The expression of HLA-G by malignant cells has been proposed as a tumor escape mechanism from immunosurveillance. However, although the inhibitory effect of HLA-G on antitumoral immune effectors has been documented in vitro, it remains to be resolved in vivo. In this context, the development of an animal model is now a priority to establish the proof of concept that an HLA-G+ tumor cell develops and tolerizes the host antitumor immune response in vivo. In the present study, we provide the first in vivo evidence of such a role by a xenotumor model in mice based on the interactions between human HLA-G and the murine paired immunoglobulin–like receptor-B (PIR-B). We demonstrate that human tumor cells expressing HLA-G grow in an immunocompetent host by affecting both innate and adaptive immunity. Expansion of blood myeloid–derived CD11b+Gr1+PIR-B+ suppressor cells, loss of peripheral T cells, and cytokinic balance in favor of Th2 versus Th1/Th17 constitute the main mechanisms by which HLA-G promotes tumor expansion. These data demonstrate for the first time that HLA-G plays a crucial role in in vivo tumor evasion. Finally, blocking HLA-G function by a specific Ab inhibits the in vivo development of the tumor, offering a new innovative therapeutic strategy in cancer.
Introduction
Although various immune effector cells are recruited to a tumor site, their antitumor functions are down-regulated largely in response to tumor-derived signals.1 Expression of the tolerogenic HLA-G molecule in the tumor milieu represents a mechanism that may favor tumor survival and drive the abortive activation of immune cells, resulting in tumor escape from the host immune system.2 Understanding such a mechanism is an important challenge in developing optimal immunotherapeutic strategies.
HLA-G differs from the classic HLA class I molecules in its genetic diversity, expression, structure, and functions. HLA-G displays a limited polymorphism and its physiologic expression is restricted to trophoblast, thymus, cornea, and erythroid and endothelial precursors, but it can be induced in various tissues under pathologic situations such as tumors and viral infections.3 A distinct feature of HLA-G is the alternative splicing of the primary transcript, which leads to 4 membrane-bound (HLA-G1 to HLA-G4) and 3 soluble isoforms (HLA-G5 to HLA-G7).3 HLA-G is known to exert inhibitory effects on both innate and adaptive effectors through direct binding to inhibitory receptors: immunoglobulin-like transcript-2 (ILT-2, also known as LILRB1 or CD85j), which is expressed by lymphoid and myeloid cells; ILT-4 (LILRB2 or CD85d), which is expressed by myeloid cells; and KIR2DL4 (specific for HLA-G), which is expressed constitutively by some CD8+ T cells and CD56bright natural killer (NK) cells and is inducible on the CD56dim subset.3 Although ILT-2 and ILT-4 can bind to other HLA class I molecules, their affinity is highest for HLA-G.4
Over the past few years, HLA-G has been shown to be expressed in many types of primary tumors and metastases and in malignant effusions,3 and it can also be found on tumor cells and tumor-infiltrating cells.5 The clinical relevance of HLA-G in cancer is supported by the following observations: (1) HLA-G expression is associated with malignant transformation and is never observed in the healthy surrounding tissues6 ; (2) HLA-G is expressed in liquid and solid tumors of high histological grades and advanced clinical stages7,8 ; and (3) the use of HLA-G as a prognostic marker has been proposed because HLA-G expression in biopsies and/or high levels of soluble HLA-G in plasma have been significantly correlated with poor prognosis in different types of cancer.7,9,,,,–14 All of these data highlight a role for HLA-G in the immune surveillance of tumors and progression of the disease.
The inhibitory effect of HLA-G on antitumoral immune effectors has now been documented in vitro,15 but the development of an animal model is a priority to establish the proof of concept that an HLA-G+ tumor cell can develop and tolerize the host antitumor immune response in vivo. This model would also provide the opportunity to test the effect of therapies interfering with HLA-G expression and/or function on the clinical course of malignant diseases. Although there is no murine homolog of HLA-G, the present study was possible because human HLA-G can bind and mediate a signal via the murine receptor paired immunoglobulin-like receptor-B (PIR-B), the homolog of human ILTs.16,17 We developed a xenotumor model in mice to show that human tumor cells expressing HLA-G can grow in an immunocompetent host and that blocking HLA-G function with a specific Ab inhibits tumor development.
Methods
Mice
Female C57BL/6, CBA/J, and Balb/c mice (6-10 weeks of age) were obtained from Charles River Laboratories. All experimental protocols were approved by the ethics review committee for animal experimentation of the Hôpital Saint-Louis (Paris, France) and followed the guiding principles for the care and use of animals approved by our local committee.
Cell culture
M8 is an HLA-A+, HLA-B+, HLA-C+, and HLA-E+ but HLA-G− tumor cell line.18 Stably transfected cells were obtained as described previously using the pcDNA3.1 vector containing HLA-G1 (M8-HLA-G1) cDNA.18 M8 cells transfected with the pcDNA3.1 vector alone were used as a negative control (M8-pcDNA), as described previously. The cell lines Fon, a tumor cell line naturally expressing HLA-G,19 and YAC-1, a murine T-lymphoma cell line (ATCC) were maintained in complete medium.
Subcutaneous tumor induction
Groups of mice were injected subcutaneously at day 0 with 10 × 106 M8-pcDNA, M8-HLA-G1, Fon−, or Fon+ cells resuspended in PBS. Where indicated, tumor cells were pretreated for 1 hour with the anti–HLA-G1 87G mAb or the isotypic control (IgG2a; EXBIO Diagnostics) before injection. The blocking anti–PIR-B Ab (clone 259.2; a kind gift of Genentech) was administered subcutaneously at days −1, 0, and 1 at 100 μg per injection per mouse. The tumor area was measured with digital calipers at the indicated time points after injection. Tumor volume was estimated by the formula: (L × W2)/2 (Figure 1A).
HLA-G1 allows tumor growth in immunocompetent mice. (A) Schematic representation of experimental procedures. (B) Mice were injected subcutaneously with 10 × 106 M8-pcDNA or M8-HLA-G1 cells and tumor growth was monitored at indicated time points. Data represent the means ± SD from 1 representative experiment with 3 mice in each group. Insert shows cell-surface expression of HLA-G1 by the M8-pcDNA and M8-HLA-G1 cell lines and the isotype control. (C) Photographs of subcutaneous tumors 3 days after injection of tumor cells. (D) Expression of HLA-G and melanoma antigens at the tumor site at day 14 (d14). (E) M8-HLA-G1 cells were pretreated with the 87G Ab (M8-HLA-G1 + anti-HLA-G) or the isotype control (M8-HLA-G1 + control Ab) before subcutaneous injection into Balb/c mice, and tumor growth was monitored at the indicated time points. Data represent the means ± SD from 1 representative experiment with 3 mice in each group.
HLA-G1 allows tumor growth in immunocompetent mice. (A) Schematic representation of experimental procedures. (B) Mice were injected subcutaneously with 10 × 106 M8-pcDNA or M8-HLA-G1 cells and tumor growth was monitored at indicated time points. Data represent the means ± SD from 1 representative experiment with 3 mice in each group. Insert shows cell-surface expression of HLA-G1 by the M8-pcDNA and M8-HLA-G1 cell lines and the isotype control. (C) Photographs of subcutaneous tumors 3 days after injection of tumor cells. (D) Expression of HLA-G and melanoma antigens at the tumor site at day 14 (d14). (E) M8-HLA-G1 cells were pretreated with the 87G Ab (M8-HLA-G1 + anti-HLA-G) or the isotype control (M8-HLA-G1 + control Ab) before subcutaneous injection into Balb/c mice, and tumor growth was monitored at the indicated time points. Data represent the means ± SD from 1 representative experiment with 3 mice in each group.
Immunohistochemistry
M8-HLA-G1 tumors were harvested 14 days after injection, embedded in paraffin, and sliced into 4-μm sections. HLA-G expression was analyzed using the pan-HLA-G MEM-G/02 mAb (EXBIO Diagnostics) and the pan-melanoma HMB45 mAb (Novus), as described previously.20
Preparation of cell suspensions and cell proliferation assay
Spleens and draining lymph nodes were removed at the indicated times. Cells were dissociated through a cell strainer (100 μm pore; BD Falcon). Peripheral blood was recovered on EDTA and centrifuged at 600g for 10 minutes to separate plasma and cells. Mononuclear cells were then isolated by density gradient centrifugation using Histopaque (Sigma-Aldrich). After washing, 105 mononuclear cells were seeded in 96-well round-bottom culture plates. Mononuclear cell proliferation was tested in response to 105 tumor cells treated with 50 μg/mL mitomycin C (Sigma-Aldrich). Where indicated, M8-HLA-G1 cells were pretreated with the anti–HLA-G1 87G mAb for 1 hour at 37°C. Proliferation was assessed after 3 days of coculture by [3H]-thymidine incorporation (1 μCi/well) during the last 16 hours of culture.
Cytotoxicity assay
Mice were implanted subcutaneously with 10 × 106 tumor cells. Eighteen hours before the in vitro assay, mice were injected IP with 200 μg of poly(I):poly(C) (Sigma-Aldrich).21 Such pretreatment was required to detect NK cytotoxic activity in vitro and did not affect the HLA-G1+ tumor growth (data not shown). Spleens were removed and NK cells were isolated from splenocytes using a mouse NK isolation kit (Dynal; Invitrogen). Total cells or purified NK cells were coincubated at different ratios with the indicated 51Cr-labeled target cells for a classic 4-hour 51Cr-release assay. Where indicated, Gr1+ cells purified from spleens using a Gr1-PE mAb (BD Biosciences) and the EasySep Mouse PE Positive Selection Kit (StemCell Technologies) were used as a third party at a ratio of 1 Gr1+ cell:1 NK. Gr1− cells were used as controls. The percentage of specific lysis was calculated as described previously.19
Antibodies and flow cytometry
The following mAbs were used in this study: purified anti–PIR-A/B (6C1), FITC–anti-CD8a (53-6.7), anti-CD40 (3/23), anti-CD80 (16-10A1), anti-CD86 (GL1), anti-CD11b (M1/70), PE–anti-CD4 (GK1.5), anti–PIR-A/B (6C1), anti–Ly-6G (1A8), and PE-Cy5 anti-CD3e (145-2C11) (all from BD Biosciences); biotin goat anti–rat IgG, FITC–anti-CD25 (7D4), PE–anti-CD28 (37.51), anti–MHC class II (NIMR-4), anti–NK1.1 (PK136), and PE-Cy5 streptavidin (from Beckman Coulter); and PE–anti–HLA-G1 (MEM-G/9; EXBIO Diagnostics). Relevant isotype controls were purchased from the corresponding supplier. Cells were incubated with the indicated fluorochrome-conjugated antibodies in PBS/1% BSA in the presence of an optimal concentration of mouse Fc Block (anti-CD16/CD32; BD Biosciences) for 30 minutes at 4°C. After washing, acquisition was performed on an Epic XL flow cytometer (Beckman Coulter) using Expo32 Version 1.2 software.
Confocal microscopy
Gr1+ cells were purified from the spleens of M8-HLA-G1–injected mice and incubated with purified HLA-G protein (180 ng/mL)22,23 for 45 minutes at 4°C to allow HLA-G binding. PIR-B expression on all Gr1+ cells was confirmed by flow cytometry. Where indicated, the cells were preincubated with 50 μg/mL blocking anti–PIR-B Ab (clone 259.2) for 45 minutes at 4°C. HLA-G was then detected with anti–HLA-G and goat anti–mouse Ab conjugated to FITC (BD Biosciences). Cells were analyzed by confocal microscopy, as described previously.24
ELISA
Plasma was harvested after centrifugation of blood for 10 minutes at 600g, aliquoted, and stored at −80°C until ELISA was performed. Cytokines were quantified using the Th1/Th2/Th17 Multi-Analyte ELISArray kit (Tebu-Bio).
Detection of the humoral response
Two different methods were used to detect the humoral response: slot-blot analysis and flow cytometry. For the former, 5 × 106 M8-pcDNA cells were washed with cold PBS, and the pellet was lysed and boiled for 5 minutes. After centrifugation, the supernatant was blotted on 10% SDS-PAGE gels, and proteins were transferred on Immobilon-P membranes (Millipore). Blots were saturated with 5% fat-free milk/PBS/0.2% Tween 20 for 1 hour and then incubated with the different mouse plasma samples using the Mini-Protean II multiscreen apparatus (Bio-Rad). Detection was performed using HRP-conjugated anti-mouse Ab (Sigma-Aldrich) with the ECL kit (Amersham). For flow cytometry, mouse plasma diluted in PBS/0.1% BSA was incubated with M8-pcDNA cells for 30 minutes at 4°C. Mouse IgG specific for melanoma antigens was then detected by incubation with PE-conjugated goat anti–mouse IgG for 30 minutes at 4°C.
Statistical analysis
Unpaired 2-tailed Student t tests were performed using Prism Version 5.01 software (GraphPad). P values < .05 were considered statistically significant.
Results
HLA-G1–expressing tumor cells grow in immunocompetent mice
Using a human tumor cell line transfected or not (M8-pcDNA) with HLA-G1 cDNA (M8-HLA-G1), we described previously that HLA-G1+ tumor cells are protected in vitro from cytotoxic effectors.25,,–28 To provide in vivo evidence of the role of HLA-G in tumor escape from the immune response, in the present study we have developed for the first time an HLA-G+ xenotumor model in mice. We first injected M8-pcDNA or M8-HLA-G1 tumor cells subcutaneously in immunocompetent mice. Whereas 106 cells were not sufficient to allow tumor growth (data not shown), 10 × 106 M8 tumor cells expressing HLA-G1 grew rapidly under the skin of Balb/c, CBA, and C57Bl/6 mice. In contrast, M8 cells transfected with the vector alone were rejected immediately (Figure 1B-C). M8-HLA-G1 tumor cells grew as rapidly as 1 day and persisted for 2 weeks before being rejected because of their xenogenic status. Therefore, 2 phases were defined: the “tum+” phase corresponds to the period when the tumor was macroscopically visible (from days 1-15), followed by the “tum−” phase, when the tumor was not visible. Immunohistochemical staining on serial sections of M8-HLA-G1 tumors showed a colocalization between HLA-G+ and melanoma antigen–positive cells, attesting to the in vivo growth of the HLA-G1+ tumor (Figure 1D). Therefore, human HLA-G1 expressed by tumor cells induces an immune state that allows tumor growth in immunocompetent mice.
M8-HLA-G1 cells were treated with an HLA-G–specific blocking Ab and then injected subcutaneously. Masking HLA-G1 on tumor cells reversed the growth-promoting effect of HLA-G1, because tumor size was comparable to that of M8-pcDNA control cells (Figure 1E). M8-HLA-G1 cells preincubated with an irrelevant Ab grew in the same way as M8-HLA-G1 cells. These results demonstrate the direct involvement of HLA-G1 in the growth of HLA-G1+ tumors.
Proliferation of mononuclear cells from tumor-bearing mice
To dissect the immune mechanisms involved in the tum+ and tum− phases, we first analyzed the ex vivo proliferative capacities of mononuclear cells isolated from the blood, spleens, and draining lymph nodes at different time points after tumor cell injection. These mononuclear cells are essentially composed of T cells (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Mononuclear cells purified from mice injected with M8-pcDNA or M8-HLA-G1 tumor cells proliferated in the same way in response to their antigenic stimuli (Figure 2A). Two patterns of proliferation were observed. First, during the tum+ phase (ie, day 6 and day 13), mononuclear cells from both groups of mice had a high proliferative capacity in response to M8-pcDNA cells. In contrast, proliferation in response to M8-HLA-G1 cells was significantly lower, which confirms the in vitro inhibitory effect of HLA-G1 toward T cells.29 This inhibitory effect was directly due to HLA-G1, because a specific HLA-G blocking mAb restored the proliferation of mononuclear cells almost completely in response to M8-HLA-G1 cells (Figure 2B). Second, 2 weeks after injection, during the tum− phase (day 20 and day 27), the proliferative ability of mononuclear cells was consistently decreased, more so in response to M8-HLA-G1 than to M8-pcDNA cells (Figure 2A). This increased response was partially blocked by a specific mAb to HLA-G (Figure 2B), showing that an immune response against HLA-G1 was mounted. A control Ab had no effect on proliferation of mononuclear cells (data not shown). Similar results were obtained at a distinct responder:stimulator ratio (supplemental Figure 1). The fact that proliferative capacities were lost along with tumor development may indicate that the immune cells have acquired effector or tolerogenic functions.
Proliferation of mononuclear cells is not affected in mice injected with M8-HLA-G1 cells. (A) Balb/c mice were injected subcutaneously with M8-pcDNA cells (left panel) or M8-HLA-G1 cells (right panel). At the indicated time points, mononuclear cells were isolated from lymph nodes (LN), spleens, or peripheral blood (PBMC), and their proliferative capacity was tested in response to M8-pcDNA or M8-HLA-G1 cells at a responder:stimulator ratio of 1:1. Mononuclear cells cultured in medium alone were used as a control. Results represent the means ± SD of triplicates from 1 representative experiment of 3. The numbers indicated between parentheses are the P values. (B) Experiments were conducted as described in panel A except that M8-HLA-G1 cells in the presence of the anti–HLA-G 87G mAb were used. Left panel shows mice injected with M8-pcDNA cells; right panel, mice injected with M8-HLA-G1 cells. Results represent the means ± SD of triplicates from 1 representative experiment of 3. *P < .05; **P < .001; ***P < .005.
Proliferation of mononuclear cells is not affected in mice injected with M8-HLA-G1 cells. (A) Balb/c mice were injected subcutaneously with M8-pcDNA cells (left panel) or M8-HLA-G1 cells (right panel). At the indicated time points, mononuclear cells were isolated from lymph nodes (LN), spleens, or peripheral blood (PBMC), and their proliferative capacity was tested in response to M8-pcDNA or M8-HLA-G1 cells at a responder:stimulator ratio of 1:1. Mononuclear cells cultured in medium alone were used as a control. Results represent the means ± SD of triplicates from 1 representative experiment of 3. The numbers indicated between parentheses are the P values. (B) Experiments were conducted as described in panel A except that M8-HLA-G1 cells in the presence of the anti–HLA-G 87G mAb were used. Left panel shows mice injected with M8-pcDNA cells; right panel, mice injected with M8-HLA-G1 cells. Results represent the means ± SD of triplicates from 1 representative experiment of 3. *P < .05; **P < .001; ***P < .005.
Cytotoxic activity of NK cells from tumor-bearing mice
Because NK cells have a pivotal role in tumor elimination, we next examined the functional involvement of such cells in the immune-mediated control of tumor growth. Purified NK cells efficiently killed the YAC-1 cell line, which is known as a target sensitive to NK-cell lysis (Figure 3A). Then we analyzed the NK-lytic activity toward M8 cells. Results showed that 3 days after injection, cytotoxic activity against M8-HLA-G1 cells was specifically enhanced in mice injected with M8-HLA-G1 compared with mice injected with M8-pcDNA tumor cells. In contrast, 11 days after injection, NK cytotoxic activity was drastically decreased in both groups of mice (Figure 3A). The same pattern was observed with total splenocytes used as effector cells (Figure 3B). Importantly, the cytotoxic activities of splenocytes isolated from M8-HLA-G1–injected mice were decreased compared with those from purified NK cells, indicating that a suppressive population was present within total splenocytes in these mice.
HLA-G1 influences cytotoxicity against tumor cells. (A-B) Balb/c mice were injected subcutaneously with M8-pDNA or M8-HLA-G1 tumor cells at day 0. A classic 4-hour 51Cr-release cytotoxic assay was conducted with NK cells purified from spleens (A) or with total splenocytes (B) as effectors, and YAC-1, M8-pcDNA, or M8-HLA-G1 cells as targets. Results are expressed as the mean percentage lysis from duplicates. Standard deviations were below 5%. Data are from 1 representative experiment of 3.
HLA-G1 influences cytotoxicity against tumor cells. (A-B) Balb/c mice were injected subcutaneously with M8-pDNA or M8-HLA-G1 tumor cells at day 0. A classic 4-hour 51Cr-release cytotoxic assay was conducted with NK cells purified from spleens (A) or with total splenocytes (B) as effectors, and YAC-1, M8-pcDNA, or M8-HLA-G1 cells as targets. Results are expressed as the mean percentage lysis from duplicates. Standard deviations were below 5%. Data are from 1 representative experiment of 3.
HLA-G1 induces the loss of peripheral T cells and the emergence of a myeloid-suppressive population
To define the immune subpopulations present in the blood, spleens, and draining lymph nodes of tumor-injected mice, mononuclear cells were isolated at different time points after injection and their phenotype analyzed (Figure 4 and supplemental Table 1). The most striking results were obtained from PBMCs, in which T cells were almost completely absent from the blood of mice injected with M8-HLA-G1 cells compared with naive mice or mice injected with M8-pcDNA cells. This was true of both CD4+ and CD8+ T cells. These differences were amplified over time (Figure 4 and supplemental Table 1). The expression of CD80, CD40, and MHC class II on CD11c+ cells was slightly increased in HLA-G1+ tumor–bearing mice, whereas the expression of CD86 was not affected. Finally, expression of the PIR-A/B receptor, the homolog of human ILTs, and the percentage of PIR-A/B+ cells were consistently augmented in HLA-G1+ tumor–bearing mice.
Loss of T cells and emergence of MDSCs in the peripheral blood of mice injected with M8-HLA-G1 tumor cells. Mice were killed after they rejected tumors completely (day 17). After purification of mononuclear cells from the peripheral blood, cells were stained with the indicated antibodies and analyzed by flow cytometry. Data are from 1 representative experiment of 3.
Loss of T cells and emergence of MDSCs in the peripheral blood of mice injected with M8-HLA-G1 tumor cells. Mice were killed after they rejected tumors completely (day 17). After purification of mononuclear cells from the peripheral blood, cells were stained with the indicated antibodies and analyzed by flow cytometry. Data are from 1 representative experiment of 3.
HLA-G has been found to induce the emergence of myeloid-derived suppressive cells (MDSCs), with a CD11b+ Gr1+ phenotype directly involved in the prolongation of allogeneic skin graft survival.30 Accordingly, a CD11bhigh Gr1high population was detected in the peripheral blood of mice injected with M8-HLA-G1 cells as early as 1 day after tumor cell injection (3.9% vs 1.05% in mice injected with M8-pcDNA cells). This subpopulation was expanded along with the growth of the M8-HLA-G1 tumor (from 3.9%-8.5%), but not in mice injected with M8-pcDNA cells (Figure 4 and supplemental Table 1). These cells also emerged to a lesser extent in the spleen, but were absent from draining lymph nodes (supplemental Table 1).
No other significant differences between mice injected with M8-pcDNA and M8-HLA-G1 tumor cells were observed in draining lymph nodes or spleens throughout the course of the study (supplemental Table 1).
HLA-G1–expanded MDSCs express PIR-B and are suppressive
To further characterize the potential involvement of MDSCs in HLA-G–mediated tumor escape, we first looked at the expression of PIR-B on MDSCs. As shown in Figure 5A-B, MDSCs purified from the blood or spleens of tumor-bearing mice and defined as CD11b+Gr1high cells were all PIR-B+, making them susceptible to the effects of HLA-G. This was confirmed by the binding of HLA-G to MDSCs, as revealed the first time by confocal microscopy (Figure 5C).
MDSCs induced by HLA-G express PIR-B and are immunosuppressive. (A-B) Mice injected with M8-pcDNA cells, M8-HLA-G1 cells, or M8-HLA-G1 cells pretreated with the 87G mAb (M8-HLA-G1 + anti–HLA-G) were killed at day 21 after injection. Mononuclear cells were purified from blood (A) or spleens (B) and stained with the indicated antibodies. Analyses were performed by flow cytometry. Data are from 1 representative mouse of 3. (C) Immunostaining of purified HLA-G protein bound to MDSCs. Gr1+ cells were purified from spleens of mice injected with M8-HLA-G1 cells. The purity (> 95%) was attested by flow cytometry (inset). Cells were incubated with HLA-G− medium (control), a purified form of HLA-G at 180 ng/mL (HLA-G), or HLA-G + anti–PIR-B (clone 259.2). HLA-G binding to cells was detected by anti–HLA-G and goat anti–mouse conjugated to FITC through confocal microscopy analysis. (D) NK cells were purified from spleens of M8-HLA-G1–injected mice 3 days after injection. Gr1+ cells were purified from spleens of mice injected with M8-pcDNA or M8-HLA-G1 cells 21 days after injection. The Gr1− counterpart was also tested. A classic 4-hour 51Cr-release cytotoxic assay was conducted with NK cells as effectors (E), M8-HLA-G1 cells as targets (T), and Gr1+ or Gr1− cells as a third party (I). Results are expressed as the mean percentage lysis from triplicates. Data are from 1 representative mouse of 3. (E) Mice were injected subcutaneously with M8-pcDNA or M8-HLA-G1 cells alone at day 0 or with an anti–PIR-B Ab (clone 259.2) at days −1, 0, and 1, and M8-HLA-G1 cells at day 0 (M8-HLA-G1 + anti-PIR-B). Tumor growth was monitored at the indicated time points. Data represent the means ± SD from 1 representative experiment with 3 mice in each group. Inset shows cell-surface expression of PIR-B on PBMCs of M8-HLA-G1–injected mice detected with the clone 259.2.
MDSCs induced by HLA-G express PIR-B and are immunosuppressive. (A-B) Mice injected with M8-pcDNA cells, M8-HLA-G1 cells, or M8-HLA-G1 cells pretreated with the 87G mAb (M8-HLA-G1 + anti–HLA-G) were killed at day 21 after injection. Mononuclear cells were purified from blood (A) or spleens (B) and stained with the indicated antibodies. Analyses were performed by flow cytometry. Data are from 1 representative mouse of 3. (C) Immunostaining of purified HLA-G protein bound to MDSCs. Gr1+ cells were purified from spleens of mice injected with M8-HLA-G1 cells. The purity (> 95%) was attested by flow cytometry (inset). Cells were incubated with HLA-G− medium (control), a purified form of HLA-G at 180 ng/mL (HLA-G), or HLA-G + anti–PIR-B (clone 259.2). HLA-G binding to cells was detected by anti–HLA-G and goat anti–mouse conjugated to FITC through confocal microscopy analysis. (D) NK cells were purified from spleens of M8-HLA-G1–injected mice 3 days after injection. Gr1+ cells were purified from spleens of mice injected with M8-pcDNA or M8-HLA-G1 cells 21 days after injection. The Gr1− counterpart was also tested. A classic 4-hour 51Cr-release cytotoxic assay was conducted with NK cells as effectors (E), M8-HLA-G1 cells as targets (T), and Gr1+ or Gr1− cells as a third party (I). Results are expressed as the mean percentage lysis from triplicates. Data are from 1 representative mouse of 3. (E) Mice were injected subcutaneously with M8-pcDNA or M8-HLA-G1 cells alone at day 0 or with an anti–PIR-B Ab (clone 259.2) at days −1, 0, and 1, and M8-HLA-G1 cells at day 0 (M8-HLA-G1 + anti-PIR-B). Tumor growth was monitored at the indicated time points. Data represent the means ± SD from 1 representative experiment with 3 mice in each group. Inset shows cell-surface expression of PIR-B on PBMCs of M8-HLA-G1–injected mice detected with the clone 259.2.
We also found that the PIR-B+ MDSC subpopulation was increased in both the blood and the spleens from M8-HLA-G1 tumor–bearing mice compared with M8-pcDNA mice. Interestingly, the expansion of PIR-B+ MDSCs was inhibited in both compartments when mice were injected with M8-HLA-G1 cells pretreated with an anti–HLA-G Ab, indicating that HLA-G is directly implicated in PIR-B+ MDSC expansion (Figure 5A-B).
We next determined whether HLA-G–expanded MDSCs were suppressive. MDSCs from M8-pcDNA or M8-HLA-G1 mice were purified according to Gr1 expression and then used as third-party cells in a NK cytotoxicity assay (supplemental Figure 2). Results showed that Gr1+ cells from M8-HLA-G1 mice inhibited the cytotoxic activity of NK cells, whereas their Gr1− counterparts did not (Figure 5D). Remarkably, such inhibition was not observed when Gr1+ cells from M8-pcDNA mice were used, showing that the suppressive activity of MDSCs is selectively acquired in an HLA-G–induced environment.
Finally, the involvement of PIR-B in the immunosuppressive function of HLA-G was investigated using the only available blocking anti–PIR-B Ab previously established for in vitro neuronal studies.31 The injection of this Ab in our in vivo tumor model did not inhibit the tumor growth of M8-HLA-G1 cells in mice (Figure 5E), nor did it inhibit the binding of HLA-G to PIR-B on MDSCs, as revealed by confocal microscopy (Figure 5C).
HLA-G1 affects the humoral response in vivo
Because PIR-B is expressed by B cells, we hypothesized that HLA-G may affect B-cell function.32 We first quantified the total amount of plasma IgG using ELISA, and found no significant differences between the groups of mice (data not shown). In contrast, we observed a dramatic decrease of the Ab response against antigens derived from the human M8 melanoma cell line in HLA-G1+ tumor–bearing mice at day 27 after injection (Figure 6A). These results were confirmed by cytometry to detect murine antibodies against antigens derived from the human M8 melanoma cell line. For this purpose, M8 cells served as targets that were incubated with the respective mouse plasma. Results showed that the plasma from M8-HLA-G1 mice was less reactive against M8 antigens compared with the plasma from control mice (Figure 6B). These data demonstrate that the antigen-specific B-cell response is affected in HLA-G1+ tumor–bearing mice.
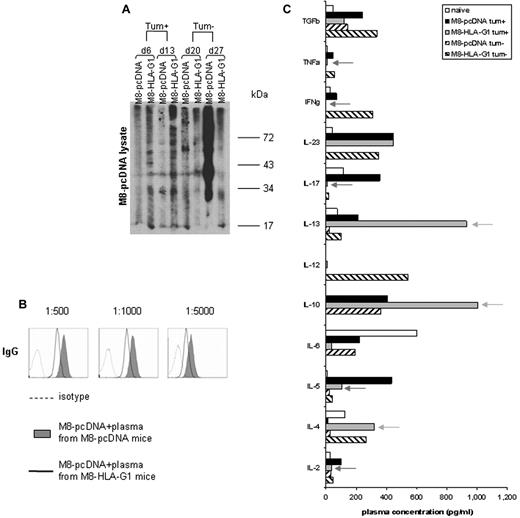
HLA-G1 influences Ab responses and the cytokine environment in vivo. (A) Presence of specific antibodies against antigens derived from the human melanoma cell line (M8-pcDNA lysate) at days 6, 13, 20, and 27 after recovery of plasma as detected by slot-blot analysis. Data are from 1 representative experiment of 3. (B) Detection of specific Ab against melanoma Ag at day 27 by flow cytometry. Plasma dilutions were 1:500, 1:1000, and 1:5000. (C) Plasma concentrations of cytokines from naive mice or mice injected with tumor cells were quantified by ELISA. Green and red arrows indicate the cytokines with levels increased or decreased by HLA-G, respectively. M8-HLA-G1 tum+ indicates plasma collected at day 6; M8-HLA-G1 tum−, plasma collected at day 20.
HLA-G1 influences Ab responses and the cytokine environment in vivo. (A) Presence of specific antibodies against antigens derived from the human melanoma cell line (M8-pcDNA lysate) at days 6, 13, 20, and 27 after recovery of plasma as detected by slot-blot analysis. Data are from 1 representative experiment of 3. (B) Detection of specific Ab against melanoma Ag at day 27 by flow cytometry. Plasma dilutions were 1:500, 1:1000, and 1:5000. (C) Plasma concentrations of cytokines from naive mice or mice injected with tumor cells were quantified by ELISA. Green and red arrows indicate the cytokines with levels increased or decreased by HLA-G, respectively. M8-HLA-G1 tum+ indicates plasma collected at day 6; M8-HLA-G1 tum−, plasma collected at day 20.
HLA-G1 influences the cytokine environment in vivo
We measured the plasma levels of different types of cytokines: Th1, Th2, and Th17. During the tum+ phase, IL-4, IL-10, and IL-13 were detected at high levels, whereas IL-6, IL-17, IFNγ, and TNFα levels were decreased in the plasma of mice injected with M8-HLA-G1 cells compared with M8-pcDNA cells (Figure 6C). IL-12 was not detected in either group of mice. This cytokine environment in mice injected with HLA-G1+ tumor cells supports a Th2 profile rather than Th1 or Th17, thereby promoting tumor growth. In contrast, during the tum− phase, high levels of IL-12, IL-23, IFNγ, TNFα, and TGFβ were detected in the plasma of these mice (Figure 6C). These data indicate that a Th1-driven immune response was present in mice injected with M8-HLA-G1 cells, which could explain the tumor rejection.
Effects of HLA-G1 after reinjection of tumor cells
We next investigated whether HLA-G could modulate the growth of tumor cells after reinjection. For this purpose, Balb/c mice were first injected with M8-pcDNA, M8-HLA-G1, or M8-HLA-G1 cells pretreated with the anti–HLA-G–specific Ab. When tumors were no longer macroscopically detectable, M8-HLA-G1 cells were reinjected in all mice. Mice injected twice with M8-HLA-G1 tumor cells did not develop tumors after reinjection (supplemental Figure 3). Interestingly, mice injected with M8-pcDNA and then with M8-HLA-G1 cells developed tumors comparable to those observed in mice injected once with M8-HLA-G1 cells. Similarly, mice injected with M8-HLA-G1 cells pretreated with the anti–HLA-G mAb and then with M8-HLA-G1 tumor cells also developed tumors (supplemental Figure 3).
Tumors expressing constitutively HLA-G grow in immunocompetent mice
The studies described thus far were conducted with tumor cell lines stably transfected with the HLA-G1 transcript. We next investigated whether similar results could be observed in tumor cells naturally expressing HLA-G. Whereas the primary culture cell line exhibited a high level of HLA-G1 cell-surface expression (Fon+ cells), this was completely lost during long-term in vitro propagation (Fon− cells; Figure 7A inset).19 We subcutaneously injected 10 × 106 Fon− or Fon+ cells and performed the same experiments as described for M8 cells. 10 × 106 Fon+ cells expressing HLA-G1 grew rapidly under the skin of Balb/c mice, whereas Fon− cells were rejected immediately (Figure 7A). In M8-HLA-G1 cells, 2 phases of development (tum+ and tum−) could be defined. These data indicate that similar HLA-G–mediated effects were observed with a second human tumor, thereby validating our results on the role of HLA-G in tumor escape.
Tumor cells expressing HLA-G constitutively grow in immunocompetent mice. (A) Groups of 3 Balb/c mice were injected subcutaneously with 10 × 106 Fon− or Fon+ cells at day 0, and tumor growth was monitored at the indicated time points. Data represent the means ± SD from 1 representative experiment of 3. Inset shows the cell-surface expression of HLA-G1 by Fon− and Fon+ cells and isotype controls. (B) Phenotypic analysis of PBMCs was performed at day 22, as described in the legend to Figure 4. Data are from 1 representative experiment of 2. (C) Proliferation of mononuclear cells of lymph nodes (LN), spleens, and peripheral blood (PBMC) was assessed as described in the legend to Figure 2 in response to Fon− or Fon+ cells. Results represent the means ± SD of triplicates from 1 representative experiment of 2. The numbers indicated between parentheses are the P values. *P < .05; **P < .001; ***P < .005. (D) Levels of cytokines in plasma of naive mice or mice injected with tumor cells were quantified by ELISA. Fon+ tum+ indicates plasma collected at day 4; Fon+ tum−, plasma collected at day 21.
Tumor cells expressing HLA-G constitutively grow in immunocompetent mice. (A) Groups of 3 Balb/c mice were injected subcutaneously with 10 × 106 Fon− or Fon+ cells at day 0, and tumor growth was monitored at the indicated time points. Data represent the means ± SD from 1 representative experiment of 3. Inset shows the cell-surface expression of HLA-G1 by Fon− and Fon+ cells and isotype controls. (B) Phenotypic analysis of PBMCs was performed at day 22, as described in the legend to Figure 4. Data are from 1 representative experiment of 2. (C) Proliferation of mononuclear cells of lymph nodes (LN), spleens, and peripheral blood (PBMC) was assessed as described in the legend to Figure 2 in response to Fon− or Fon+ cells. Results represent the means ± SD of triplicates from 1 representative experiment of 2. The numbers indicated between parentheses are the P values. *P < .05; **P < .001; ***P < .005. (D) Levels of cytokines in plasma of naive mice or mice injected with tumor cells were quantified by ELISA. Fon+ tum+ indicates plasma collected at day 4; Fon+ tum−, plasma collected at day 21.
As observed with M8-HLA-G1 tumor cells, peripheral T cells were almost completely absent from blood of mice injected with Fon+ cells compared with naive mice or mice injected with Fon− cells. The expression of CD80, CD86, MHC class II, and PIR-A/B was not affected (Figure 7B). Remarkably, the CD11bhigh Gr1high population was detected in the peripheral blood of mice injected with Fon+ cells (7.1% vs 1.2% in mice injected with Fon− cells), but not in the spleens or draining lymph nodes (data not shown).
Cells from mice injected with Fon+ cells proliferated less than those purified from naive mice or mice injected with Fon− cells (Figure 7C), showing that the immune cells had lost their proliferative capacity after an in vivo contact with Fon+ cells. The inhibitory effect of HLA-G1 in vitro was also demonstrated by the fact that the proliferation of mononuclear cells in response to Fon+ cells was significantly inhibited compared with Fon− cells (Figure 7C).
No significant differences in the cytokines present in the plasma of mice injected with Fon− or Fon+ cells during the tum+ phase were observed (Figure 7D). In contrast, during the tum− phase, high levels of IL-2, IL-23, IFNγ, TNFα, and TGFβ were detected in the plasma of mice injected with Fon+ cells (Figure 7D). IL-12 was never detected.
Discussion
The results of some in vitro studies have suggested that because the engagement of HLA-G generates inhibitory signals in various immune cells, this may represent a mechanism used by tumor cells to escape from immunosurveillance, and blocking this molecule may therefore constitute a novel innovative antitumoral approach.2,33 Testing the validity of these possibilities will benefit from the development of an animal model. Addressing these questions was the goal of our present study.
We initiated this work by postulating that, although xenogenic, a human tumor expressing HLA-G could grow in vivo in an immunocompetent mouse. Our results confirmed this hypothesis, because human tumor M8 cells expressing HLA-G at their cell surface could progress in vivo, whereas their HLA-G− counterparts were rejected by the murine immune system immediately. The direct contribution of HLA-G in tumor progression in vivo was definitively provided by masking HLA-G with a specific Ab, which led to tumor rejection.
In humans, HLA-G mediates these activities through interaction with immunoreceptor tyrosine–based inhibitory motif–bearing receptors such as ILT2 and ILT4.3 Whereas ILT2 is expressed by all immune cells, ILT4 expression is restricted to myeloid cells. Interestingly, HLA-G can also interact with the murine receptor PIR-B,16,34 which is expressed by murine myeloid- and lymphoid-lineage cells, contains 3 immunoreceptor tyrosine–based inhibitory motif domains, and shares sequence homology with the human ILT receptors.16 The difference between PIR-B and ILT is the absence of PIR-B expression on murine T and NK cells, whereas ILT-2 is expressed on both subsets. Therefore, unlike the effects on human cells, the HLA-G–mediated effects on murine T and NK cells may be indirect.
Supporting the involvement of PIR-B, we found that the percentage of PIR-B+ cells and the surface level expression of PIR-B were consistently increased when HLA-G was present compared with controls. Such PIR-B expression increased rapidly by day 1 and continued to increase until day 20, strongly suggesting that PIR-B is involved in the inhibitory effect of HLA-G. To investigate this possibility, we used the only available blocking anti–PIR-B Ab, which was previously established for in vitro neuronal studies.31 The injection of this Ab in the present in vivo tumor model did not reverse tumor growth in HLA-G+ tumor–bearing mice. Therefore, we cannot exclude the possibility that another, as-yet-unknown murine receptor(s) capable of interacting with HLA-G may also be involved in HLA-G–mediated immunosuppressive effects—or that this Ab was not functional in our system.
Surprisingly, purified NK cells from HLA-G1+ tumor–bearing mice exhibited normal in vitro cytotoxic activity (Figure 3A). Nevertheless, because HLA-G1+ tumor cells grew in vivo, we expected that NK cells would be indirectly inhibited through HLA-G–driven mechanisms because they do not express PIR-B. In agreement with this, increased levels of IL-10, IL-4, and IL-13, and expansion of peripheral CD11b+Gr1+ regulatory cells (also called MDSCs) were observed in HLA-G1+ tumor–bearing mice. In particular, IL-10 is a potent regulator of NK-cell functions.35,36 Supporting a potential role of HLA-G1+ tumor–expanded CD11b+Gr1+ MDSCs, decreased NK cytotoxic activity was observed when total mononuclear cells from HLA-G1+ tumor–bearing mice were used as effectors or when Gr1+ MDSCs from HLA-G1+ tumor–bearing mice were added as a third party in the cytotoxic assay (Figure 3B and 5C). Remarkably, such tumor-derived suppressor cells have been described as inducing anergy of NK cells through membrane-bound TGF-β1.37 In addition, HLA-G inhibits the transendothelial migration of NK cells, which may account for the inhibition of NK-cell recruitment at the tumor site in vivo.38 All of these mechanisms could contribute to the escape of HLA-G+ tumor cells from NK-cell lysis in vivo, although NK cells were found to be functional in vitro. In the human context, HLA-G is known to inhibit NK-cell cytotoxicity directly through binding to ILT-2. Nevertheless, indirect effects of HLA-G, such as the inhibition of human NK cells by HLA-G–induced tolerogenic dendritic cells,39 have been also described in humans, which parallels our explanation of the suppressive role of HLA-G1+ tumor–expanded CD11b+Gr1+ MDSCs on murine NK cells.
Regarding adaptive immunity, analysis of T cells showed that, regardless of whether they derived from HLA-G− or HLA-G+ tumor–bearing mice, their proliferative capacities toward tumor cells were not affected in vitro. As discussed for NK cells, T cells are likely to be indirectly inhibited by HLA-G in vivo through HLA-G–induced mechanisms. In vivo levels of CD4+ and CD8+ peripheral T cells from HLA-G+ mice were significantly lower than those observed in HLA-G− mice. This T-cell loss at the periphery did not result from a compartmental redistribution, because no concomitant T-cell increase was observed in the spleens and draining lymph nodes, but may instead have been due to T-cell apoptosis in vivo. This T-cell loss occurred concomitantly with the increase in the peripheral CD11b+Gr1+–regulatory population in HLA-G1+ tumor–bearing mice. These myeloid suppressor cells have been described previously as triggering apoptosis in antigen-activated T cells.40 In addition, we found a decreased Ab response against the antigens derived from the human M8 melanoma cell line in HLA-G+ tumor–bearing mice compared with their HLA-G− counterparts (Figure 6A-B). These data demonstrate the inhibitory effects of HLA-G toward murine B cells that express the PIR-B receptor.32
HLA-G+ tumor growth was associated with Th2 cytokine production. Whereas Th1/Th17 cytokines were highly detected in HLA-G− tumor–bearing mice, they were absent in HLA-G+ mice, showing an inhibitory effect of HLA-G toward Th1/Th17 cytokine secretion. In the present study, we show for the first time that the secretion of the antitumoral IL-17 cytokine41,42 is down-regulated by HLA-G. Such inhibition is in total agreement with the B-cell response defect observed in HLA-G+ tumor–bearing mice, because IL-17 was shown to positively control B-cell biology, especially the differentiation of B cells into Ig-secreting cells.43 Conversely, during the tum− phase, the cytokine milieu favored Th1, because high levels of IL-12, IL-23, IFNγ, TNFα, and TGFβ were detected in the plasma of HLA-G+ tumor–bearing mice. This suggests the presence of a Th1-driven immune response in mice injected with HLA-G1+ tumor cells, which could explain the tumor rejection.
Horuzsko et al investigated the suppressive function of HLA-G–induced MDSCs in the context of allogeneic skin transplantation in mice. Their results clearly demonstrated that both the expansion and the suppressive function of MDSCs are mediated by engagement of inhibitory receptors on MDSCs by HLA-G, and that HLA-G–induced MDSCs are directly involved in long-term survival of allografts.30 Interestingly, the expression of IL-4 and IL-13 by MDSCs was up-regulated after treatment with HLA-G, strongly suggesting enhanced suppressive activity of HLA-G–induced MDSCs.30 Moreover, transplantation-activated or inhibitory receptor–activated MDSCs have signatures similar to those of tumor-bearing MDSCs.30,44 We observed a similar IL-4 and IL-13 cytokine pattern in HLA-G+ tumor–bearing mice during the tumor-growth phase, strengthening the suppressive activity of the expanding myeloid cells in mice challenged with HLA-G+ tumors. Interestingly, MDSCs expressed PIR-B and were expanded in M8-HLA-G1 tumor–bearing mice. HLA-G was directly involved in the expansion of this population, because the use of an anti–HLA-G mAb inhibited the expansion of PIR-B+ MDSCs and purified HLA-G protein bound to this cell population.
Whereas HLA-G− tumors may recruit essentially innate effectors during the primary response, HLA-G+ tumors may instead mobilize adaptive effectors, as shown by the kinetics of rejection of each tumor type. In mice injected first with HLA-G− M8-pcDNA tumor cells, subsequent injection of M8-HLA-G1 tumor cells led to tumor growth, supporting the inhibition of innate responses by HLA-G. Furthermore, M8-HLA-G1 cells were immediately rejected after a second injection of these cells, showing that HLA-G+ tumor cells do not escape from adaptive secondary responses. Indeed, the specific primary response against the xenogenic HLA-G1+ tumor cells may be potent enough to bypass the secondary HLA-G1–mediated inhibition.
These results were confirmed using a second xenomodel of tumor cells expressing HLA-G1 constitutively. These cells progressed in immunocompetent mice and led to myeloid-derived suppressor cell expansion, whereas their HLA-G− counterparts did not.
In conclusion, we provide the first proof of concept that HLA-G constitutes a relevant in vivo tumor escape mechanism by showing that blocking HLA-G function with a specific Ab inhibits the development of tumors. These findings should be taken into consideration in the design of future therapeutic strategies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Isabelle Poras for technical assistance, Irène Krawice-Radanne for immunohistochemical staining, Dr Benoit Favier for expertise in confocal microscopy, and Drs Marika Pla and Martine Chopin for the use of animal care facilities. We also thank Genentech for providing us with the anti–PIR-B Ab.
This study was supported by the Commissariat a l'Energie Atomique.
Authorship
Contribution: S.A., E.D.C., and N.R-F. designed and analyzed the experiments; S.A. and N.R.-F. wrote the manuscript; and S.A. performed experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nathalie Rouas-Freiss, CEA, I2BM, Service de Recherches en Hémato-Immunologie, Hôpital Saint-Louis, Institut Universitaire d'Hématologie, 1, avenue Claude Vellefaux, 75010 Paris, France; e-mail: Nathalie.Rouas-Freiss@cea.fr.