Abstract
The transcription factor (TF) RUNX1 cooperates with lineage-specifying TFs (eg, PU.1/SPI1) to activate myeloid differentiation genes, such as macrophage and granulocyte macrophage colony-stimulating factor receptors (MCSFR and GMCSFR). Disruption of cooperative gene activation could contribute to aberrant repression of differentiation genes and leukemogenesis initiated by mutations and translocations of RUNX1. To investigate the mechanisms underlying cooperative gene activation, the effects of Runx1 deficiency were examined in an in vitro model of Pu.1-driven macrophage differentiation and in primary cells. Runx1 deficiency decreased Pu.1-mediated activation of Mcsfr and Gmcsfr, accompanied by decreased histone acetylation at the Mcsfr and Gmcsfr promoters, and increased endogenous corepressor (Eto2, Sin3A, and Hdac2) coimmunoprecipitation with Pu.1. In cotransfection experiments, corepressors were excluded from a multiprotein complex containing full-length RUNX1 and PU.1. However, corepressors interacted with PU.1 if wild-type RUNX1 was replaced with truncated variants associated with leukemia. Histone deacetylase (HDAC) enzyme activity is a major component of corepressor function. HDAC inhibition using suberoylanilide hydroxamic acid or MS-275 significantly increased MCSFR and GMCSFR expression in leukemia cell lines that express PU.1 and mutated or translocated RUNX1. RUNX1 deficiency is associated with persistent corepressor interaction with PU.1. Thus, inhibiting HDAC can partly compensate for the functional consequences of RUNX1 deficiency.
Introduction
One of the most frequent genetic abnormalities in myelodysplastic syndrome and acute myeloid leukemia (AML) is mutation or translocation of RUNX1 (AML1, CBFA2).1,2 These genetic abnormalities decrease RUNX1 function by haploinsufficiency or by generating loss of function or dominant negative mutants.1,2 RUNX1 is necessary for definitive hematopoiesis: Runx1 knockout murine embryos have no detectable definitive erythrocytes or myeloid cells in their circulation or livers and die in utero at embryonic day 12.5 (E12.5).3 However, RUNX1 is insufficient for hematopoiesis; hematopoietic lineage specification and differentiation require and are driven by key lineage-specifying transcription factors (TFs), such as members of the ETS (including PU.1), CEBP, and GATA families. RUNX1 synergistically increases transcriptional activation by ETS1, PU.1(SPI1), CCAAT/enhancer binding protein-α (CEBPA), GATA1, GATA2, and FLI1.4-10 (RUNX factors alone are relatively weak activators of transcription.4,5,7,8,11 ). The mechanisms by which RUNX1 cooperates with these lineage-specifying TFs could be a key to understanding the altered hematopoietic differentiation and leukemia initiated by RUNX1 deficiency. A number of aspects of RUNX1 cooperation with lineage-specifying TFs are known. Response elements for RUNX1 and PU.1 and/or CEBPA are located in proximity in the promoters of key myeloid differentiation genes, such as those for macrophage colony-stimulating factor receptor (MCSFR) and granulocyte macrophage colony-stimulating factor receptor α-chain (GMCSFR).7,8,12 RUNX1 physically interacts with PU.1, CEBPA, GATA1, GATA2, PAX5, FLI1, and ETS1.4-10,13 RUNX1 can directly regulate expression of PU.1.14 Nonetheless, as shown by Western blot15 and in publicly available microarray gene expression data, PU.1 and CEBPA are expressed at high levels in leukemia cells containing mutated and translocated RUNX1. Despite the high PU.1 and CEBPA expression in these AML cells, the differentiation gene targets of PU.1 and CEBPA are aberrantly repressed, possibly because of impairment of the usual assistance from RUNX1.
Although the aforementioned evidence suggests that cooperation between RUNX1 and lineage-specifying TF is an important aspect of normal and leukemic hematopoiesis, the specific molecular mechanisms underlying such cooperation have only been well characterized for RUNX1 cooperation with ETS1.9,10 Both RUNX1 and ETS1 autoinhibit their own DNA binding.9,10 Mutual interaction of the autoinhibitory domains of RUNX1 and ETS1 mutually relieves the autoinhibition of DNA binding and leads to synergistic gene activation.9,10 However, the heterodimer β-subunit of RUNX1 and CBFB can also relieve autoinhibition of RUNX1 DNA binding,16 and autoinhibition of DNA binding is not known to be a general feature of lineage-specifying TFs. Therefore, other mechanisms for cooperation probably operate.
A key determinant of gene activation versus repression by DNA-binding factors is coactivator versus corepressor recruitment. There is limited understanding of the mechanisms that determine coactivator versus corepressor recruitment by RUNX1 and PU.1.17-20 In the present studies, the possibility that RUNX1 and PU.1 cooperate to regulate corepressor recruitment is addressed. PU.1 is a key lineage-specifying TF. There is a complete absence of macrophages, B cells, mature T cells, and a decrease in neutrophils and hematopoietic stem cells in Pu.1−/− mice.21 PU.1 can interact with coactivators or corepressors to function as either a transcriptional activator or a transcriptional repressor.17-19 In mice, a decrease in Pu.1-mediated differentiation gene activation causes leukemia.22 In human AML, however, PU.1 mutations are not frequent, although they have been reported.23-25 Therefore, in AML cells, which express mutated or translocated RUNX1 and wild-type PU.1, a possible alternative route to decreased activation of PU.1 target genes is disrupted RUNX1/PU.1 cooperation. Here, to understand the mechanics of RUNX1/PU.1 cooperation and the impact of leukemia-associated abnormalities on these mechanics, the effects of Runx1 deficiency were examined in a cell-line model of Pu.1-driven macrophage differentiation, in murine Runx1 haploinsufficient (Runx1+/−) primary cells, and in human AML cell lines containing translocated or mutated RUNX1. The generated observations are the first description, to our knowledge, of cooperation by 2 DNA binding TFs to exclude corepressors. This cooperation is disrupted by RUNX1 deficiency or by RUNX1 variants associated with leukemia. Histone deacetylase (HDAC) enzyme activity is a major aspect of corepressor function. These results provide a mechanistic rationale for using inhibitors of HDAC to compensate for the functional consequences of RUNX1 deficiency.
Methods
Generation of PUER cells with stable suppression of Runx1 expression
A lentiviral vector pLenti6-DEST (Invitrogen) was used to construct short hairpin (sh) RNA for Runx1. Three shRNA oligos specific to 19-bp target sequences of mouse Runx1 (shRUNX1-clone 1, 5′-GGCACTCTGGTCACCGTCA-3′; shRUNX1-clone 2, 5′-GGCCATGAAGAACCAGGTA-3′; and shRUNX1-clone 3, 5′GGCAAGAGCTTCACTCTGA-3′) were designed using Invitrogen's BLOCK-iT RNAi Designer and synthesized in sense and antisense orientation by integrated DNA technology. The single-strand oligos were then annealed to form double-strand oligos and subsequently ligated with pENTRY vector (Invitrogen) downstream of an RNA promoter. The ligated constructs were transformed into Escherichia coli TOPO10. Positive clones were verified by DNA sequencing. The verified clones were then recombined into pLenti6-DEST vector using Invitrogen's ViralPack kit, resulting in pLenti6-shRunx1. The pLenti6-shRunx1 or empty vector pLenti6 (to generate PUER control cells) was then transfected together with envelop encoding plasmid (VSVG) into 293FT packaging cell line to produce lentivirus. The supernatant-containing lentivirus was harvested at 48 hours after transfection. Titers were determined on NIH3T3 cells as transducing units using serial dilutions of vector stocks with 8 μg/mL polybrene (Sigma-Aldrich). PUER cells (gift of Dr Harinder Singh26 ) are murine hematopoietic precursor cells that have been retrovirally transduced to express PU.1 fused to the ER. PUER cells were grown in Iscove modified Eagle medium, without phenol-red, with 10% fetal bovine serum, 5 ng/mL murine interleukin-3, 1μg/mL puromycin, 55μM β-mercaptoethanol, 1% penicillin/streptomycin at 37°C in a humidified atmosphere with 5% CO2 in air. The lentivirus-containing supernatant was added to the cell culture at appropriate 4 particles/cell concentration with 8 μg/mL polybrene. Twenty-four hours after infection, 4 μg/mL of blasticidin was added to the cell culture for positive clone selection. The blasticidin-resistant cells were analyzed for Runx1 by quantitative RT-PCR and Western blot. Addition of 4-hydroxy-tamoxifen (OHT) to PUER triggers their terminal differentiation into macrophages.26 Differentiation status was analyzed by: (1) presence of adherent cells by light microscopy, (2) morphologic changes in Giemsa-stained cytospin preparations, (3) quantitative RT-PCR for stem cell and differentiation gene expression, and (4) flow-cytometry for c-Kit and F4/80 protein expression.
AML cell lines containing translocated and mutated RUNX1
Kasumi-1 cells were obtained from the DSMZ. CG-SH cells were characterized as previously described.27
Murine Runx1 haploinsufficient (+/−) cells
Runx1 haploinsufficient mice were a generous gift of the Jim Downing laboratory. Runx1+/− mice and wild-type littermate controls were genotyped as previously described.28 Whole bone marrow for immunoprecipitation, Western blot, and in vitro culture was harvested from the hind limb bones after death. Lineage-negative cells were isolated after depletion of lineage-positive cells using a lineage cell depletion kit (MACS, Miltenyi Biotec). All experiments and procedures were approved by the Cleveland Clinic Institutional Animal Care and Use Committees.
Quantitative RT-PCR
mRNA levels were assayed using quantitative RT-PCR. Briefly, total cellular RNA was isolated from 5 × 105 cells using RNeasy Plus (QIAGEN), according to the manufacturer's protocol. For cDNA synthesis, after a denaturation step of 5 minutes at 65°C, 1 μg of RNA was reverse-transcribed to single-stranded cDNA using a mix of random hexamers and oligo dT primers and Moloney murine leukemia virus reverse transcriptase for first-strand synthesis (Promega). Real-time PCR was performed with Real-time PCR Master Mix containing SYBR Green I and HotStart Taq DNA polymerase (Takara). Glyceraldehyde-3-phosphate dehydrogenase was amplified as internal control. Real-time detection of the emission intensity of SYBR Green bound to double-stranded DNA was detected using the 7500 Fast Real-time PCR System (Applied Biosystems). Data are reported as relative expression value, which was determined by raising 2 to the power of the negative value of δδCt for each sample. Primer sequences for murine genes were as follows: Bmi-1: forward 5′-AATTAGTCCCAGGGCTTTTCAA-3′, reverse 5′-TCTTCTCCTCATCTGCAACTTCTC-3′; HoxB4: forward 5′-CCAGAATCGGCGCATGA-3, reverse 5′-CCCGAGCGGATCTTGGT-3′; c-Kit forward 5′-GCCCACCCTGGTCATTACAGAA-3′, reverse 5′-CTTCCTTGATCATCTTGTAGAACT-3′; F4/80: forward 5′-AGATGGGGGATGACCACACTTC-3′, reverse 5′-TGTTCAGGGCAAACGTCTCG-3′; Gm-csfr: forward 5′-GCTCGCCCTGCTCTTCTCCA-3′, reverse 5′-CGACCGTGCCATTGACATCCA-3′; M-csfr: forward 5′-TCCCCCAGAGGTCAGTGTTAC-3′, reverse 5′-CTTCAGGGTGGGTGTCATTCC-3′; actin: forward 5′-CTGTCCCTGTATGCCTCTGGT-3′, reverse 5′-CATGAGGTAGTCTGTCAGGTC-3′. Primer sequence for human genes were as follows: hGM-CSFR: forward 5′-AAACAGCCCAGGACCTATCAG-3′, reverse 5′-GACAAGGGTTCCCACGATTAG-3′; hM-CSFR: forward 5′-CAATGGCAGCGTGGAATGG-3′, reverse 5′-GCAGTAGTGCGTCCTGGTCCTC-3′.
ChIP assays
Chromatin immunoprecipitation (ChIP) assays were performed with the acetyl-histone H3 immunoprecipitation Assay Kit (Millipore, catalog no. 17–245) according to the manufacturer's instructions. Briefly, 2 × 106 cells were crosslinked with 1% formaldehyde for 10 minutes at 37°C and subsequently harvested, washed with ice-cold PBS, then lysed with SDS lysis buffer with fresh protease inhibitors (1mM phenylmethylsulfonyl fluoride [PMSF], 1 μg/mL aprotinin, and 1 μg/mL pepstatin A), followed by sonication for a total of 120 seconds at 20-second intervals using Fisher Scientific Sonic Dismembrator 550 equipped with microtip (setting 4; 30% input). Gel electrophoresis was used to confirm that fragments were less than 500 bp in size. The sonicated chromatin lysates were then used for immunoprecipitations with anti-acetyl-histone H3 antibody (Millipore), with normal IgG as control. Lysates were incubated with antibodies at 4°C overnight with rotation. Immunoprecipitates were then collected with salmon sperm DNA/Protein A agarose slurry beads (Millipore, catalog no. 16–157C). Histone/DNA crosslinks were reversed by incubating at 65°C for 4 hours, followed by incubation with 0.05 μg/mL protease K for 1 hour at 45°C. DNA was recovered by phenol/chloroform extraction and ethanol precipitation, and purified using QIAGEN PCR purification kit (QIAGEN). ChIP DNA containing gene promoters associated with acetyl-histone H3 was analyzed by quantitative RT-PCR. Primers targeting the proximal promoters of M-csf receptor, G-csf receptor, and Gm-csf receptor were as follows: M-CSFR: forward 5′-GGAGTGATTTGTCTACAGG-3′, reverse 5′-ATCTGCCCTTAAGGCAGAAGG-3′ (M-csfr promoter from −260 to −105). Gm-csfr: forward 5′-GCGTATACTTACTGTGCGCAT-3′, reverse 5′-CTTCCTCTTCCTCATCTGCAG-3′ (Gm-csfr promoter from −216 to −60). G-csfr: forward 5′-TAAGACCCCTGAGGCAGGAA-3′, reverse 5′-CTAGCCCCGTCGTTAATGACA-3′ (G-csfr promoter from −222 to −37).
Cell fractionation and nuclear protein extraction
Approximately 100 million each PUER, PUER shRunx1, or 50 million each Runx1 haploinsufficient and wild-type littermate control bone marrow cells (pooled from multiple mice) were used in the preparation. After removal of the medium, cells were transferred to 15-mL conical tubes and washed twice with 10 mL ice-cold 1 times PBS. Cells were resuspended in 500 μL of 1 times hypotonic buffer containing 10mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 1.5mM MgCl2, 10mM KCl, 0.5mM dithiothreitol, 10mM PMSF, and protease inhibitor cocktail (Sigma-Aldrich, A8340), and incubated for 10 minutes on ice. A total of 20 μL of 10% NP-40 was added to cell suspensions to break the cell membrane. After another 10-minute incubation on ice, cell suspensions were centrifuged at 344g for 10 minutes. The supernatant was transferred to clean 1.5-mL Eppendorf tubes and labeled as the cytoplasm fraction. Nuclear pellets were washed twice with ice-cold 1 times PBS, and resuspended in 100 μL of 50mM Tris-HCl, pH 8.0, 1mM MgCl2, 10mM PMSF, protease inhibitor cocktail (Sigma-Aldrich, A8340) and Benzonase (Sigma-Aldrich, D5915, 250 units). The nuclear suspensions were incubated on ice for 90 minutes with vigorous vortex every 5 minutes. At the end of incubation, 500 μL protein extraction buffer containing 1.5% NP-40, 500mM NaCl, 5mM dithiothreitol, 10mM PMSF, and 5 μL of protease inhibitor cocktail (Sigma-Aldrich, A8340) in 50mM phosphate buffer (pH 7.4) was added. After 30-minute incubation on ice with vortexing every 5 minutes, the mixture was centrifuged at 12396g for 10 minutes. The supernatant containing nuclear proteins was transferred to clean tubes, and protein concentration was determined by BCA assay.
Covalent bound antibody to protein G beads
Goat anti-PU.1 (SCBT, sc-5949) and control goat IgG were covalently coupled to Sepharose-protein G beads using dimethylpimelimidate. Briefly, 25 mg of protein G-Sepharose CL-4B (GE Healthcare, 17-0780-01) was swelled in 1 mL 1 times PBS overnight and incubated with 200 μL of antibody (50 μg) solution (1 times PBS) for 1 hour at room temperature. Antibody bound protein G beads were then incubated with 1% chicken egg ovalbumin in PBS for another hour to block nonspecific binding sites. After 3 washes with 1 times PBS, 25 mg of dimethylpimelimidate in 1 mL of 200mM triethanyl amine was added, and coupling reaction was proceeded at room temperature for 30 minutes. The reaction was repeated 2 more times with fresh addition of dimethylpimelimidate and quenched with 50mM ethanol amine. The reacted protein G beads were washed extensively with 1 times PBS before immunoprecipitation.
Immunoprecipitation
Nuclear protein extracts (∼ 1 mg protein) were transferred to tubes with antibody bound protein G beads and rocked gently at 4°C overnight. Nonspecific bound proteins were removed with 5 washes of 1 times PBS containing 1% NP-40. Immunoprecipitation products were extracted from the protein G beads using Laemmli sample buffer.
1D SDS-polyacrylamide gel electrophoresis and Western blot analysis
Immunoprecipitation products, control immunprecipitation products, 25 μg of nuclear protein extracts from PUER and PUER shRunx1, together with molecular weight markers, were subjected to 1D SDS-polyacrylamide gel electrophoresis electrophoresis on precast 4% to 20% NuPAGE gels (Invitrogen). After electrophoresis per the manufacturer's manual (Invitrogen), proteins were transferred to polyvinylidene difluoride membranes (Millipore) at 35 constant voltage for 1 hour using Invitrogen's XCell II Blot module. Primary antibodies included anti-PU.1 (Cell Signaling, 2266), anti-Runx1 (SCBT, sc-101146), anti-ETO2 (SCBT, sc-28694), anti-Sin3A (SCBT, sc-994), anti-HDAC2 (SCBT, sc-7899), and anti–β-actin peroxidase (Sigma-Aldrich, A3854). Secondary antibodies, anti–rabbit (GE Healthcare, NA934) and anti–mouse (GE Healthcare, NXA931), were used at 1:5000 and 1:10 000 dilutions, respectively.
Protein identification by LC-MS/MS
Anti-Pu.1 and isotype antibody immunoprecipitation products were subjected to SDS-polyacrylamide gel electrophoresis and stained with colloidal Coomassie Blue (Gel Code Blue, Pierce Chemical). Gel slices were excised from the top to the bottom of the lane; proteins were reduced with dithiothreitol (10mM), alkylated with iodoacetamide (55mM), and digested in situ with trypsin. Peptides were extracted from gel pieces 3 times using 60% acetonitrile and 0.1% formic acid/water. The dried tryptic peptide mixture was redissolved in 15 μL of 0.1% formic acid and 5% acetonitrile for mass spectrometric analysis. Tryptic peptide mixtures were analyzed by on-line LC-coupled tandem mass spectrometry (LC-MS/MS) on a QTOF2 mass spectrometer (Waters) using a Cap LC XE system (Waters), a 0.3 × 5-mm trapping column (C18 PepMap 100, LC Packings), a reverse phase separating column (75 μm × 5 cm, Vydac C18), and a flow rate of 250 nL/min. Gradient LC separation was achieved with aqueous formic acid/acetonitrile solvents. The QTOF2 mass spectrometer was operated in standard MS/MS switching mode with the 4 most intense ions in each survey scan subjected to MS/MS analysis. Instrument operation and data acquisition used MassLynx Version 4.1 software (Waters). Initial protein identifications from MS/MS data used the online Mascot search engine and the Swiss Protein database. The Swiss Protein database search parameters included Mouse, 4 missed tryptic cleavage sites allowed, precursor ion mass tolerance of 1.2 Da, fragment ion mass tolerance of 0.8 Da, protein modifications for Met oxidation, and Cys carbamidomethylation. Error tolerance is included. MS/MS datasets were also analyzed by the same search engine and parameters against all mammals. A minimum Mascot ion score of 20 was used for accepting all peptide MS/MS spectra.
Vectors
Microarray gene expression data analysis
Quality controlled raw data (Affymetrix CEL files or SOFT files) from previously published experiments (GSE1447130 ) were downloaded from Gene Expression Omnibus datasets (www.ncbi.nlm.nih.gov/geo) and subject to RMA normalization supplemented with the median over the entire array method (BRM-ArrayTools Version 3.7.1, developed by Dr Richard Simon and BRB-ArrayTools Development Team). Heat-maps were generated using ArrayStar Version 3.0 (DNASTAR).
Treatment of AML cells with HDAC inhibitors
A total of 100mM stock solutions for MS275 (Alexis Corporation) and Zolinza (Vorinistat, suberoylanilide hydroxamic acid [SAHA], Merck) were prepared by reconstituting the lyophilized drugs in 100% dimethyl sulfoxide and storage at −20°C. Working solution was generated by diluting the stock solution 1:100 in PBS immediately before addition to the cells to intended final concentration. CG-SH,27 Kasumi-1, and murine Runx1+/− primary cells were treated with 0.5μM of MS275 and 1μM of SAHA with timings as designated per the figure legends. Dimethyl sulfoxide/PBS vehicle to same concentrations were added to control cells.
Results
AML cells containing RUNX1 abnormalities express PU.1 and CEBPA but low levels of MCSFR and GMCSFR
In cotransfection experiments, RUNX1, PU.1, and CEBPA play essential roles in activation of the hematopoietic differentiation genes MCSFR and GMCSFR.7,8,31 To evaluate whether RUNX1 abnormality is associated with high level expression of PU.1 and CEBPA yet decreased MCSFR and GMCSFR expression, a public database of gene expression in primary AML cells (GSE1447130 ) was analyzed. AML cells containing the leukemia fusion protein RUNX1-ETO expressed PU.1 and CEBPA at high levels similar to other AML subtypes (PU.1 and CEBPA expression was notably decreased only in megakaryocytic leukemia cells; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Nonetheless, the RUNX1-ETO AML cells expressed lower levels of MCSFR and GMCSFR compared with most other AML subtypes (supplemental Figure 1). High PU.1 protein expression in AML cells, including primary M0 and M2 AML cells, has been previously reported.15
PU.1-mediated repression of stem cell genes is preserved, but activation of differentiation genes is impaired, in Runx1 deficient cells
PUER cells are murine Pu.1−/− hematopoietic precursors, which have been transduced with a retroviral vector to express Pu.1 fused to the ER.26 These cells were additionally transduced with lentivirus to suppress Runx1 expression with shRNA (PUER shRunx1). Runx1 suppression was more than 50% by quantitative RT-PCR (supplemental Figure 2A-B) and by Western blot (supplemental Figure 2C). Pu.1-ER protein expression was not affected by the shRunx1 (supplemental Figure 2C). Control cells were transduced with empty lentiviral vector (PUER control).
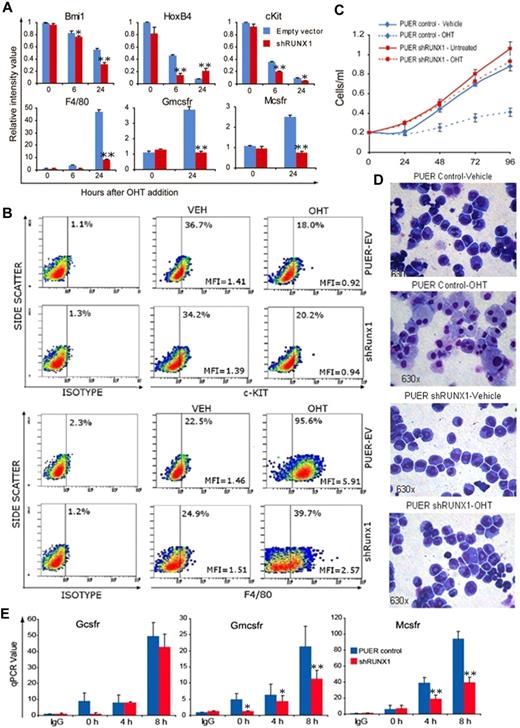
In cell culture with murine interleukin-3, PUER cells self-renew indefinitely. Addition of OHT to these cells (as an estrogen agonist) causes functional reintroduction of Pu.1 by translocation of Pu.1-ER into the nucleus, and triggers terminal macrophage differentiation.26 PUER control self-renewing in mIL3 expressed high levels of the self-renewal/stem cell promoting factors HoxB4, Bmi-1, and c-Kit (Figure 1A). After addition of OHT to PUER control, HoxB4, Bmi-1, and c-Kit were repressed followed by activation of the macrophage differentiation genes Mcsfr, Gmcsfr, and F4/80 (Figure 1A). Similar to PUER control, in PUER shRunx1, OHT induced repression of stem cell genes (HoxB4, Bmi-1, and c-Kit). However, the subsequent activation of macrophage differentiation genes (Mcsfr, Gmcsfr, and F4/80) was impaired. This pattern was also seen at the protein level: OHT suppressed c-Kit protein expression in both control and shRunx1 cells (Figure 1B); however, F4/80 protein up-regulation was impaired in shRunx1 cells (Figure 1B). In PUER control, OHT induced morphologic changes of macrophage differentiation (increase in cell size, decrease in nuclear:cytoplasmic ratio, clumping of nuclear chromatin, adherence to culture plates, cytoplasmic vacuolization), and a decrease in proliferation (Figure 1C-D). However, in PUER shRunx1 treated with OHT, morphologic changes of macrophage differentiation did not occur (Figure 1D), and the cells continued to expand exponentially (Figure 1C), with only a small decrease in proliferation compared with shRunx1 cells without OHT (Figure 1C). OHT treatment did not increase Gr-1/Ly6G (a granulocyte marker) protein expression in either control or shRunx1 cells (supplemental Figure 3).
In Runx1-deficient (shRunx1) cells, Pu.1 (OHT) repressed stem cell genes but myeloid differentiation gene activation was impaired. (A) The pattern of mRNA expression in PUER shRunx1 compared with PUER empty vector (control) cells treated with OHT. Gene expression after OHT addition was measured by quantitative RT-PCR. Stem cell genes are Bmi-1, Hoxb4, and c-Kit. Differentiation genes are F4/80, Mcsfr, and Gmcsfr. Data are mean ± SD. shRunx1 versus control for each time point: *P < .05, **P < .01 (Student t test). Experiments were performed in triplicate. (B) The pattern of c-Kit and F4/80 protein expression. Measured by flow cytometry 48 hours after OHT. MFI indicates mean fluorescence intensity of all cells. (C) OHT effect on cell proliferation of shRunx1 and control cells. Cell counts were measured daily with an automatic cell counter. Experiments performed in triplicate. Error bars represent SE. (D) OHT effect on cell morphology of shRunx1 and control cells. Cell morphology was evaluated on Giemsa-stained cytospin slides 48 hours after OHT. Microscope: Leica DMR, 63×/1.32 oil PH3 type N immersion oil. Image capture: CRI Nuance NzMSI-FX with Nuance 2.8 software. (E) Decreased histone acetylation at Gmcsfr and Mcsfr proximal promoters in shRunx1 compared with control cells. The Gcsfr promoter, which is not known to be regulated by Runx1/PU.1, was analyzed as control. ChIP performed with anti-H3Ac. Coimmunoprecipitation of promoter regions was analyzed by quantitative RT-PCR. Data are mean ± SD. shRunx1 vs control for each time point after OHT: *P < .05, **P < .01 (Student t test). Experiments performed in triplicate.
In Runx1-deficient (shRunx1) cells, Pu.1 (OHT) repressed stem cell genes but myeloid differentiation gene activation was impaired. (A) The pattern of mRNA expression in PUER shRunx1 compared with PUER empty vector (control) cells treated with OHT. Gene expression after OHT addition was measured by quantitative RT-PCR. Stem cell genes are Bmi-1, Hoxb4, and c-Kit. Differentiation genes are F4/80, Mcsfr, and Gmcsfr. Data are mean ± SD. shRunx1 versus control for each time point: *P < .05, **P < .01 (Student t test). Experiments were performed in triplicate. (B) The pattern of c-Kit and F4/80 protein expression. Measured by flow cytometry 48 hours after OHT. MFI indicates mean fluorescence intensity of all cells. (C) OHT effect on cell proliferation of shRunx1 and control cells. Cell counts were measured daily with an automatic cell counter. Experiments performed in triplicate. Error bars represent SE. (D) OHT effect on cell morphology of shRunx1 and control cells. Cell morphology was evaluated on Giemsa-stained cytospin slides 48 hours after OHT. Microscope: Leica DMR, 63×/1.32 oil PH3 type N immersion oil. Image capture: CRI Nuance NzMSI-FX with Nuance 2.8 software. (E) Decreased histone acetylation at Gmcsfr and Mcsfr proximal promoters in shRunx1 compared with control cells. The Gcsfr promoter, which is not known to be regulated by Runx1/PU.1, was analyzed as control. ChIP performed with anti-H3Ac. Coimmunoprecipitation of promoter regions was analyzed by quantitative RT-PCR. Data are mean ± SD. shRunx1 vs control for each time point after OHT: *P < .05, **P < .01 (Student t test). Experiments performed in triplicate.
Histone 3 acetylation (H3Ac) is a histone modification associated with transcriptional activation. H3Ac at the Mcsfr and Gmcsfr proximal promoters was examined by chromatin immunoprecipitation. The granulocyte colony stimulating factor receptor (Gcsfr) promoter was analyzed as a control (Gcsfr is regulated by Pu.1 in cooperation with CEBPA12 ). Although OHT treatment increased H3Ac at the Gcsfr promoter to a similar extent in both PUER control and PUER shRunx1 cells, in the shRunx1 cells, the H3Ac increase at the Mcsfr and Gmcsfr promoters was significantly smaller (Figure 1E).
Coimmunoprecipitation of endogenous corepressor (Eto2, Sin3A, and Hdac2) with Pu.1
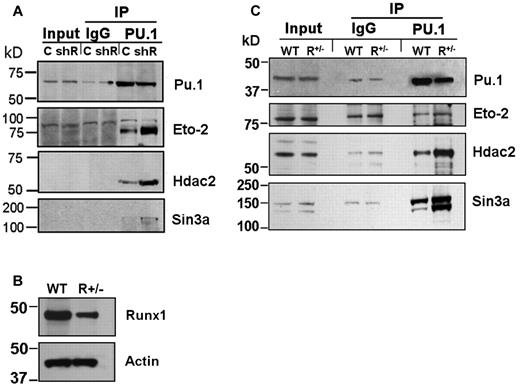
A possible explanation for intact Pu.1-mediated repression of stem cell genes in shRunx1 cells, but impairment of subsequent differentiation gene activation, accompanied by decreased H3Ac at the differentiation gene promoters, is persistent corepressor interaction with Pu.1. Corepressors with a well-established role in hematopoiesis include Eto2 and Sin3a, both of which associate with Hdac2 (Hdac2 is an HDAC that is highly expressed in hematopoietic cells). Therefore, coimmunoprecipitation of endogenous Eto2, Sin3a, and Hdac2 with Pu.1 from PUER control and PUER shRunx1 cells was examined. Substantially more Eto2, Sin3a, and Hdac2 coimmunoprecipitated with Pu.1 from PUER shRunx1 than from PUER control (Figure 2A). The Sin3a and Hdac2 bands were relatively weak compared with the Eto2 bands. Therefore, to confirm the results obtained by Western blot, in separate experiments, proteins coimmunoprecipitating with Pu.1 were analyzed for Sin3a and Hdac2 using LC-MS/MS. LC-MS/MS identified Sin3A peptides in the immunoprecipitate from PUER shRunx1 cells only, and not in the immunoprecipitate from PUER control (Table 1; supplemental Figure 4B). Although Hdac2 peptides were detected in both immunoprecipitates, these peptides were substantially enriched in the immunoprecipitate from shRunx1 cells (Table 1; supplemental Figure 4A).
Increased coimmunoprecipitation (co-IP) of endogenous corepressor (Eto2, Sin3A, and Hdac2) with Pu.1 in Runx1-deficient cells. (A) Increased co-IP of endogenous corepressor with Pu.1 from PUER shRunx1 compared with PUER empty vector (control) cells. Input indicates nonimmunoprecipitated cell lysate; IgG, control IP with isotype antibody; C, PUER control; and shR, PUER shRunx1. Cells were lysed 4 hours after addition of OHT. (B) Runx1 protein expression is decreased in primary Runx1 haploinsufficient (Runx1+/−) compared with wild-type littermate control (WT) bone marrow cells. Mice were genotyped as previously described.28 (C) Co-IP of endogenous corepressor with endogenous Pu.1 from Runx1+/− compared with WT bone marrow. Lysates were generated from freshly harvested bone marrow.
Increased coimmunoprecipitation (co-IP) of endogenous corepressor (Eto2, Sin3A, and Hdac2) with Pu.1 in Runx1-deficient cells. (A) Increased co-IP of endogenous corepressor with Pu.1 from PUER shRunx1 compared with PUER empty vector (control) cells. Input indicates nonimmunoprecipitated cell lysate; IgG, control IP with isotype antibody; C, PUER control; and shR, PUER shRunx1. Cells were lysed 4 hours after addition of OHT. (B) Runx1 protein expression is decreased in primary Runx1 haploinsufficient (Runx1+/−) compared with wild-type littermate control (WT) bone marrow cells. Mice were genotyped as previously described.28 (C) Co-IP of endogenous corepressor with endogenous Pu.1 from Runx1+/− compared with WT bone marrow. Lysates were generated from freshly harvested bone marrow.
The PUER experiments were complemented by experiments using primary cells. Coimmunoprecipitation of endogenous corepressor with endogenous Pu.1 was examined in Runx1+/− and wild-type littermate bone marrow cells (Figure 2B). Substantially more Sin3a and Hdac2 coimmunoprecipitated with Pu.1 from Runx1+/− bone marrow compared with wild-type control (Figure 2C). Eto2 expression was not as prominent in the primary cells as in PUER cells; nonetheless, coimmunoprecipitation of Eto2 with Pu.1 was also greater in Runx1+/− bone marrow cells (Figure 2C).
Exclusion of SIN3A and ETO2 from the RUNX1/PU.1 complex but not RUNX1-ETO/PU.1 or RUNX1ΔC-terminus/PU.1 complexes
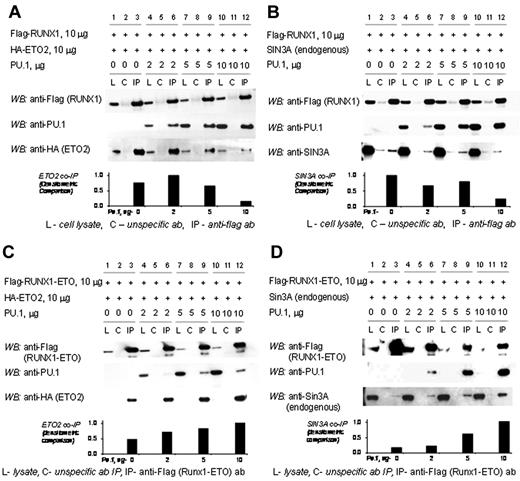
RUNX1 and PU.1 physically interact with each other and with corepressors. Therefore, we examined whether RUNX1, PU.1, and ETO2 or SIN3A can form a tripartite complex. In transiently transfected 293T cells, ETO2 coimmunoprecipitates with RUNX1 (Figure 3A, lane 3). However, as PU.1 concentrations are increased in the cells by transient transfection, PU.1 preferentially coimmunoprecipitates with RUNX1 with a corresponding decrease in coimmunoprecipitated ETO2 (Figure 3A, lanes 6, 9, and 12). Similarly, endogenous SIN3A coimmunoprecipitates with RUNX1 (Figure 3B, lane 3). However, as PU.1 concentrations are increased, PU.1 preferentially coimmunoprecipitates with RUNX1 with a corresponding decrease in coimmunoprecipitated SIN3A (Figure 3B, lanes 6, 9, and 12).
The corepressors SIN3A and ETO2 are excluded from the RUNX1/PU.1 complex but not the RUNX1-ETO/PU.1 complex. (A) Increasing concentrations of PU.1 decrease the amount of ETO2 coimmunoprecipitated with RUNX1. 293T cells were transfected with expression vectors for flag-RUNX1, HA-ETO2, and PU.1 in increasing amounts. In the absence of PU.1, ETO2 co-IPs with RUNX1 (lane 3). As PU.1 amounts are increased, PU.1 preferentially co-IPs with the RUNX1 and ETO2 is progressively excluded (lanes 6, 9, and 12). (B) Increasing concentrations of PU.1 decrease the amount of endogenous SIN3A coimmunoprecipitated with RUNX1. Sin3A co-IPs with RUNX1 in the absence of PU.1 (lane 3). As PU.1 concentrations are increased, PU.1 preferentially co-IPs with the RUNX1 and Sin3A is progressively excluded (lanes 6, 9, and 12). (C) Addition of PU.1 to RUNX1-ETO increases ETO2 recruitment. 293-T cells were transiently transfected with expression vectors for flag-RUNX1-ETO, ETO2, and increasing amounts of PU.1. Increasing amounts of PU.1 increased the amount of ETO2 in the RUNX1-ETO/PU.1 complex (lanes 6, 9, and 12). (D) Addition of PU.1 to RUNX1-ETO increases SIN3A recruitment. 293-T cells were transiently transfected with expression vectors for flag-RUNX1-ETO and increasing amounts of PU.1. Increasing amounts of PU.1 increased the amount of SIN3A in the RUNX1-ETO/PU.1 complex.
The corepressors SIN3A and ETO2 are excluded from the RUNX1/PU.1 complex but not the RUNX1-ETO/PU.1 complex. (A) Increasing concentrations of PU.1 decrease the amount of ETO2 coimmunoprecipitated with RUNX1. 293T cells were transfected with expression vectors for flag-RUNX1, HA-ETO2, and PU.1 in increasing amounts. In the absence of PU.1, ETO2 co-IPs with RUNX1 (lane 3). As PU.1 amounts are increased, PU.1 preferentially co-IPs with the RUNX1 and ETO2 is progressively excluded (lanes 6, 9, and 12). (B) Increasing concentrations of PU.1 decrease the amount of endogenous SIN3A coimmunoprecipitated with RUNX1. Sin3A co-IPs with RUNX1 in the absence of PU.1 (lane 3). As PU.1 concentrations are increased, PU.1 preferentially co-IPs with the RUNX1 and Sin3A is progressively excluded (lanes 6, 9, and 12). (C) Addition of PU.1 to RUNX1-ETO increases ETO2 recruitment. 293-T cells were transiently transfected with expression vectors for flag-RUNX1-ETO, ETO2, and increasing amounts of PU.1. Increasing amounts of PU.1 increased the amount of ETO2 in the RUNX1-ETO/PU.1 complex (lanes 6, 9, and 12). (D) Addition of PU.1 to RUNX1-ETO increases SIN3A recruitment. 293-T cells were transiently transfected with expression vectors for flag-RUNX1-ETO and increasing amounts of PU.1. Increasing amounts of PU.1 increased the amount of SIN3A in the RUNX1-ETO/PU.1 complex.
The chromosomal translocation t(8;21) is a recurrent abnormality in AML that replaces the RUNX1 C-terminal transactivation domain with most of ETO.2 In contrast to the results with RUNX1, increasing PU.1 increased the amount of ETO2 or SIN3A corepressor that coimmunoprecipitated with RUNX1-ETO (Figure 3C-D, lanes 6, 9, and 12). Because the RUNX1-ETO concentrations were held stable, the increasing corepressor coimmunoprecipitation was presumably the result of the increasing amounts of PU.1 in the complex.
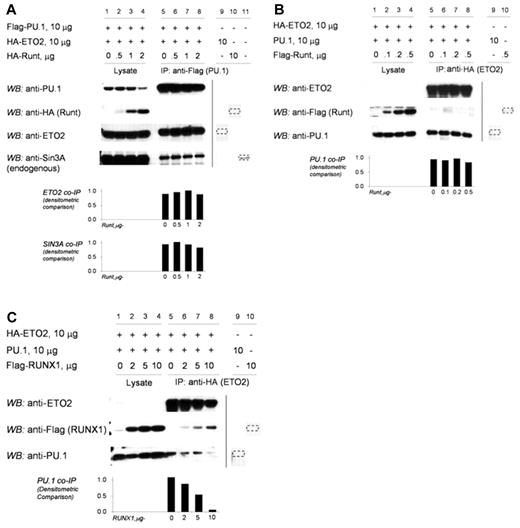
In RUNX1-ETO, the RUNX1 C-terminus is replaced by ETO. Therefore, we examined whether RUNX1ΔC-terminus (RUNT) produces a similar result as RUNX1-ETO. In contrast to full-length RUNX1, increasing concentrations of RUNT did not decrease coimmunoprecipitation of ETO2 or SIN3A with PU.1 (Figure 4A, lanes 5-8). In reverse coimmunoprecipitation experiments, increasing concentrations of RUNT did not prevent the coimmunoprecipitation of PU.1 with ETO2 (Figure 4B, lanes 5-8). Increasing concentrations of full-length RUNX1, as expected from the earlier results, decreased the coimmunoprecipitation of PU.1 with ETO2 (Figure 4C, lanes 5-8).
The RUNX1 C-terminus is required to exclude corepressors from the RUNX1/PU.1 complex. (A) Increasing concentrations of RUNT did not prevent the co-IP of ETO2 or SIN3A with PU.1. 293T cells were transfected with HA-ETO2, Flag-PU.1, and increasing amounts of HA-RUNT. Increasing amounts of RUNT did not decrease co-IP of ETO2 or endogenous SIN3A with PU.1 (lanes 5-8). The vertical line separates repositioned gel lanes (lanes 9-11) that showed anti-Flag did not IP HA-RUNT, HA-ETO2, or SIN3A in the absence of Flag-PU.1. Dashed outline shows expected position of protein in these lanes. (B) In the reverse co-IP experiment, increasing amounts of RUNT did not prevent the co-IP of PU.1 with ETO2. 293T cells were transfected with HA-ETO2, PU.1, and increasing amounts of Flag-RUNT. Increasing amounts of RUNT did not decrease the co-IP of PU.1 with ETO2 (lanes 5-8). The vertical line separates repositioned gel lanes (lanes 9 and 10), which showed that anti-HA did not immunoprecipitate PU.1 or Flag-RUNT in the absence of HA-ETO2. Dashed outline shows expected position of protein in these lanes. (C) In contrast, increasing amounts of RUNX1 decreased co-IP of PU.1 with ETO2. 293T cells were transfected with expression vectors for HA-ETO2, PU.1, and flag-RUNX1. PU.1 co-IPs with ETO2 (lane 5). Increasing amounts of RUNX1 decreased co-IP of PU.1 (lanes 6-8). The vertical line separates repositioned gel lanes (lanes 9 and 10) that showed anti-HA did not IP PU.1 or Flag-RUNX1 in the absence of HA-ETO2. Dashed outline indicates expected position of protein in these lanes.
The RUNX1 C-terminus is required to exclude corepressors from the RUNX1/PU.1 complex. (A) Increasing concentrations of RUNT did not prevent the co-IP of ETO2 or SIN3A with PU.1. 293T cells were transfected with HA-ETO2, Flag-PU.1, and increasing amounts of HA-RUNT. Increasing amounts of RUNT did not decrease co-IP of ETO2 or endogenous SIN3A with PU.1 (lanes 5-8). The vertical line separates repositioned gel lanes (lanes 9-11) that showed anti-Flag did not IP HA-RUNT, HA-ETO2, or SIN3A in the absence of Flag-PU.1. Dashed outline shows expected position of protein in these lanes. (B) In the reverse co-IP experiment, increasing amounts of RUNT did not prevent the co-IP of PU.1 with ETO2. 293T cells were transfected with HA-ETO2, PU.1, and increasing amounts of Flag-RUNT. Increasing amounts of RUNT did not decrease the co-IP of PU.1 with ETO2 (lanes 5-8). The vertical line separates repositioned gel lanes (lanes 9 and 10), which showed that anti-HA did not immunoprecipitate PU.1 or Flag-RUNT in the absence of HA-ETO2. Dashed outline shows expected position of protein in these lanes. (C) In contrast, increasing amounts of RUNX1 decreased co-IP of PU.1 with ETO2. 293T cells were transfected with expression vectors for HA-ETO2, PU.1, and flag-RUNX1. PU.1 co-IPs with ETO2 (lane 5). Increasing amounts of RUNX1 decreased co-IP of PU.1 (lanes 6-8). The vertical line separates repositioned gel lanes (lanes 9 and 10) that showed anti-HA did not IP PU.1 or Flag-RUNX1 in the absence of HA-ETO2. Dashed outline indicates expected position of protein in these lanes.
Inhibition of HDAC rescues MCSFR and GMCSFR expression in RUNX1-deficient human AML cells
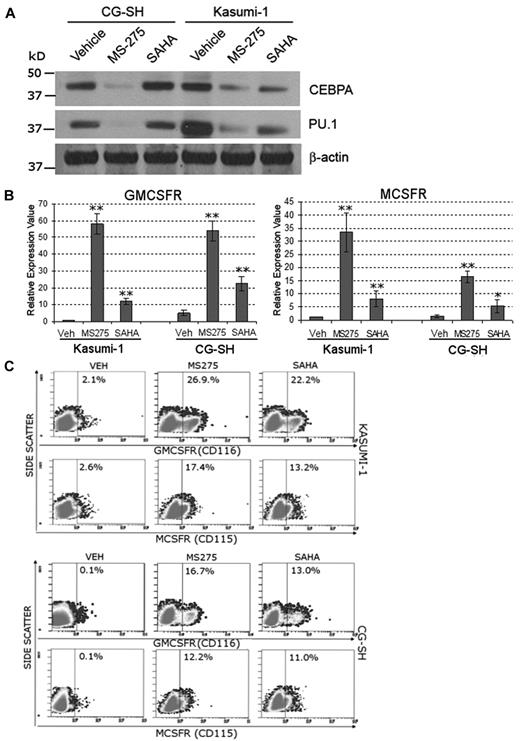
HDAC enzymatic activity is a major component of corepressor function. To examine whether inhibition of HDAC can mitigate the functional consequences of RUNX1 deficiency, human AML cells containing RUNX1-ETO (Kasumi-1 cells) or mutated RUNX1 and normal cytogenetics (CG-SH cells27 ) were treated with the HDAC inhibitors SAHA (a broad HDAC inhibitor) and MS-275 (an inhibitor of HDAC1, 2, 3, and 8). Western blot was used to confirm that Kasumi-1 and CG-SH express PU.1 and CEBPA (Figure 5A). Treatment with MS-275 (0.5μM) and SAHA (1μM) significantly increased MCSFR and GMCSFR mRNA (Figure 5B) and protein (Figure 5C) expression in Kasumi-1 and CG-SH cells. The increase in MCSFR and GMCSFR expression did not appear to be secondary to an increase in PU.1 or CEBPA protein expression (Figure 5A). HDAC inhibitor treatment decreased Kasumi1 and CG-SH cell proliferation (supplemental Figure 5A). MS-275 and SAHA also increased Mcsfr and Gmcsfr expression by primary murine Runx1+/− hematopoietic cells (supplemental Figure 5B).
Inhibition of HDAC rescues MCSFR and GMCSFR expression in AML cells that express lineage-specifying TFs and contain mutated or translocated RUNX1. Kasumi-1 cells contain RUNX1-ETO. CG-SH cells contain mutated RUNX1 and normal cytogenetics. (A) AML cells containing abnormal RUNX1 express high levels of the lineage-specifying TFs PU.1 and CEBPA. Western blot 72 hours after addition of drug. (B-C) Treatment with HDAC inhibitors significantly increased MCSFR and GMCSFR mRNA and protein expression. Kasumi-1 and CG-SH were treated once with SAHA 1μM or MS-275 0.5μM. mRNA expression was measured by quantitative RT-PCR 72 hours after HDAC inhibitor treatment. Data are mean ± SD. HDAC inhibitor treatment versus vehicle treatment: *P < .05, **P < .01 (Student t test). Protein expression was measured by flow cytometry. Experiments were performed in triplicate.
Inhibition of HDAC rescues MCSFR and GMCSFR expression in AML cells that express lineage-specifying TFs and contain mutated or translocated RUNX1. Kasumi-1 cells contain RUNX1-ETO. CG-SH cells contain mutated RUNX1 and normal cytogenetics. (A) AML cells containing abnormal RUNX1 express high levels of the lineage-specifying TFs PU.1 and CEBPA. Western blot 72 hours after addition of drug. (B-C) Treatment with HDAC inhibitors significantly increased MCSFR and GMCSFR mRNA and protein expression. Kasumi-1 and CG-SH were treated once with SAHA 1μM or MS-275 0.5μM. mRNA expression was measured by quantitative RT-PCR 72 hours after HDAC inhibitor treatment. Data are mean ± SD. HDAC inhibitor treatment versus vehicle treatment: *P < .05, **P < .01 (Student t test). Protein expression was measured by flow cytometry. Experiments were performed in triplicate.
Discussion
Physiologic gene activation is most probably cooperative involving multiple TFs collaborating to bind DNA, exclude corepressors, recruit coactivators, and recruit the basal TF complex.32 In keeping with this paradigm, RUNX1 alone is a relatively weak activator of transcription.4,5,7,8,11 However, RUNX1 potently enhances gene activation by multiple key hematopoietic lineage-specifying TFs.4-10 Corepressor versus coactivator interaction could be an important determinant of repression versus activation by TF complexes. However, the mechanisms that regulate corepressor versus coactivator interactions of RUNX1 and PU.1 are incompletely understood.17-20 One possible mechanism is cooperation to recruit or exclude coactivators or corepressors. This has been described for other TF. Nuclear factor-κB, IRF1, and ATF2/c-Jun cooperatively recruit the coactivators CBP and p300 at the interferon-β (IFN-β) promoter.33 Multiprotein complexes that include IRF2 or OCT-1 cooperatively exclude coactivators at the IFN-β and HLA-DRA promoters, respectively.34,35 The B-cell lineage-specifying TF PAX5 cooperates with PU.1 to recruit the corepressor GRG4 at the MCSFR promoter.36 The present study (summarized in Figure 6) describes cooperative exclusion of corepressors by 2 TFs. RUNX1 and PU.1 independently interact with the corepressors ETO2 and SIN3A. However, ETO2 and SIN3A did not bind to the combined RUNX1/PU.1 complex. The RUNX1 C-terminus is highly conserved from amino acids 291 to 411.37 Corepressor exclusion required the RUNX1 C-terminus because RUNT and the leukemia fusion protein RUNX1-ETO, which lacks the RUNX1 C-terminus, allowed corepressor interaction with PU.1. Loss of this function of the RUNX1 C-terminus could contribute to the inability of RUNT to synergize with PU.1 to transactivate the MCSFR promoter,8 and the impaired differentiation and leukemogenesis associated with expression of RUNX1a (a RUNX1 isoform that lacks the RUNX1 C-terminus).38,39
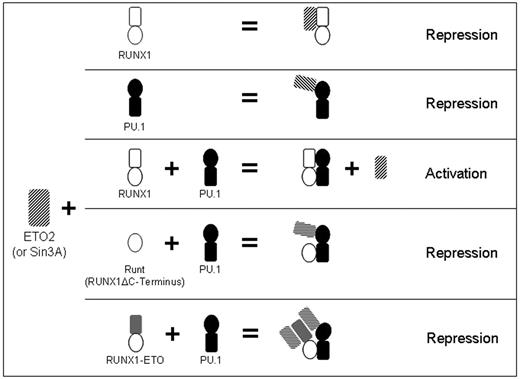
Model for RUNX1 regulation of PU.1 interaction with corepressors. Proposed model for RUNX1-mediated regulation of the transcriptional activity of PU.1 in normal hematopoiesis, and effects of RUNX1 deletions and translocations on this cooperation. RUNX1 and PU.1 independently interact with SIN3A or ETO2 (rows 1 and 2). When both RUNX1 and PU.1 are present, ETO2 and SIN3A are excluded from the RUNX1:PU.1 complex (row 3). Corepressor exclusion requires the RUNX1 C-terminus. Therefore, RUNT does not inhibit PU.1 interaction with corepressor (row 4). Similarly, corepressor is recruited to the RUNX1-ETO/PU.1 complex (RUNX1-ETO lacks the C-terminus). In this complex, corepressor is recruited to both the ETO moiety of the leukemia fusion protein and to PU.1 (row 5).
Model for RUNX1 regulation of PU.1 interaction with corepressors. Proposed model for RUNX1-mediated regulation of the transcriptional activity of PU.1 in normal hematopoiesis, and effects of RUNX1 deletions and translocations on this cooperation. RUNX1 and PU.1 independently interact with SIN3A or ETO2 (rows 1 and 2). When both RUNX1 and PU.1 are present, ETO2 and SIN3A are excluded from the RUNX1:PU.1 complex (row 3). Corepressor exclusion requires the RUNX1 C-terminus. Therefore, RUNT does not inhibit PU.1 interaction with corepressor (row 4). Similarly, corepressor is recruited to the RUNX1-ETO/PU.1 complex (RUNX1-ETO lacks the C-terminus). In this complex, corepressor is recruited to both the ETO moiety of the leukemia fusion protein and to PU.1 (row 5).
Aberrant corepressor recruitment by the ETO moiety in RUNX1-ETO has been proposed as a rationale for HDAC inhibitor treatment of this disease.40 However, the ETO domains that recruit corepressors are frequently deleted from the RUNX1-ETO leukemia fusion protein seen in primary AML cells.41,42 The present observations suggest that these truncated forms of RUNX1-ETO may continue to cause aberrant repression of late differentiation genes because loss of the RUNX1 C-terminus is sufficient to cause persistent corepressor recruitment to PU.1. Frame-shift mutations in the C-terminus exons of RUNX1 are frequently seen in myelodysplastic syndrome and AML.1 Transcripts from these mutated RUNX1 genes can escape nonsense-mediated decay, resulting in expression of RUNX1 variants that lack the C-terminus.43 The observations here suggest the mechanistic rationale for HDAC inhibitor therapy may also apply to cases with these RUNX1 abnormalities.
Hematopoietic precursors subject to a cytokine differentiation stimulus rapidly repress key stem cell genes.44 This process is reiterated here in the model of Pu.1-triggered macrophage differentiation, with rapid repression of Hoxb4, c-Kit, and Bmi-1 after introduction of Pu.1 into the nucleus. Runx1 deficiency allowed these Pu.1-triggered repression events and instead selectively impaired activation of the differentiation genes Mcsfr, Gmcsfr, and F4/80. Therefore, Runx1 deficiency allowed some Pu.1-mediated differentiation events. Runx1 deficiency permissiveness for partial differentiation has been seen in other models. In Runx1+/− mice, hematopoietic stem cells are decreased but myeloid progenitors are increased.28 In primary AML cells with translocated RUNX1, there is high expression of PU.115 and CEBPA (indicating lineage-commitment of the cells), but relatively low expression of MCSFR and GMCSFR. An important translational goal is to find differences between self-renewing leukemia cells (leukemia-initiating cells) and normal stem cells that can be used to selectively terminate the growth of the leukemia-initiating cells. A partially differentiated phenotype of leukemia-initiating cells, also suggested by some surface markers of differentiation (supplementary references), is potentially such a difference. The high PU.1 and CEBPA expression, but persistent corepressor recruitment that impairs PU.1 and CEBPA-mediated activation of late differentiation genes that terminate proliferation, could explain why antagonizing corepressor function with HDAC inhibitors can resume differentiation and terminate proliferation.40,45-47 In contrast, treatment of normal hematopoietic stem cells with HDAC inhibitors has the opposite effect, maintaining self-renewal,44,48-50 possibly by preventing repression of stem cell genes by differentiation stimuli.44
These results suggest a molecular mechanism for cooperative differentiation gene activation by RUNX1/PU.1, and for aberrant repression of late differentiation genes in AML cells containing mutated or translocated RUNX1. Inhibitors of HDAC are known to differentiate AML cells.40,45-47 These observations provide a mechanistic explanation for this effect of HDAC inhibitors and augment the rationale for development of corepressor inhibitors as differentiation therapy agents for myelodysplastic syndrome and AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Harinder Singh laboratory (University of Chicago) for the gift of PUER cells, the Jim Downing laboratory (St Jude Children's Research Hospital) for the gift of Runx1 haploinsufficient mice, John W. Crabb for assistance with LC-MS/MS analyses, and Quteba Ebrahem for assistance with murine primary cell experiments.
S.A. is supported by the National Institutes of Health (RO1AI033043). Y.S. was supported by the National Institutes of Health (1R01CA138858), the Department of Defense (PR081404), and the Cleveland Clinic Foundation.
National Institutes of Health
Authorship
Contribution: Z.H., X.G., K.B., V.I., A.S., and S.K. performed experiments (with the majority of experiments performed by Z.H., X.G., K.B., and V.I.); R.M. provided CG-SH cells; S.A. and G.N. provided critical commentary on the data analyses, presentation, and manuscript; and Y.S. supervised experiments and developed hypotheses.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yogen Saunthararajah, Taussig Cancer Institute, 9500 Euclid Ave, R40, Cleveland, OH 44195; e-mail: saunthy@ccf.org.
References
Author notes
Z.H. and X.G. contributed equally to this study.