with 500mM MMC for 24 hours and assessed for FANCD2 and FANCI monoubiquitylation by Western blot. FANCD2 and FANCI monoubiquitylation was restored in untreated RAD18-deficient cells compared with the expression of green fluorescent protein as a control. Topo II, tubulin, and Ku86 western blots represent loading controls. The ratio of FANCD2-L:FANCD2-S was calculated from densitometry of each Western blot using ImageJ software Version 1.44. (E) Endogenous PCNA was depleted from MRC-5 cells expressing a ubiquitylation-resistant form of PCNA, followed by treatment of the cells with 500mM MMC over several time points for 24 hours. Immunoblotting shows that FANCD2 monoubiquitylation is unaffected by PCNA status, indicating that modification of PCNA does not regulate monoubiquitylation of FANCD2. (F) HeLa cells were depleted of PCNA by siRNA transfection and then treated with 500mM MMC over several time points for 24 hours. Immunoblotting for FANCD2 shows no delay in the monoubiquitylation of FANCD2 compared with control knockdown.
RAD18 regulates FANCD2 and FANCI monoubiquitylation. (A) H1299 cells were depleted of RAD18 by siRNA transfection and then treated with 500mM MMC over several time points for 24 hours. Immunoblotting for FANCD2 shows a 6- to 8-hour delay in the monoubiquitylation of FANCD2 compared with control knockdown. (B) Rad18-deficient and wild-type cells were treated with 500mM MMC over several time points for 24 hours and assessed for FANCD2 and FANCI monoubiquitylation by Western blot. FANCD2 and FANCI monoubiquitylation is delayed by 6-8 hours in RAD18-deficient cells compared with wild-type cells. (C) RAD18-deficient and wild-type cells were treated with 75uM cisplatin over several time points for 24 hours. Immunoblotting reveals that FANCD2 and FANCI monoubiquitylation was again delayed by nearly 6-8 hours in RAD18-deficient cells. (D) HA-tagged RAD18 was expressed in Rad18-deficient and wild-type cells, which were then treated with 500mM MMC for 24 hours and assessed for FANCD2 and FANCI monoubiquitylation by Western blot. FANCD2 and FANCI monoubiquitylation was restored in untreated RAD18-deficient cells compared with the expression of green fluorescent protein as a control. Topo II, tubulin, and Ku86 western blots represent loading controls. The ratio of FANCD2-L:FANCD2-S was calculated from densitometry of each Western blot using ImageJ software Version 1.44. (E) Endogenous PCNA was depleted from MRC-5 cells expressing a ubiquitylation-resistant form of PCNA, followed by treatment of the cells with 500mM MMC over several time points for 24 hours. Immunoblotting shows that FANCD2 monoubiquitylation is unaffected by PCNA status, indicating that modification of PCNA does not regulate monoubiquitylation of FANCD2. (F) HeLa cells were depleted of PCNA by siRNA transfection and then treated with 500mM MMC over several time points for 24 hours. Immunoblotting for FANCD2 shows no delay in the monoubiquitylation of FANCD2 compared with control knockdown.
To study this phenomenon further, we next turned to human RAD18-knockout cells to determine whether the residual FANCD2 monoubiquitylation seen in H1299 cells was due to incomplete depletion of RAD18 by siRNA transfection. Analysis of human RAD18-knockout cells and their wild-type counterparts revealed a similar delay in FANCD2 monoubiquitylation after MMC treatment (compare lanes 1-4 and 7-10 in Figure 2B). In addition, FANCI monoubiquitylation was delayed until approximately 8 hours after treatment with MMC. Similar results were obtained when these cells were treated with cisplatin over the same time course (compare lanes 1-4 and 7-10 in Figure 2C). The expression of RAD18 in HCT116 RAD18−/− cells using adenoviral vectors resulted in partial correction of the defect in FANCD2 monoubiquitylation in untreated cells (compare lanes 2 and 4 in Figure 2D).
Because RAD18 is known to monoubiquitylate PCNA, and it has recently been reported that PCNA interacts with FANCD2,12 we next sought to determine whether the effect of RAD18 on FANCD2 monoubiquitylation was an indirect result of modification of PCNA. Using MRC-5 cells that express a ubiquitylation-resistant form of PCNA harboring silent mutations that render it resistant to depletion by siRNA,23 we depleted endogenous PCNA by siRNA transfection. Treatment of these cells for 24 hours with MMC followed by immunoblotting revealed that FANCD2 monoubiquitylation was intact both in cells expressing mainly the mutant form of PCNA and in cells expressing endogenous PCNA (Figure 2E). Furthermore, HeLa cells were transfected with siRNA directed toward PCNA or a nontargeting siRNA directed toward luciferase. Treatment of these cells over several time points for 24 hours followed by immunoblotting showed that FANCD2 was monoubiquitylated even at early time points when PCNA was depleted (Figure 2F). These data indicate a significant role for RAD18 in the monoubiquitylation of FANCD2 and FANCI even in the absence of PCNA, and discount the necessity of ubiquitylated PCNA in the monoubiquitylation of FANCD2.
RAD18-deficient cells are hypersensitive to DNA cross-linking agents
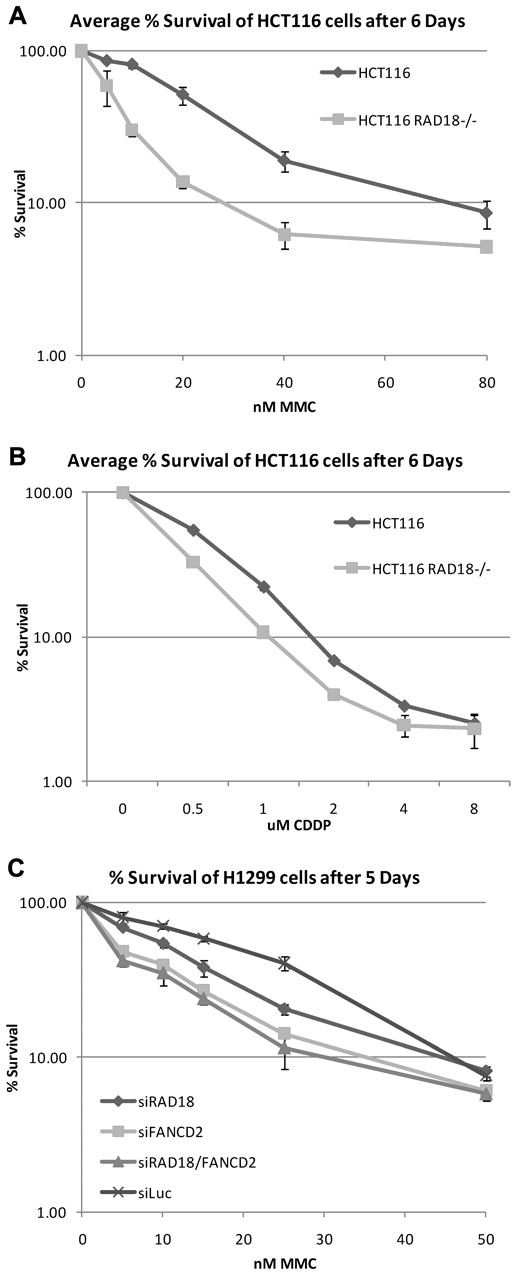
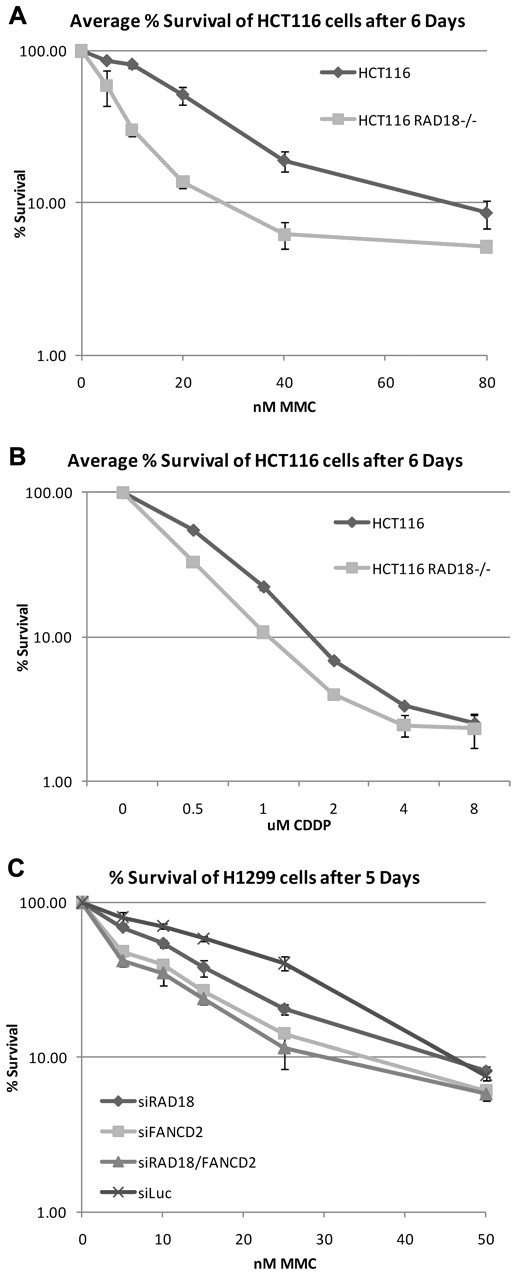
Because RAD18 is required for efficient monoubiquitylation of FANCD2 and FANCI, we next determined whether RAD18 was required for normal cell survival after treatment with DNA cross-linking agents. RAD18-knockout and wild-type cells were seeded into 6-well plates, treated with increasing concentrations of MMC and cisplatin, and left to grow for 6 days. As shown in Figure 3A-B, RAD18-deficient cells were hypersensitive to MMC and cisplatin. When RAD18 was partially depleted from H1299 cells by siRNA transfection, these cells were more sensitive to MMC treatment than cells depleted of luciferase as a control (Figure 3C). In addition, depletion of FANCD2 from these cells and the double knockdown of both RAD18 and FANCD2 resulted in comparable hypersensitivity to MMC. These data indicate that RAD18 is epistatic to FANCD2 and is likely involved in the same pathway in the repair of DNA ICLs.
RAD18-deficient cells are hypersensitive to DNA cross-linking agents. (A-B) RAD18-deficient and wild-type cells were plated into 6-well plates, treated with increasing amounts of MMC (A) or cisplatin (B), and allowed to grow for 6 days. Crystal violet survival assays show that RAD18-deficient cells are hypersensitive to both MMC and cisplatin. (C) RAD18, FANCD2, luciferase, or RAD18 and FANCD2 were depleted from H1299 cells by siRNA transfection and plated for cell survival assays. Single knockdown of RAD18 or FANCD2 results in hypersensitivity to MMC, whereas double knockdown of RAD18 and FANCD2 reveals an epistatic relationship.
RAD18-deficient cells are hypersensitive to DNA cross-linking agents. (A-B) RAD18-deficient and wild-type cells were plated into 6-well plates, treated with increasing amounts of MMC (A) or cisplatin (B), and allowed to grow for 6 days. Crystal violet survival assays show that RAD18-deficient cells are hypersensitive to both MMC and cisplatin. (C) RAD18, FANCD2, luciferase, or RAD18 and FANCD2 were depleted from H1299 cells by siRNA transfection and plated for cell survival assays. Single knockdown of RAD18 or FANCD2 results in hypersensitivity to MMC, whereas double knockdown of RAD18 and FANCD2 reveals an epistatic relationship.
RAD18 regulates chromatin loading of FANCD2 and FANCI
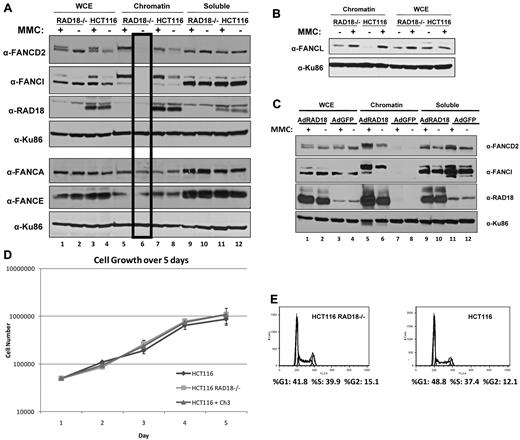
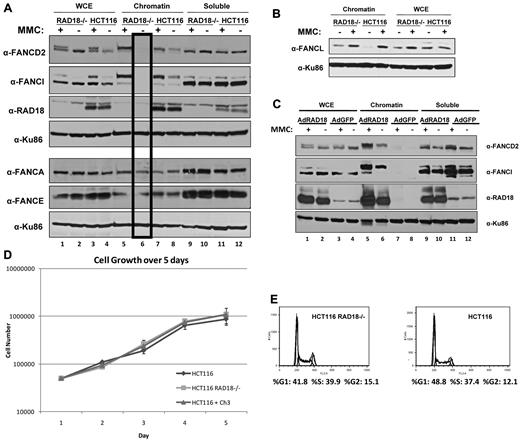
Because RAD18 plays a role in FANCD2 and FANCI monoubiquitylation, which in turn leads to chromatin loading, we next sought to examine the localization of FA proteins in RAD18-knockout cells in response to DNA damage. Cellular fractionation of RAD18-knockout cells and their RAD18 wild-type counterparts revealed that although FANCD2 and FANCI were successfully loaded onto chromatin after treatment with MMC for 24 hours in both cell types, no FANCD2 or FANCI was detectable in the chromatin fraction of untreated RAD18-knockout cells (Figure 4A lane 6). In contrast, untreated RAD18 wild-type cells showed detectable FANCD2 and FANCI in the chromatin fraction (lane 8) and an induction in the loading of these proteins after treatment with MMC (lane 7). However, core complex proteins such as FANCA and FANCE were found in the chromatin fraction of both cell types in similar amounts, indicating that RAD18 only affects chromatin loading of FANCD2 and FANCI. Recent reports have indicated a role for RAD18 in the recruitment of FANCL to chromatin.24 Contrary to these results, our data show that, similar to FANCA and FANCE, FANCL efficiently loads onto chromatin in RAD18-deficient cells (Figure 4B). The expression of RAD18 in HCT116 RAD18−/− cells using adenoviral vectors resulted in correction of the defect in FANCD2 and FANCI chromatin loading in untreated cells (Figure 4C lane 6).
RAD18 regulates chromatin loading of FANCD2 and FANCI. (A) RAD18-deficient and wild-type cells were treated with 500mM MMC for 24 hours and fractionated into chromatin and soluble fractions. Immunoblotting revealed that FANCD2 and FANCI are successfully loaded onto chromatin by 24 hours after treatment, but are undetectable in the chromatin fraction of untreated RAD18-deficient cells (lane 6). The same extracts run on a parallel gel were immunoblotted for core complex proteins FANCA and FANCE and show no difference in chromatin loading of these proteins in RAD18-deficient versus wild-type cells. (B) RAD18-deficient and wild-type cells were exposed to 500nM MMC for 24 hours and then fractionated into chromatin and soluble fractions. Western blotting shows no difference in chromatin loading of FANCL in RAD18-deficient versus wild-type cells or FANCL chromatin in RAD18-knockout cells. (C) HA-tagged RAD18 was expressed in HCT116 RAD18−/− cells using adenoviral vectors, treated with 500nM MMC for 24 hours, and then fractionated into chromatin and soluble fractions. Western blotting shows that FANCD2 and FANCI chromatin loading is restored in RAD18-deficient cells (lanes 5-6). (D) RAD18-deficient and wild-type cells were plated into 6 well plates and then trypsinized and counted every 24 hours for 5 days. Growth of each cell type was similar. (E) Cell-cycle analysis of RAD18-deficient and wild-type cells shows similar cell-cycle profiles.
RAD18 regulates chromatin loading of FANCD2 and FANCI. (A) RAD18-deficient and wild-type cells were treated with 500mM MMC for 24 hours and fractionated into chromatin and soluble fractions. Immunoblotting revealed that FANCD2 and FANCI are successfully loaded onto chromatin by 24 hours after treatment, but are undetectable in the chromatin fraction of untreated RAD18-deficient cells (lane 6). The same extracts run on a parallel gel were immunoblotted for core complex proteins FANCA and FANCE and show no difference in chromatin loading of these proteins in RAD18-deficient versus wild-type cells. (B) RAD18-deficient and wild-type cells were exposed to 500nM MMC for 24 hours and then fractionated into chromatin and soluble fractions. Western blotting shows no difference in chromatin loading of FANCL in RAD18-deficient versus wild-type cells or FANCL chromatin in RAD18-knockout cells. (C) HA-tagged RAD18 was expressed in HCT116 RAD18−/− cells using adenoviral vectors, treated with 500nM MMC for 24 hours, and then fractionated into chromatin and soluble fractions. Western blotting shows that FANCD2 and FANCI chromatin loading is restored in RAD18-deficient cells (lanes 5-6). (D) RAD18-deficient and wild-type cells were plated into 6 well plates and then trypsinized and counted every 24 hours for 5 days. Growth of each cell type was similar. (E) Cell-cycle analysis of RAD18-deficient and wild-type cells shows similar cell-cycle profiles.
Because FANCD2 and FANCI are known to be monoubiquitylated not only in response to DNA damage, but also on entry into S phase, we investigated the possibility that RAD18 is required for the monoubiquitylation and chromatin loading of these proteins during S phase. As shown in Figure 4D, RAD18-knockout cells grew at similar rates as their wild-type counterparts. In addition, cell-cycle analysis of both RAD18-deficient and wild-type cells demonstrated that these cells have similar cell-cycle profiles (Figure 4E). These data indicate that RAD18 is likely required for the monoubiquitylation and chromatin loading of FANCD2 and FANCI during S phase.
RAD18 regulates FANCD2 foci formation
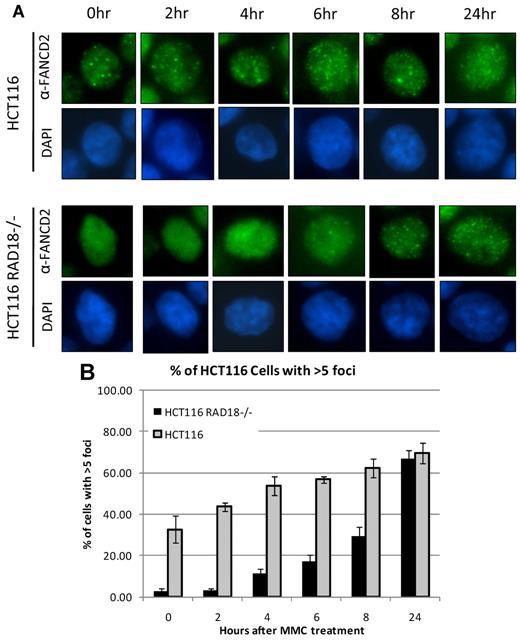
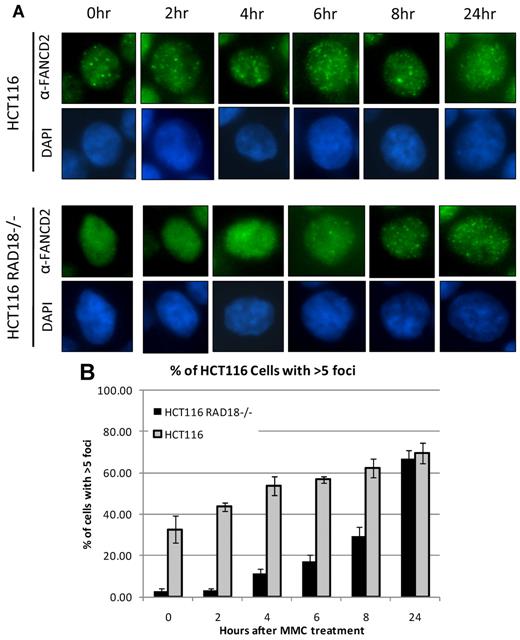
FANCD2 has been shown to form nuclear foci in response to treatment with various DNA-damaging agents, and these nuclear foci are generally considered to represent chromatin-bound FANCD2.10-13 Because RAD18 regulates chromatin loading of FANCD2 and FANCI, we next decided to examine FANCD2 foci formation in RAD18-deficient cells. RAD18-deficient and wild-type cells were seeded onto chamber slides and treated with MMC for the durations indicated. Slides were then examined by immunofluorescence microscopy after staining for FANCD2. As shown in Figure 5A, RAD18-knockout cells displayed diffuse nuclear staining in untreated cells and at early time points, indicating a delay in FANCD2 foci formation. This is in contrast to RAD18 wild-type cells, in which some FANCD2 foci were already visible in untreated cells and increased in quantity with longer exposures to MMC. Quantitation of cells positive for FANCD2 foci showed a clear delay in foci formation in RAD18-knockout cells versus wild-type counterparts (Figure 5B). Similar data were observed in H1299 cells in which RAD18 was depleted by siRNA transfection (data not shown).
RAD18 regulates FANCD2 foci formation. (A) RAD18-deficient and wild-type cells were plated into chamber slides and treated over several time points with 500mM MMC. Staining for FANCD2, followed by analysis by immunofluorescence microscopy, shows a delay in FANCD2 foci formation for nearly 8 hours in RAD18-deficient cells compared with wild-type cells. (B) Approximately 200 cells from each condition in panel A were assessed for the presence of FANCD2 foci. Graph represents the percentage of cells positive for FANCD2 foci, with cells containing more than 5 foci considered positive.
RAD18 regulates FANCD2 foci formation. (A) RAD18-deficient and wild-type cells were plated into chamber slides and treated over several time points with 500mM MMC. Staining for FANCD2, followed by analysis by immunofluorescence microscopy, shows a delay in FANCD2 foci formation for nearly 8 hours in RAD18-deficient cells compared with wild-type cells. (B) Approximately 200 cells from each condition in panel A were assessed for the presence of FANCD2 foci. Graph represents the percentage of cells positive for FANCD2 foci, with cells containing more than 5 foci considered positive.
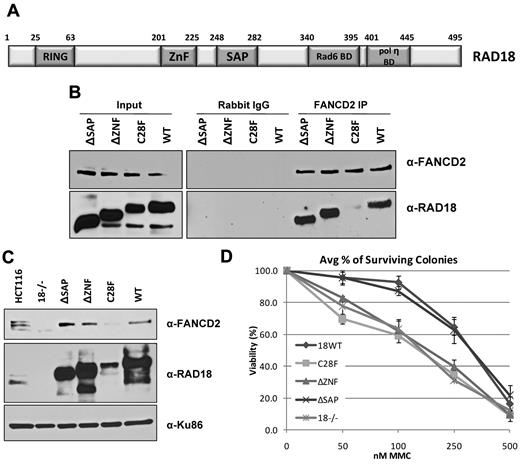
The E3 ligase activity of RAD18 is required for FANCD2 chromatin loading
RAD18 has known functions in several different DNA-repair pathways, including roles that are independent of its ubiquitylation activity.20 Therefore, we next sought to determine which regions within RAD18 were important for activation of the FA pathway during S phase. Several RAD18 mutants were expressed in human RAD18-knockout cells and then endogenous FANCD2 was immunoprecipitated from whole-cell extracts. As shown in Figure 6B, only the RING domain point mutant (C28F) did not bind to FANCD2, whereas the wild-type, ΔZNF, and ΔSAP domain deletion mutant forms of RAD18 all interacted with FANCD2 to similar extents. When these mutants were again expressed in human RAD18-knockout cells and subjected to cellular fractionation, FANCD2 was found in the chromatin fraction of untreated cells expressing the wild-type, ΔZNF, ΔSAP, and endogenous (HCT116 cells) forms of RAD18, but was absent in RAD18-deficient cells and those expressing the C28F mutant (Figure 6C). FANCD2 was loaded onto chromatin at similar levels when the ΔSAP mutant was expressed as when wild-type RAD18 was expressed. Because the SAP domain is required for the ubiquitylation of PCNA,20 this confirms that the modification of PCNA is not required for the ubiquitylation of FANCD2 during S phase. The ZNF domain of RAD18 is responsible for binding to ubiquitin25 and targeting RAD18 to sites of double-strand breaks.20 Cells expressing the ΔZNF mutant showed no defect in FANCD2 chromatin loading in untreated cells, but were hypersensitive to DNA cross-linking agents (Figure 6D), indicating a role for RAD18 in multiple steps during the repair of ICLs. As expected, RAD18-deficient cells and cells expressing the C28F mutant also displayed hypersensitivity to MMC, whereas expression of wild-type RAD18 resulted in resistance to MMC (Figure 6D). Surprisingly, cells expressing the ΔSAP mutant were also resistant to MMC, suggesting that RAD18-dependent monoubiquitylation of PCNA is not required for ICL repair. These results suggest a key role for the E3 ubiquitin ligase activity of RAD18 in the chromatin localization of FANCD2 during S phase that is independent of the ubiquitylation of PCNA, and a separate role for RAD18 downstream of FANCD2 chromatin loading in ICL repair.
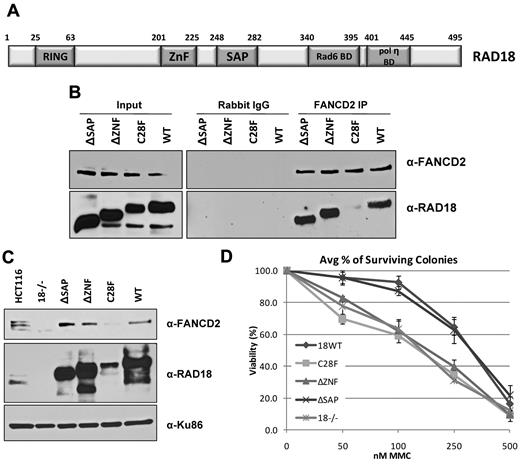
The E3 ligase activity of RAD18 is required for FANCD2 chromatin loading. (A) The domain structure of human RAD18 is shown. HCT116 RAD18−/− cells were transfected with plasmids expressing domain deletion, point mutation, and wild-type forms of RAD18. (B) Immunoprecipitation of endogenous FANCD2 from untreated cells expressing RAD18 mutants shows that FANCD2 binds all forms of RAD18 except the RING domain point mutant C28F. (C) Cellular fractionation of cells expressing RAD18 mutants shows that FANCD2 is loaded onto chromatin during S phase in all except RAD18-deficient cells and cells expressing the C28F mutant. (D) Colony survival assay of cells expressing RAD18 mutants shows that RAD18-deficient cells and cells expressing the C28F and ΔZNF mutants are hypersensitive to MMC. Cells expressing wild-type RAD18 and the ΔSAP mutant are resistant to MMC.
The E3 ligase activity of RAD18 is required for FANCD2 chromatin loading. (A) The domain structure of human RAD18 is shown. HCT116 RAD18−/− cells were transfected with plasmids expressing domain deletion, point mutation, and wild-type forms of RAD18. (B) Immunoprecipitation of endogenous FANCD2 from untreated cells expressing RAD18 mutants shows that FANCD2 binds all forms of RAD18 except the RING domain point mutant C28F. (C) Cellular fractionation of cells expressing RAD18 mutants shows that FANCD2 is loaded onto chromatin during S phase in all except RAD18-deficient cells and cells expressing the C28F mutant. (D) Colony survival assay of cells expressing RAD18 mutants shows that RAD18-deficient cells and cells expressing the C28F and ΔZNF mutants are hypersensitive to MMC. Cells expressing wild-type RAD18 and the ΔSAP mutant are resistant to MMC.
Discussion
Monoubiquitylation and the subsequent chromatin loading of FANCD2 and FANCI are considered hallmarks of FA pathway activation and crucial steps in the repair of ICLs. In this study, we describe a core-complex independent interaction between FANCD2 and RAD18, finding that this interaction occurs both in the presence and absence of MMC. FANCD2 is able to bind to several different RAD18 domain deletion mutants, but not the RING domain point mutant C28F, which is unable to execute ubiquitylation of substrates. In addition, we found that RAD18 is required for FANCD2 and FANCI monoubiquitylation and chromatin loading in untreated cells, and RAD18 depletion resulted in a delay in these phenomena after treatment with DNA cross-linking agents. Again, only the C28F mutant form of RAD18 lacked the ability to load FANCD2 onto chromatin during S phase. Our results indicate an S phase–specific role for the interaction between RAD18 and FANCD2.
Previous studies have generally shown various distinctions in posttranslational modifications of FA proteins during S phase compared with after DNA damage. For example, the phosphorylation of FANCG on serine 7,26 the phosphorylation of FANCD2 on several sites,27,28 and the monoubiquitylation of FANCD2 and FANCI have all been shown to occur during S phase29 and after DNA damage. However, recently published evidence from our laboratory indicates that activation of the FA pathway can be differentiated during these separate events. FANCA is phosphorylated on serine 1449 in an ATR-dependent manner after DNA damage. However, phosphorylated FANCA could not be detected either during S phase or in asynchronous cells, indicating that this is a DNA damage–specific event.30 These data suggest the possibility that early signaling events in the FA pathway differ in the context of DNA damage and S phase, and lend credence to our proposal that monoubiquitylation of FANCD2 and FANCI may be regulated by different upstream signaling pathways.
Additional evidence for the differential regulation of DNA repair proteins of interest during S phase has been provided by Shiomi et al,31 who demonstrated an S phase–specific function for RAD18 in DNA single-strand break repair. This function of RAD18 was independent of PCNA monoubiquitylation and polymerase switching, indicating distinct roles for RAD18 during S phase and after replication fork stalling. Our present data show that FANCD2 and FANCI are not monoubiquitylated or loaded onto chromatin in asynchronous cells lacking RAD18 (Figure 2A-C and Figure 4A), and that this deficiency is not the result of a defect in the monoubiquitylation of PCNA (Figure 2E). Because data have shown both differential modification of FA proteins and differential functions for RAD18, we therefore suggest that the RAD18-dependent modification of FANCD2 and FANCI and their subsequent chromatin localization is an S phase–specific event. This hypothesis can further be supported by the recent report that the E3 ubiquitin ligase complex CRL4Cdt2 monoubiquitylates PCNA in nondamaged proliferating cells.32 Similar to our results seen with FANCD2 and FANCI in RAD18-deficient cells, cells depleted of components of this complex display delayed kinetics of PCNA monoubiquitylation after DNA damage.
Song et al33 demonstrate that FANCD2 interacts with exogenously expressed RAD18 after treatment with the genotoxin benzo[a]pyrene dihydrodiol epoxide (BPDE). RAD18 also regulates ubiquitylation and chromatin loading of FANCD2 after treatment with BPDE; however, in contrast to our results, they suggested that the mechanism is TLS dependent and requires the ubiquitylation of PCNA. We have shown that PCNA modification is not required for the ubiquitylation and chromatin loading of FANCD2 during S phase in 3 ways. First, cells expressing a ubiquitylation-resistant form of PCNA show similar levels of FANCD2 ubiquitylation as cells expressing wild-type PCNA. Second, expression of the RAD18 SAP domain deletion mutant in RAD18-knockout cells supports chromatin loading of FANCD2 at levels similar to cells expressing wild-type and endogenous RAD18. Third, depletion of PCNA by siRNA in HeLa cells does not affect levels of monoubiquitylation of FANCD2. This discrepancy could be the result of the difference in genotoxic agents used in these studies, because BPDE and UV form bulky DNA adducts, whereas MMC and CDDP induce DNA ICLs. In addition, our study examines the effect on FANCD2 and FANCI ubiquitylation and chromatin loading over several time points, so this discrepancy may also be a matter of kinetics.
Geng et al24 recently reported that RAD18 regulates FANCD2 monoubiquitylation and FANCL chromatin loading in a manner dependent on PCNA monoubiquitylation. We show that, in contrast to these reports, several members of the FA core complex, including FANCL, are efficiently loaded onto chromatin in RAD18-deficient cells (Figure 4A-B). Our data therefore argue for a role of RAD18 in the regulation of monoubiquitylation and chromatin loading of FANCD2 and FANCI in a FANCL–independent manner.
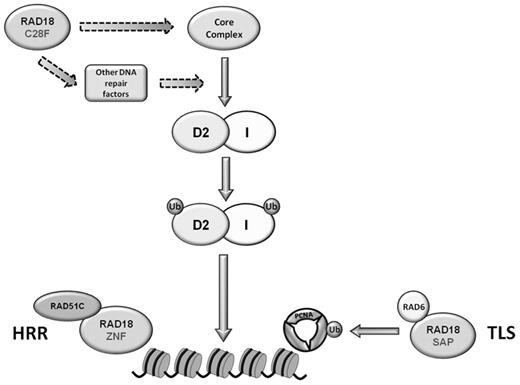
RAD18 is best known for its function as an E3 ubiquitin ligase, but it has been shown that FANCL performs the monoubiquitylation reaction for FANCD2 and FANCI. RAD18/RAD6 could not efficiently ubiquitylate FANCD2 and FANCI in vitro (Williams et al, unpublished data). However, this does not preclude the possibility that RAD18 is responsible for modifying members of the core complex or other proteins important in the FA pathway (Figure 7), or that these modifications may be necessary for the monoubiquitylation of FANCD2 and FANCI solely during S phase. Alternatively, RAD18 could exert its effect through USP1, the de-ubiquitylating enzyme for FANCD2 and FANCI.34
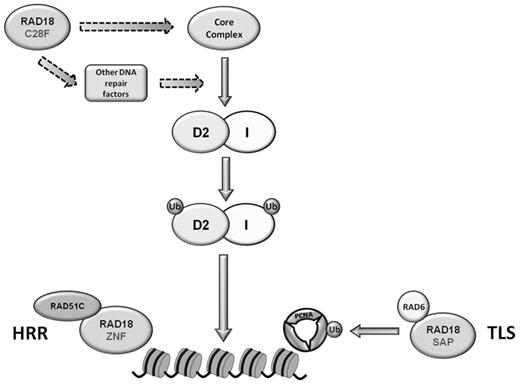
Model of possible roles for RAD18 during the S phase and after ICL damage. The E3 ubiquitin ligase activity of RAD18 is required for the ubiquitylation and chromatin loading of FANCD2 and FANCI. RAD18 may modify members of the core complex or other unidentified DNA repair proteins upstream of FANCD2 and FANCI monoubiquitylation and FA pathway activation. RAD18 is also required for ICL repair in steps downstream of FANCD2 and FANCI monoubiquitylation, such as the recruitment of RAD51C in homologous recombinatorial repair through its ZNF domain or possibly the monoubiquitylation of PCNA in TLS.
Model of possible roles for RAD18 during the S phase and after ICL damage. The E3 ubiquitin ligase activity of RAD18 is required for the ubiquitylation and chromatin loading of FANCD2 and FANCI. RAD18 may modify members of the core complex or other unidentified DNA repair proteins upstream of FANCD2 and FANCI monoubiquitylation and FA pathway activation. RAD18 is also required for ICL repair in steps downstream of FANCD2 and FANCI monoubiquitylation, such as the recruitment of RAD51C in homologous recombinatorial repair through its ZNF domain or possibly the monoubiquitylation of PCNA in TLS.
Other recent reports have shown that RAD18 can function in the coordination of early events involved in homologous recombination.20 Huang et al demonstrated that RAD18 is recruited to sites of DNA damage and then serves as an adaptor for recruitment of other DNA repair proteins. This function is independent of its role as an E3 ubiquitin ligase. Similarly, it is also possible that RAD18 is responsible for the recruitment of FANCD2 and FANCI to sites of DNA damage mediated solely through the binding of RAD18 and FANCD2. However, we demonstrate that the ubiquitylation activity of RAD18 is necessary for chromatin loading of FANCD2 in untreated cells, suggesting that RAD18 modifies proteins upstream of FANCD2 and FANCI monoubiquitylation.
Huang et al demonstrated that RAD18 functions as the adaptor for the recruitment of RAD51C. These data are particularly interesting in light of the fact that RAD51C was recently identified to be mutated in an FA-like disorder.35 Cells deficient in RAD51C display increased sensitivity and radial formation in response to treatment with ICL agents. Our data show that mutation of both the ZNF and C28F domains of RAD18 leads to hypersensitivity to MMC, suggesting a model in which RAD18 plays multiple roles in ICL repair. First, RAD18 plays a role in the regulation of FANCD2 and FANCI that is dependent on its E3 ubiquitin ligase activity. Second, because the ZNF domain of RAD18 is required for recruitment of RAD51C to chromatin, RAD18 performs a separate function in downstream ICL repair. Because deletion of the SAP domain results in resistance to MMC, it is unclear what role RAD18-mediated PCNA modification may play in ICL repair. Regardless, the data presented here identify a novel role for RAD18 in the FA pathway that is important for the monoubiquitylation and proper cellular localization of FANCD2 and FANCI during S phase.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank J. Chen for HCT116 RAD18−/− cells and RAD18 deletion/point mutants and A. Lehmann for MRC-5 PCNA K164R cells.
This work was supported by National Institutes of Health grant R01 HL063776 to G.M.K, ES015252 to P.S., and ES09558/12917 to C.V.
National Institutes of Health
Authorship
Contribution: S.A.W., P.S., C.V., and G.M.K. conceived and designed experiments; S.A.W. and S.L. performed research; S.L., P.S., and C.V. contributed vital new reagents; and S.A.W. and G.M.K. analyzed data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gary M. Kupfer, Department of Pediatrics, Division of Pediatric Hematology-Oncology, Yale University School of Medicine, 333 Cedar St, LMP 2073, New Haven, CT 06520; e-mail: Gary.Kupfer@yale.edu.