Abstract
Hematopoiesis is evolutionarily conserved from zebrafish to mammals, and this includes both primitive and definitive waves during embryogenesis. Primitive hematopoiesis is dominated by erythropoiesis with limited myelopoiesis. Protein sumoylation, a ubiquitination-like posttranslational protein modification, is implicated in a variety of biochemical processes, most notably in transcriptional repression. We show here that the loss of 6 small ubiquitin-related modifier (SUMO) paralogs triggers a sharp up-regulation of the myeloid-specific marker mpo and down-regulation of the erythroid-specific marker gata1 in myelo-erythroid progenitor cells (MPCs) in the intermediate cell mass (ICM) during primitive hematopoiesis. Accordingly, in transgenic zebrafish lines, hyposumoylation expands myelopoiesis at the expense of erythropoiesis. A SUMO–CCAAT/enhancer–binding protein α (SUMO-C/ebpα) fusion restores the normal myelopoiesis/erythropoiesis balance, suggesting that sumoylation status of C/ebpα contributes to myelo-erythroid lineage determination. Our results therefore implicate sumoylation in early lineage determination and reveal the possible molecular mechanism underlying the puzzling biased primitive hematopoiesis in vertebrates.
Introduction
Vertebrate hematopoiesis is a successive process involving primitive and definitive waves occurring in anatomically distinct sites. Whereas primitive hematopoiesis predominantly produces erythrocytes and some macrophages, definitive hematopoiesis generates self-renewing pluripotent hematopoietic stem cells capable of giving rise to all blood cell types.1 The zebrafish (Danio rerio) has emerged as an excellent model organism for the study of hematopoiesis because it combines many advantages of other model organisms.2,3 In zebrafish, primitive wave originates in 2 intraembryonic sites known as the anterior lateral plate mesoderm (A-LPM) and the posterior lateral plate mesoderm (P-LPM) that subsequently forms the intermediate cell mass (ICM).4 Whereas the A-LPM is the primary site of production of primitive macrophages, the ICM is the location of multipotential blood progenitors containing predominantly primitive erythrocytes and a few granulocytes.5
The molecular mechanisms of multipotential progenitor commitment in the ICM remain poorly understood. However, the respective specification of myeloid and erythroid cell lineages of zebrafish myelo-erythroid progenitor cells (MPCs) appear to be controlled by the antagonistic regulation of Gata1 and Pu.1.6,7 The zebrafish CCAAT/enhancer–binding protein α (C/ebpα), a master regulator of granulopoiesis,8 is also expressed, along with pu.1 and gata1, in the hematopoietic progenitors of P-LPM during primitive hematopoiesis.9 C/ebpα is the founding member of a family of transcription factors containing a basic leucine zipper domain.10 Loss of C/ebpα in mice results in the complete absence of mature neutrophils and eosinophils and increased numbers of erythroid progenitors and erythroid cells in the fetal liver.11,12 C/ebpα not only acts as a cell fate switch to induce granulocyte differentiation from bipotential granulocyte-macrophage progenitors,8 but also determines the hematopoietic cell fate in multipotential progenitor cells by inhibiting erythroid development while promoting myeloid differentiation.12 In addition, megakaryocyte/erythroid progenitors and common lymphoid progenitors could be redirected to myeloid lineage by ectopic activation of C/ebpα.13 C/ebpα can be posttranslationally modified by small ubiquitin-related modifier (SUMO), which controls its transactivation activity.14,15
SUMO is highly conserved throughout evolution, and SUMO conjugation is carried out by a multistep enzymatic cascade reaction consisting of a SUMO-activating enzyme (E1), Aos1/Uba2, a unique SUMO-conjugating enzyme (E2), Ubc9, and 3 families of SUMO ligases (E3), which eventually attach SUMO to the substrates.16 Protein sumoylation plays important roles in a wide variety of processes, including transcription regulation, genome integrity, and subcellular localization.17
It has long been observed that the primitive hematopoiesis in vertebrates is dominated by erythropoiesis with limited myelopoiesis.18,19 Although C/ebpα and Pu.1, the master regulators of myelopoiesis, are coexpressed with erythropoietic Gata1 in the hematopoietic progenitors, the underlying molecular mechanism responsible for the biased hematopoiesis is unexploited. In the present study, we demonstrate that impairment of sumoylation favors myelopoiesis of MPCs at the expense of erythropoiesis in the ICM during primitive hematopoiesis of zebrafish. Sumoylation of C/ebpα accounts, at least in part, for this myelo-erythroid lineage commitment process.
Methods
Zebrafish
Zebrafish maintenance and staging were performed as described previously.20 The transgenic lines Tg(gata1:EGFP),21 Tg(mpo:EGFP), and Tg(lyz:EGFP),22 were established in our laboratory. The zebrafish facility and study were approved by the Institutional Review Board of the Institute of Health Sciences, Shanghai Institutes of Biologic Sciences, Chinese Academy of Sciences (Shanghai, China).
Generation of constructs
The zebrafish cebpa, ΔC/ebpα, gata1, and morpholino-unmatched SUMO2 and SUMO2 ΔGG were amplified by RT-PCR with the indicated primers (supplemental Table 2, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and cloned into pCS2+ vector. Lysine 125 of zebrafish C/ebpα and lysine 157 of zebrafish Gata1 were mutated to arginine. The Ubc9 (C93A) mutant was generated with the QuickChange Site-Directed Mutagenesis Kit (Stratagene) with the indicated primers (supplemental Table 2). The SUMO2, ubiquitin, or POZ domain (1-120 amino acids) of PLZF was fused in-frame to the N-terminus of C/ebpα. SUMO2-ΔC/ebpα was generated with the indicated primers (supplemental Table 2).
Morpholinos and mRNA microinjection
Morpholino oligonucleotides (MOs) were purchased from Gene Tools. SUMO1 MO GTCTCCGTGTCTGACATGATATTCC; SUMO2 MO CATGGTTATTGTATTTGCGCTTCTC; SUMO3 MO TAGGCTTGTCTTCGGACATTTTTGC; Ubc9 MO TCAGAGCAATGCCAGACATGACCAC; The doses of injection per embryo were: SUMO1 MO, 12.46 ng; SUMO2 MO, 12.46 ng; SUMO3 MO, 12.46 ng; Ubc9 MO, 12.46 ng; and the SUMO1-SUMO2-SUMO3 MO combination, 4.15 ng each. Capped mRNAs were transcribed from linearized pCS2+ plasmids (mMessage Machine; Ambion), purified, and diluted to 100 ng/μL (SUMO2, SUMO2 ΔGG, and Ubc9 (C93A) mRNA) or 50 ng/μL (SUMO2-C/ebpα, SUMO2-ΔC/ebpα, POZ-C/ebpα, and ubiquitin-C/ebpα mRNA) for microinjection.
Whole-mount mRNA in situ hybridization
Digoxigenin-labeled or fluorescein-labeled antisense RNA probes were transcribed from linearized constructs using T3 or T7 polymerase (Roche). Whole-mount mRNA in situ hybridization (WISH) was performed as described previously.23 The probes were detected using alkaline phosphatase–coupled anti-digoxigenin Fab fragment antibody (Roche) or anti-fluorescein Fab fragment antibody (Roche) with BCIP/NBT staining (Vector Laboratories) or Fast Red staining (Roche).
Phosphorylated histone H3 labeling and TUNEL assay
Terminal transferase UTP nick end labeling (TUNEL) was performed using the In Situ Cell Death Detection Kit and TMR Red (Roche) according to the manufacturer's recommendations. Phosphorylated histone H3 labeling of fixed embryos was performed with the rabbit anti–phosphohistone H3 antibody (Santa Cruz Biotechnology) at 4°C overnight and revealed with Alexa Fluor 488 goat anti–rabbit secondary antibody (Invitrogen).
Cell sorting and cytology
gata1-EGFP transgenic embryos at 22 hours postfertilization (hpf) were dissected and the trunk regions were digested with 0.5% trypsin (GIBCO) for 15 minutes at 37°C. Single-cell suspension was obtained by centrifugation at 400g for 5 minutes, washing twice with PBS, and passing through a 40μm nylon mesh filter. FACS was performed with MoFlo (DakoCytomation) to obtain homogenous enhanced green fluorescent protein (EGFP)–positive cells, which were subsequently subjected to cytospinning at 40g for 5 minutes, followed by myeloperoxidase cytochemistry and Wright staining.
Immunoblotting and immunoprecipitation
A rabbit polyclonal antiserum against zebrafish C/ebpα was generated using C/ebpα N-terminal amino acids (1-204 aa) as an antigen source (AbMART). 293T cells were transfected with indicated plasmids using Effectene Transfection Reagent (QIAGEN). Cells were harvested 48 hours after transfection and subjected to immunoblotting analysis, or cell lysates were purified by anti-Flag M2 beads (Sigma-Aldrich), followed by Western blotting with GFP-specific (Roche), SUMO2-specific (Invitrogen), and hemagglutinin-specific (Santa Cruz Biotechnology) antibodies. The blots were detected by SuperSignal West Pico Substrate (Pierce).
Luciferase reporter assay
293T cells were transfected with indicated plasmids using Effectene Transfection Reagent (QIAGEN). Cells were harvested 36 hours after transfection and luciferase activities were analyzed using the Dual Luciferase Reporter Assay Kit (Promega) according to the manufacturer's protocols. Luciferase activity was normalized to Renilla activity.
Results
Loss of SUMO paralogs leads to an increase of mpo+ cells and a decrease of gata1+ cells in the ICM
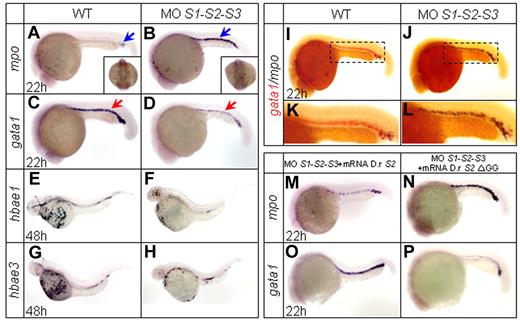
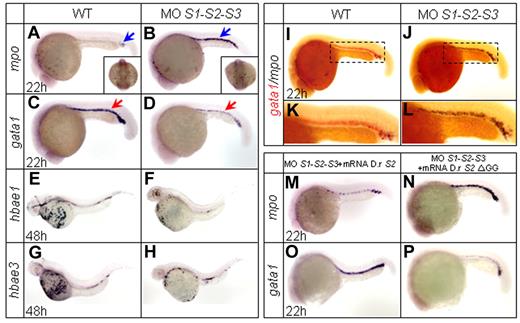
Zebrafish SUMO paralogs are expressed ubiquitously, including in hematopoietic regions during early embryogenesis.24 Many critical transcription factors involved in hematopoiesis, such as Gata1 and C/ebpα, can be sumoylated.14,15,25 To evaluate the effects of sumoylation on hematopoietic differentiation and lineage commitment, we first carried out antisense MO-mediated knock-down experiments. SUMO paralog MOs were injected at the one-cell stage, and the specificities and efficacies of the MOs have been confirmed previously.24 The temporal and spatial expression patterns of a series of transcription factors and key genes involved in embryonic hematopoiesis were then examined by WISH. Individual loss of SUMO1, SUMO2, or SUMO3 had no overt effect on the expression of the detected hematopoietic markers (supplemental Figure 1A-H and data not shown), confirming the previously reported functional redundancy of SUMO paralogs.24 Therefore, simultaneous knock-down of all 3 SUMO paralogs was carried out. Zebrafish myeloperoxidase (mpo) is normally expressed in granulocytes and observed in both the A-LPM and ICM at 20 hpf.26,27 In SUMO-deficient embryos, mpo expression was strikingly enhanced in the ICM from 20-30 hpf (supplemental Figure 1I-P), with a peak at 22 hpf (Figure 1A-B and supplemental Table 1). Interestingly, no such increase was observed within the A-LPM region (Figure 1A-B square bracket), suggesting that the molecular regulatory pathway of granulopoiesis within A-LPM and P-LPM might be somehow different. In contrast, expression of the erythroid progenitor marker gata128 was drastically decreased at 22 hpf (Figure 1C-D and supplemental Table 1). This decrease was subsequently followed by a sharply reduced expression of the gata1 downstream genes hbae1 and hbae3, markers of mature erythrocytes29 at 48 hpf (Figure 1E-H). These results were confirmed by double WISH assays for mpo and gata1 (Figure 1I-L). The SUMO deficiency–mediated phenotype could be significantly reversed by coinjection of zebrafish SUMO2 mRNA (Figure 1M,O and supplemental Table 1). To confirm that the effect of SUMO knock-down represents a lack of sumoylation rather than of free-floating SUMO protein, the nonconjugated C-terminal double-glycine–truncated SUMO2 ΔGG mutant was constructed and shown to be ineffective in a rescue assay (Figure 1N,P and supplemental Table 1). This result demonstrated that the phenotype was indeed sumoylation dependent.
WISH assays. (A-D) WISH assays of mpo and gata1 at 22 hpf. Knock-down of SUMO paralogs by MOs results in an increased expression of the granulocytic-specific marker mpo (A-B) and a decreased expression of the erythroid-specific marker gata1 (C-D). Blue arrows indicate mpo+ cells and red arrows indicate gata1+ cells. Note that mpo expression in the A-LPM does not show any significant change (square bracket in panels A and B). (E-H) WISH assays of hbae1 and hbae3 at 48 hpf. Expression of hemoglobin hbae1 and hbae3 decreases in SUMO-deficient embryos (F and H) compared with those of WT (E and G). (I-L) Two-color WISH analysis of gata1 (red) and mpo (black) at 22 hpf. Panels K and L are highly magnified images of corresponding boxed regions from I and J, respectively. (M-P) Whereas zebrafish SUMO2 mRNA specifically reverses the SUMO-deficient phenotypes (M and O), SUMO2 ΔGG mRNA becomes ineffective (N and P). D.r indicates Danio rerio.
WISH assays. (A-D) WISH assays of mpo and gata1 at 22 hpf. Knock-down of SUMO paralogs by MOs results in an increased expression of the granulocytic-specific marker mpo (A-B) and a decreased expression of the erythroid-specific marker gata1 (C-D). Blue arrows indicate mpo+ cells and red arrows indicate gata1+ cells. Note that mpo expression in the A-LPM does not show any significant change (square bracket in panels A and B). (E-H) WISH assays of hbae1 and hbae3 at 48 hpf. Expression of hemoglobin hbae1 and hbae3 decreases in SUMO-deficient embryos (F and H) compared with those of WT (E and G). (I-L) Two-color WISH analysis of gata1 (red) and mpo (black) at 22 hpf. Panels K and L are highly magnified images of corresponding boxed regions from I and J, respectively. (M-P) Whereas zebrafish SUMO2 mRNA specifically reverses the SUMO-deficient phenotypes (M and O), SUMO2 ΔGG mRNA becomes ineffective (N and P). D.r indicates Danio rerio.
Overexpression of Ubc9 (C93A), a dominant-negative mutant of wild-type (WT) Ubc9 that lacks E2 activity but competes with endogenous Ubc9 for substrate binding,30 triggered a phenotype similar to the one observed in SUMO-deficient embryos (data not shown and supplemental Table 1). To investigate whether this observed SUMO-deficiency–mediated phenotype was due to abnormal apoptosis or cell proliferation, the TUNEL assay and anti-phosphorylated histone H3 (pH3) immunostaining, a mitotic marker, were performed. No significant difference in proliferative or apoptotic cell number was observed within the ICM between SUMO morphants and controls (supplemental Figure 1Q-T).
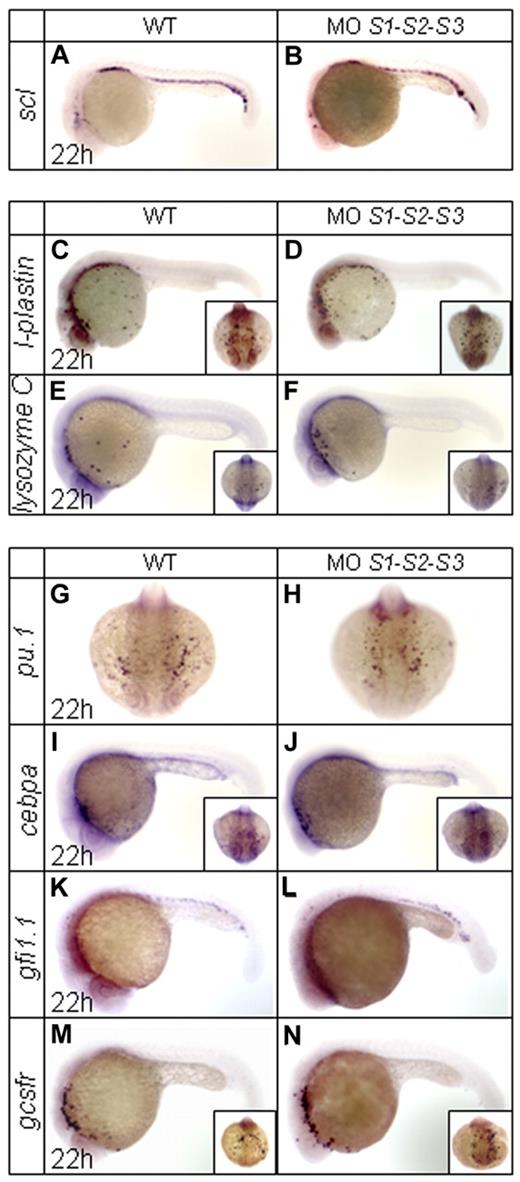
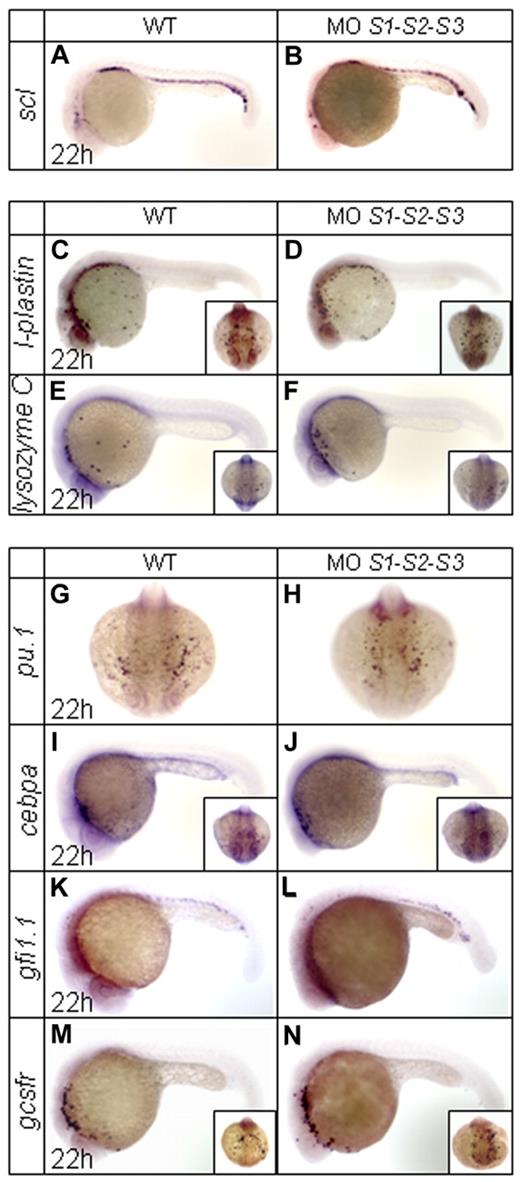
Expression of scl, a key transcription factor for the specification of all blood lineages,31 was not affected in SUMO morphants (Figure 2A-B). Similarly, there were no overt changes in the rostral expression of the macrophage markers l-plastin and lysozyme C32,33 (Figure 2C-F). Concerning granulopoiesis, key markers of early myeloid cells such as pu.1,34 cebpa,8 and gfi1.135 were unaffected in SUMO-deficient embryos (Figure 2G-L). Consistently, gcsfr, a marker of granulocytes that is mainly expressed in the A-LPM during primitive hematopoiesis,36 remained unchanged (Figure 2M-N). These results strongly suggest that SUMO deficiency only affects granulopoiesis within the ICM region.
WISH assays of other key primitive hematopoietic markers in WT and SUMO-deficient embryos at 22 hpf. (A-N) Expression profiles of scl (A-B), l-plastin (C-D), lysozyme C (E-F), pu.1 (G-H), cebpa (I-J), gfi1.1 (K-L), and gcsfr (M-N). Note that no obvious changes are observed for these tested markers.
WISH assays of other key primitive hematopoietic markers in WT and SUMO-deficient embryos at 22 hpf. (A-N) Expression profiles of scl (A-B), l-plastin (C-D), lysozyme C (E-F), pu.1 (G-H), cebpa (I-J), gfi1.1 (K-L), and gcsfr (M-N). Note that no obvious changes are observed for these tested markers.
SUMO deficiency favors myelopoiesis at the expense of erythropoiesis during primitive hematopoiesis in transgenic zebrafish
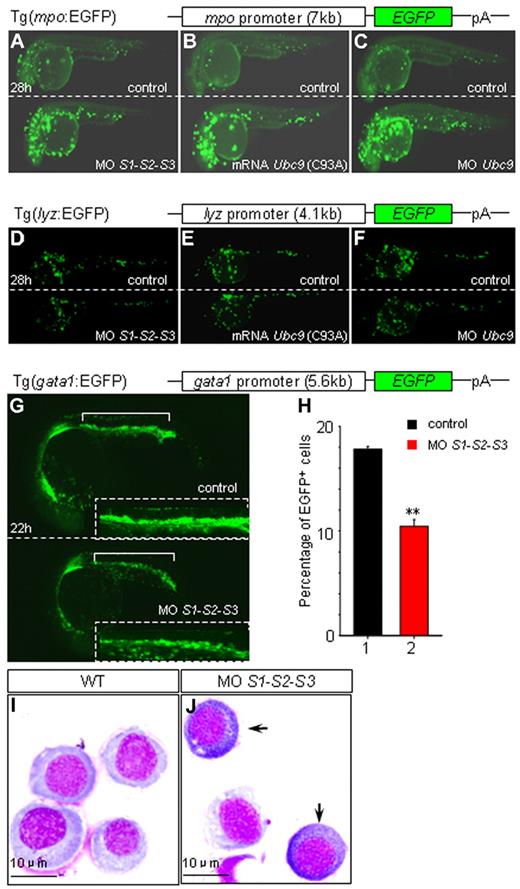
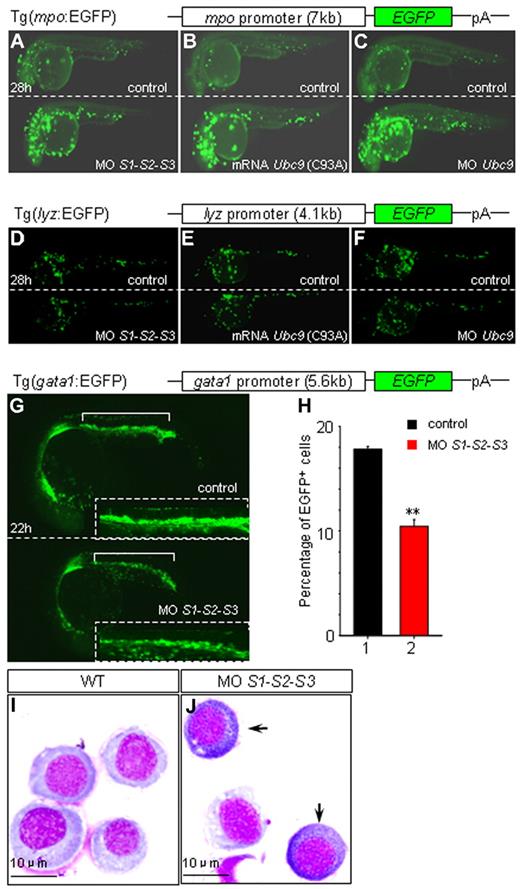
To confirm that the drastic increase in mpo expression was due to hyposumoylation, a stable zebrafish transgenic line expressing EGFP under the control of the mpo promoter was analyzed. As expected, EGFP expression in the embryos injected with SUMO or Ubc9 MOs, as well as Ubc9 (C93A) mRNAs, were strikingly induced from 28 hpf on (Figure 3A-C and supplemental Figure 2A-F). Moreover, in agreement with our WISH data, loss of SUMOs did not lead to the induction of EGFP expression in a lysozyme C promoter–driven zebrafish transgenic line (Figure 3D-F and supplemental Figure 2G-L).
Impacts of SUMO deficiency in different zebrafish transgenic lines. (A-F) Knock-down SUMOs, Ubc9, or overexpression of Ubc9 (C93A) results in an increase of EGFP in Tg(mpo:EGFP) embryos (A-C), and no such changes are observed in Tg(lyz:EGFP) embryos (D-F) at 28 hpf. Sibling controls and embryos injected with SUMO1-SUMO2-SUMO3 MOs (A and D), Ubc9 (C93A) mRNA (B and E), and Ubc9 MO (C and F) of Tg(mpo:EGFP) or Tg(lyz:EGFP), respectively. (G-J) MPCs undergo myelopoiesis in SUMO-deficient embryos. SUMO deficiency leads to a down-regulation of EGFP resulting from a decreased number of EGFP+ cells (10.45% ± 0.636%, n = 150) compared with that of sibling control (17.8% ± 0.283%, n = 140) in Tg(gata1:EGFP) at 22 hpf (G-H). Insets are the amplified views of corresponding ICM. **P < .01. (I-J) Myeloperoxidase staining of sorted EGFP+ cells from ICM of sibling controls (I) and SUMO-deficient embryos (J) of Tg(gata1:EGFP) at 22 hpf. Arrows indicate mpo+ cells.
Impacts of SUMO deficiency in different zebrafish transgenic lines. (A-F) Knock-down SUMOs, Ubc9, or overexpression of Ubc9 (C93A) results in an increase of EGFP in Tg(mpo:EGFP) embryos (A-C), and no such changes are observed in Tg(lyz:EGFP) embryos (D-F) at 28 hpf. Sibling controls and embryos injected with SUMO1-SUMO2-SUMO3 MOs (A and D), Ubc9 (C93A) mRNA (B and E), and Ubc9 MO (C and F) of Tg(mpo:EGFP) or Tg(lyz:EGFP), respectively. (G-J) MPCs undergo myelopoiesis in SUMO-deficient embryos. SUMO deficiency leads to a down-regulation of EGFP resulting from a decreased number of EGFP+ cells (10.45% ± 0.636%, n = 150) compared with that of sibling control (17.8% ± 0.283%, n = 140) in Tg(gata1:EGFP) at 22 hpf (G-H). Insets are the amplified views of corresponding ICM. **P < .01. (I-J) Myeloperoxidase staining of sorted EGFP+ cells from ICM of sibling controls (I) and SUMO-deficient embryos (J) of Tg(gata1:EGFP) at 22 hpf. Arrows indicate mpo+ cells.
It has been reported recently that MPCs within ICM could potentially differentiate into either erythrocytes or myelocytes.6,7 Because hyposumoylation favors expression of myeloid genes within MPCs, we wondered whether loss of sumoylation could actually promote myelopoiesis at the expense of erythropoiesis. To address this question, gata1 promoter–driven EGFP-transgenic embryos were injected with SUMO MOs. A clear decrease in the number of EGFP-positive cells in SUMO-deficient embryos was observed (Figure 3G-H), demonstrating a global down-regulation of erythropoiesis. These EGFP-positive cells from ICM were then sorted and myeloperoxidase staining was carried out. Intriguingly, in SUMO-deficient embryos only, some gata1-expressing progenitors turned out to be peroxidase positive (Figure 3I-J). These experiments demonstrate that hyposumoylation not only decreases the relative abundance of gata1-expressing erythroid progenitors, but also actually allows some of them to acquire myeloid features. Therefore, sumoylation plays a role in the cell fate decisions during hematopoietic differentiation.
Sumoylation of C/ebpα is implicated in the transcriptional regulation of myelopoiesis of MPCs during primitive hematopoiesis
C/ebpα activates myelopoiesis through the regulation of the promoters of several myeloid granule proteins, including Mpo,37 and is also implicated in lineage conversion.12,13,38 C/ebpα is sumoylated on a highly conserved lysine residue, triggering transcriptional repression.14,15 This prompted us to investigate the potential role of C/ebpα sumoylation in the lineage determination of MPCs during primitive hematopoiesis of zebrafish.
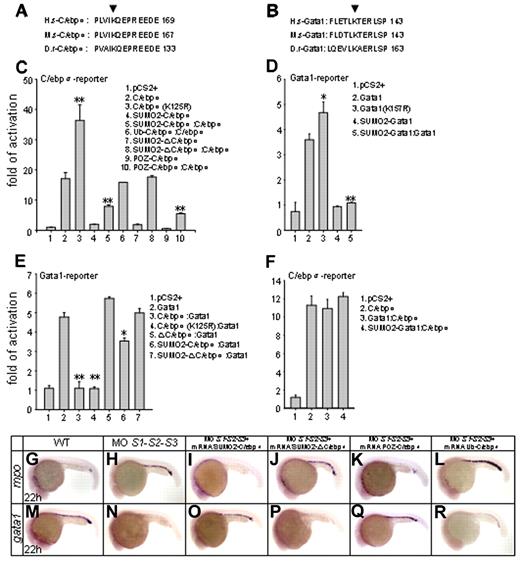
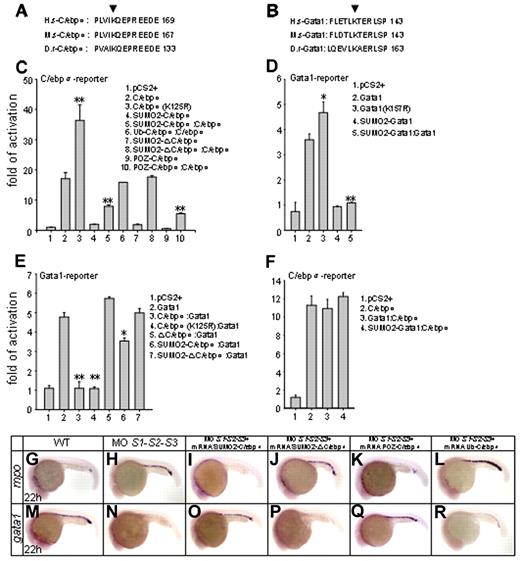
Homology blasting analysis indicated that lysine 125 of zebrafish C/ebpα was the evolutionarily conserved sumoylation modification site (Figure 4A). The 293T cells, which lack the endogenous C/ebpα,39 were cotransfected with the expression vectors for C/ebpα, unsumoylatable mutant C/ebpα (K125R) and SUMO1 or SUMO2, followed by immunoblotting with C/ebpα antibody. No sumoylated species were detected for the C/ebpα (K125R) mutant compared with the WT C/ebpα (supplemental Figure 3A). Immunoprecipitation and immunoblot analysis further revealed that the mutation of lysine 125 prevented conjugation of Flag-C/ebpα with GFP-SUMO2 (supplemental Figure 3B), indicating that this residue is indeed the target for modification by SUMO. 293T cells were transfected with a luciferase reporter containing a tetramer of C/ebpα–binding sites, along with various expression plasmids. The C/ebpα (K125R) mutant exhibited a significantly enhanced transactivation activity on the minimal C/EBP promoter compared with WT C/ebpα (Figure 4C lines 2 and 3), confirming that sumoylation of zebrafish C/ebpα at lysine 125 is critical for transcriptional repression. We also fused zebrafish SUMO2 in-frame with C/ebpα (SUMO2-C/ebpα) to mimic the constitutively sumoylated form of C/ebpα. In parallel, the POZ repression domain of the PLZF gene was fused to C/ebpα (POZ-C/ebpα) to mimic SUMO-mediated repression. As expected, the ability of the 2 mutants to activate transcription was significantly inhibited (Figure 4C lines 4 and 9) and the transcriptional activity of WT C/ebpα was significantly inhibited in the presence of either of these 2 fusions (Figure 4C lines 5 and 10). To verify that not just any fusion repressed C/ebpα, a ubiquitin-C/ebpα fusion was also constructed and had no effect on the transcriptional activity of WT C/ebpα (Figure 4C line 6). Critically, in vivo rescue assays showed that SUMO2-C/ebpα or POZ-C/ebpα mRNA could rescue SUMO-deficient phenotypes (Figure 4I,O,K,Q; supplemental Figure 4C,I,E,K; and supplemental Table 1), whereas ubiquitin-C/ebpα mRNA was ineffective (Figure 4L,R, supplemental Figure 4F,L, and supplemental Table 1). These results strongly suggest that sumoylation of C/ebpα plays an important role in transcription regulation and lineage determination of MPCs during zebrafish primitive hematopoiesis.
Ex vivo and in vivo biologic effects of C/ebpα sumoylation. (A-B) Alignment of conserved amino acid sequences of SUMO attachment of C/ebpα (A) and Gata1 (B). H.s indicates homo sapiens; M.s, Mus musculus; D.r, Danio rerio. Arrowheads indicate the evolutionarily conserved lysine residue within the consensus. (C-D) Sumoylation of C/ebpα (C) and Gata1 (D) is implicated in transcriptional repression. Luciferase activity assays were performed in 293T cells using various C/ebpα or Gata1 constructs indicated. The Renilla plasmid was used as an internal control. (E-F) Luciferase activity assays of transcriptional interplay between C/ebpα and Gata1. Note that whereas C/ebpα functions normally in the presence of Gata1 (F), it strongly inhibits the transcriptional activity of Gata1 (E). SD is derived from 3 independent transfection experiments. *P < .05; **P < .01. (G-R) In vivo rescue assay of C/ebpα mutants. Whereas SUMO2-C/ebpα (I and O) and POZ-C/ebpα (K and Q) fusions restore normal expression patterns of mpo and gata1 in SUMO-deficient embryos, SUMO2-ΔC/ebpα (J and P) becomes incapable, in agreement with its default in transcriptional activation (panel C lines 7 and 8). Ub-C/ebpα, serving as a negative control, does not rescue SUMO-deficient phenotypes (L and R). Ub indicates ubiquitin.
Ex vivo and in vivo biologic effects of C/ebpα sumoylation. (A-B) Alignment of conserved amino acid sequences of SUMO attachment of C/ebpα (A) and Gata1 (B). H.s indicates homo sapiens; M.s, Mus musculus; D.r, Danio rerio. Arrowheads indicate the evolutionarily conserved lysine residue within the consensus. (C-D) Sumoylation of C/ebpα (C) and Gata1 (D) is implicated in transcriptional repression. Luciferase activity assays were performed in 293T cells using various C/ebpα or Gata1 constructs indicated. The Renilla plasmid was used as an internal control. (E-F) Luciferase activity assays of transcriptional interplay between C/ebpα and Gata1. Note that whereas C/ebpα functions normally in the presence of Gata1 (F), it strongly inhibits the transcriptional activity of Gata1 (E). SD is derived from 3 independent transfection experiments. *P < .05; **P < .01. (G-R) In vivo rescue assay of C/ebpα mutants. Whereas SUMO2-C/ebpα (I and O) and POZ-C/ebpα (K and Q) fusions restore normal expression patterns of mpo and gata1 in SUMO-deficient embryos, SUMO2-ΔC/ebpα (J and P) becomes incapable, in agreement with its default in transcriptional activation (panel C lines 7 and 8). Ub-C/ebpα, serving as a negative control, does not rescue SUMO-deficient phenotypes (L and R). Ub indicates ubiquitin.
The transcriptional activity of Gata1 is regulated by sumoylation and interaction with C/ebpα
Gata1, a well-known key regulator of erythropoiesis and megakaryopoiesis, also contains a conserved sumoylation site25 (Figure 4B). A transient luciferase reporter assay showed that the unsumoylatable mutant, Gata1 (K157R), had a slightly stronger transactivation activity (Figure 4D lines 2 and 3). Conversely, the transcriptional activity of the SUMO2-Gata1 fusion mutant, which mimics the constitutively sumoylated form of Gata1, was significantly decreased (Figure 4D line 4). Moreover, the fusion mutant also antagonized the WT Gata1 activity (Figure 4D line 5). These results suggest that sumoylation of Gata1 is associated with transcriptional repression.
The leucine zipper domain of C/ebpα not only mediates DNA binding, but also interacts with other transcription factors involved in lineage-specific gene regulation.40 To investigate a possible mutual interference of C/ebpα and Gata1, expression vectors for C/ebpα, C/ebpα (K125R) ΔC/ebpα (which lacks the leucine zipper domain of C/ebpα), SUMO2-C/ebpα, and SUMO2-ΔC/ebpα were cotransfected with Gata1 in the presence of a Gata1-responsive reporter. The transcriptional activity of Gata1 was significantly inhibited by C/ebpα or C/ebpα (K125R; Figure 4E lines 3-4), but not by ΔC/ebpα (Figure 4E line 5). SUMO2-C/ebpα still suppressed the transcriptional activity of Gata1, but to a much lower extent (Figure 4E line 6). This repressive effect was abolished when SUMO2-ΔC/ebpα was examined (Figure 4E line 7). These data suggest that C/ebpα participates in Gata1 regulation, most likely through its C-terminal leucine zipper domain. We then performed immunoprecipitation analysis and revealed that C/ebpα interacted directly with Gata1 through the leucine zipper domain (supplemental Figure 3C). Conversely, the transcriptional activity of C/ebpα was not affected by Gata1 (Figure 4F). Moreover, in an in vivo rescue setting, the SUMO2-ΔC/ebpα was unable to reverse the SUMO deficiency–triggered phenotypes (Figure 4J,P, supplemental Figure 4D,J, and supplemental Table 1), whereas SUMO2-C/ebpα and POZ-C/ebpα did reverse these phenotypes.
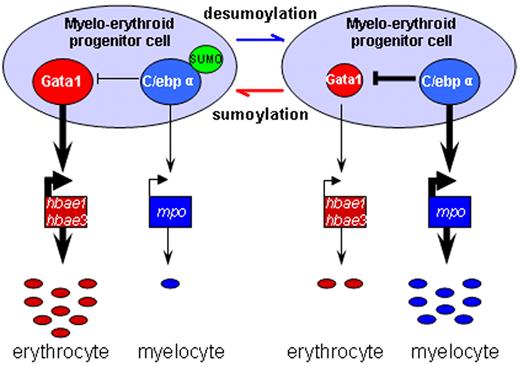
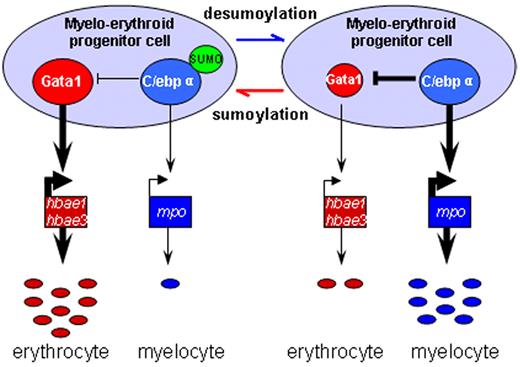
Our data suggest that C/ebpα, through sumoylation, may participate in lineage determination of MPCs in the ICM and act downstream or parallel to Pu.1 to affect Gata1, defining a new layer of complexity in the myelo-erythroid commitment cascade (Figure 5).
Schematic depiction of myelo-erythroid regulation of MPCs in the ICM. During normal primitive hematopoiesis, MPCs in the ICM predominantly differentiate into erythrocytes together with a few myelocytes (left). In SUMO-deficient embryos, there is an increased myelopoiesis of MPCs mediated by augmented transcriptional activity of desumoylated C/ebpα and its inhibition effect over Gata1 (right).
Schematic depiction of myelo-erythroid regulation of MPCs in the ICM. During normal primitive hematopoiesis, MPCs in the ICM predominantly differentiate into erythrocytes together with a few myelocytes (left). In SUMO-deficient embryos, there is an increased myelopoiesis of MPCs mediated by augmented transcriptional activity of desumoylated C/ebpα and its inhibition effect over Gata1 (right).
Discussion
In the P-LPM/ICM, MPCs are likely to represent the functional equivalent of mammalian common myeloid progenitors.6,7 Blood progenitors within the ICM are multipotent cells that differentiate into erythrocytes, neutrophils, and thrombocytes.5 To date, the balance between myelopoiesis and erythropoiesis of MPCs during primitive hematopoiesis has been proposed to be the outcome of antagonistic interactions between Gata1 and Pu.1.6,7 Our findings show that a loss of all SUMO paralogs favors myelopoiesis of MPCs in the ICM, and that C/ebpα sumoylation plays a pivotal role in the process of MPC lineage specification. SUMO may recruit HDACs17 and small interfering RNA–mediated knockdown of HDAC1 promotes myeloid differentiation in immature hematopoietic cell lines and inhibits erythroid differentiation in progenitor cells.41 Our rescue experiments demonstrate that the SUMO deficiency–triggered phenotype can be reversed by overexpressing constitutively sumoylated C/ebpα, revealing the critical role of C/ebpα sumoylation in lineage commitment. To directly address the role of C/ebpα in vivo, we tried to knock down endogenous zebrafish C/ebpα using MO but were not successful. The expressions of the C/ebpα downstream target genes showed no obvious changes when we injected C/ebpα MO, even at a high dose. This was not unexpected because several different C/ebpα MOs had been tested without success previously.9 We also tried the most recently developed approach, custom-made zinc-finger nucleases made by OPEN (Oligomerized Pool ENgineering)–mediated gene knockout,42 but failed to get any working zinc-finger nucleases (T.X.L., unpublished data). Such a technical obstacle may have been due to the fact that C/ebpα is a single-exon gene that is very rich in GC.
Among the important transcription factors related to the myelo-erythroid differentiation cascade, Gata1 may also be sumoylated,25 although the impact of Gata1 sumoylation is still under debate.25,43,44 Our luciferase data indicate that the nonsumoylatable Gata1 (K157R) mutant has slightly increased transcriptional activity, which is in agreement with the fact that Gata1 sumoylation inhibits erythropoiesis in SENP1-null mice.44 However, in SUMO-deficient embryos, although Gata1 is normally positively auto-regulated,45 gata1 expression was reduced and this was followed by down-regulation of its downstream target genes. Accordingly, expression of EGFP in gata1-EGFP–transgenic embryos was reduced when SUMOs were knocked down. These observations suggest that the transcriptional consequences of hyposumoylation of Gata1 transcription regulation were overwhelmed by those of C/ebpα. Indeed, our findings demonstrate that the transcriptional activity of Gata1 could be inhibited by C/ebpα through a direct protein-protein interaction.
Whereas the prevailing model of MPCs commitment may not perfectly explain why primitive hematopoiesis is dominated by erythropoiesis in the presence of abundant C/ebpα, our results provide the first evidence that the sumoylation of C/ebpα might contribute to the cell fate decision of MPCs in the ICM during primitive hematopoiesis in zebrafish.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Nelly Kieffer and Dr Hanyi Zhuang for critical reading of the manuscript; Dr Shuo Lin, Dr Alister C. Ward, and Dr Giannino Del Sal for providing gfi1.1, gcsfr, and gata1 luciferase reporter plasmids, respectively; and Yan Fang Fu, Ting Ting Du, and Wu Zhang for their excellent technical support.
This work was supported by grants from National Natural Science Foundation of China (30525006, 31071290, and 30525019), by the Oriental Scholar Program of Shanghai Municipal Education Commission, and by the Actions Thématiques et Incitatives sur Programme of the Centre National de la Recherche Scientifique and BNP Paribas.
Authorship
Contribution: H.Y. and J.Z. performed experiments and analyzed data; M.D., Y.Z., Y.C., and Y.J. performed experiments; J.Z., S.J.C., H.d.T., Z.C., and T.X.L. designed experiments and analyzed data; and J.Z. designed experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Jun Zhu, Rui-Jin Hospital, 197 Rui-Jin Rd II, Shanghai 200025, China; e-mail: jzhu1966@yahoo.com.cn or jun.zhu@paris7.jussieu.fr; Dr Ting Xi Liu, Institute of Health Sciences, 225 South Chong Qing Rd, Shanghai 200025, China; e-mail: txliu@sibs.ac.cn; or Dr Zhu Chen, Rui-Jin Hospital, 197 Rui-Jin Rd II, Shanghai 200025, China; e-mail: zchen@stn.sh.cn.
References
Author notes
H.Y. and J.Z. contributed equally to this study.