The platelet glycoprotein Ib-IX-V complex (GPIb-IX-IV) is the receptor for VWF and is responsible for VWF-mediated platelet activation and aggregation. Loss of the GPIb-IX-V complex is pathogenic for Bernard-soulier Syndrome (BSS), which is characterized by macrothrombocytopenia and impaired platelet function. It remains unclear how the GPIb-IX-V complex is assembled and whether there is a role for a specific molecular chaperone in the process. In the present study, we report that the assembly of the GPIb-IX-V complex depends critically on a molecular chaperone in the endoplasmic reticulum (ER): gp96 (also known as grp94 and HSP90b1). gp96/grp94 deletion in the murine hematopoietic system results in thrombocytopenia, prolonged bleeding time, and giant platelets that are clinically indistinguishable from human BSS. Loss of gp96/grp94 in vivo and in vitro leads to the concomitant reduction in GPIb-IX complex expression due to ER-associated degradation. We further demonstrate that gp96/grp94 binds selectively to the GPIX subunit, but not to gpIbα or gpIbβ. Therefore, we identify the platelet GPIX subunit of the GPIb-IX-V complex as an obligate and novel client of gp96/grp94.
Introduction
The molecular chaperone gp96,1 which is also known as grp942 or HSP90b1, is the paralog of heat-shock protein 90 (HSP90) in the endoplasmic reticulum (ER). Like other HSPs, gp96/grp94 is induced by the accumulation of misfolded proteins.3 gp96/grp94 binds and hydrolyzes ATP,4,–6 is the most abundant protein in the ER lumen, and is ubiquitously expressed in all nucleated cells. Genetic studies have begun to clarify and expand the role of gp96/grp94 as the critical chaperone for multiple TLRs and integrins7,,,–11 and in the unfolded protein response (UPR).12 Without gp96/grp94, the majority of integrins and TLRs are unable to fold properly and thus fail to exit the ER to traffic to the proper post-ER compartment. Using a Cre/loxP–mediated conditional deletion of gp96/grp94 in mice, we recently discovered that gp96/grp94 selectively regulates lymphopoiesis but not myelopoiesis.10 However, it remains unclear whether gp96/grp94 chaperones other, as-yet-unidentified client proteins in the hematopoietic system.
The platelet glycoprotein Ib-IX-V (GPIb-IX-V) complex consists of 4 transmembrane proteins: GPIbα, GPIbβ, GPIX, and GPV13 and functions as a receptor for VWF for platelet activation.14 Defects in the biogenesis of this complex result in BSS. At the molecular and structural level, GPIbα and GPIbβ are covalently linked by a disulfide bond; these proteins are noncovalently associated with GPIX and GPV.13,15 The GPIb-IX complex belongs to proteins of the leucine-rich repeat (LRR) family.16 Like other LRR-motif–containing proteins, GPIbα adopts a “horseshoe-like” shape that is thought to be involved in intermolecular interactions and ligand binding.14 In support of this notion, several mutations in the LRR domain of GPIbα, GPIbβ, and GPIX result in loss of GPIb function and the development of BSS.17,,,–21
Folding, assembly, and maturation of the GPIb-IX complex begins in the ER, where it undergoes both N-linked and O-linked glycosylation before reaching the plasma membrane.22 Lack of the GPIbα, GPIbβ, or GPIX subunit results in the abnormal processing, assembly, and cell-surface expression of the GPIb-IX complex. Protein folding of multisubunit complexes likely involves multiple checks and balances before they leave the ER in a process known as ER quality control.23 Although the folding and assembly of the wild-type (WT) and the mutant GPIb-IX complex has been studied, a role for molecular chaperones in this process has not been determined.
In the present study, we found that gp96/grp94 is critically required for the assembly of the GPIb-IX complex. Genetic knockout (KO) of gp96/grp94 in mice completely abrogated the expression of the surface GPIb-IX complex in megakaryocytes and platelets. Moreover, loss of gp96/grp94 in the hematopoietic system resulted in prolonged bleeding times, thrombocytopenia, and giant platelet disorder that were clinically and hematologically indistinguishable from human BSS. We also demonstrated that assembly of the GPIb-IX complex is highly sensitive to general ER stress. Our results reveal a novel role for gp96/grp94 in the assembly of the platelet GPIb-IX complex, and suggest the possible importance of dysregulated ER protein homeostasis in platelet disorders.
Methods
Mice
Hsp90b1flox/floxR26Rcre-ERT2 (tamoxifen-inducible gp96/grp94 KO) mice, their control littermates, and bone marrow chimeras used here were described previously.10 All other mice were obtained from The Jackson Laboratory. Mouse experiments were carried out according to established guidelines and approved protocols.
Cloning of GPIX
Human GPIX-FLAG was cloned from genomic DNA from HEK293T cells using the following primer set: forward, ATTAAGATCTGCCACCATGCCTGCCTGGGGA GCCCTG and reverse, TATAGAATTCTCACTTGTCGTCATCGTCTTTG TAGTCATCCAGGGCCTCTGTGGTGGC containing BglII and EcoRI restriction enzymes, respectively. The PCR product was digested directly and cloned into MigR retroviral vector. The correct sequence was verified by DNA sequencing.
Antibodies and other reagents
Antibodies against grp78, calreticulin, and gp96/grp94 were obtained from StressGen Biotechnologies. Antibodies against mouse GPIbα (Xia.G7) were obtained from Emfret Analytics. Antibodies against mouse CD41 and Ter-119 were obtained from eBioscience.
Bleeding time
Mice were restrained in a 50-mL Falcon tube. A superficial incision was made approximately 1 inch from the base of the tail. The incision was then blotted with Whatman paper every 10 seconds until bleeding ceased. The time from the incision and bleeding cessation was recorded.
CBC
Fifty microliters of peripheral blood was collected through tail nicking in 10 μL of acid citrate dextrose (ACD) buffer. Ten microliters of anticoagulated blood was analyzed for complete blood count (CBC) using a Beckman Coulter counter. Samples were read in duplicate.
Flow cytometry
Cell staining and flow cytometry were as described previously.9 For platelet analysis, one-tenth volume of whole blood was collected in ACD buffer and platelets were stained at room temperature. Platelets were defined as CD41+Ter119− by a low forward-scatter profile. For analysis of the cell-surface GPIb-IX complex, care was taken to avoid trypsin to prevent cleavage of the complex.
Methylcellulose CFU assay
After red blood cell lysis, 1 × 104 bone marrow cells were cultured in duplicate in MethoCult GF M3434 methylcellulose medium (StemCell Technologies). Seven days later, colonies were counted and identified by light microscopy.
Platelet adhesion
The methods we used to measure platelet adhesion were similar to those described previously.24,25 Briefly, glass slides were coated with VWF (30 μg/mL) in 0.1M NaHCO3, pH 8.3, at 4°C overnight. A fluorescence dye (10μM mepacrine; Sigma-Aldrich) was added to the same number of WT or KO platelets before the application of a constant shear rate (800/s) for 5 minutes using a cone-plate rheometer (Thermo Scientific Haake). Slides were washed and viewed with a DMI RB inverted fluorescence microscope (Leica Microsystems) using an N PLAN L 40×/0.55 numeric aperture (infinity)/0-2/C objective lens. Images were acquired using a Cool SNAP HQ CCD Camera (Photometrics) and processed with RS Image version 1.4 software (Photometrics). Stable adherent platelets were counted in 10 randomly selected microscope fields to obtain the average numbers (± SD) of adherent platelets per field.
Platelet aggregation to VWF
The method we used to measure platelet aggregation to VWF was described previously.24 Briefly, blood collection from the abdominal aorta of isoflurane-anesthetized WT or KO mice (n = 5), 0.1 μg/mL of prostaglandin E1, and 1 U/mL of apyrase were added to the blood before platelet isolation by differential centrifugation. Platelets were isolated from platelet-rich plasma and washed in modified Tyrode buffer (12mM NaHCO3, 138mM NaCl, 5.5mM glucose, 2.9mM KCl, 2mM MgCl2, 0.42mM NaH2PO4, 10mM HEPES, pH 7.4) containing 5mM EDTA. Washed platelets were resuspended in the modified Tyrode buffer and placed in a turbidimetric platelet aggregometer (Chronolog) at 37°C with stirring at 1000 rpm. VWF (Sigma-Aldrich) and botrocetin (kindly provided by Dr Michael C. Berndt, Monash University, Melbourne, Australia) were added to the washed platelets to induce aggregation.
Density enrichment of megakaryocytes
Long bones were isolated and bone marrow was flushed out with PBS using a 25-gauge syringe. Red blood cells were lysed in ammonium chloride lysis buffer, and bone marrow cells were resuspended in PBS. Bone marrow cells were carefully layered on top of a 1.5%/3.0% BSA/PBS discontinuous gradient, and megakaryocytes were allowed to sediment for 30-45 minutes. The top 2 layers were carefully removed using a Pasteur pipette and discarded. The bottom layer enriched to > 80% megakaryocytes was washed and used for immunofluorescence, flow cytometric, and/or Western blot analysis. Enrichment was routinely checked by cytospin and/or flow cytometry for CD41/61 expression.
Ploidy analysis
Mouse bone marrow was flushed in ACD/PBS buffer and megakaryocytes were harvested by slow-speed centrifugation (80g). After blocking of the Fc receptor with anti–mouse CD16/32 (clone 93; eBiosciences), cells were stained with anti-CD41 biotin antibody for 20 minutes at room temperature. Cells were washed once in ACD/PBS at 300g and stained with streptavidin Alexa Fluor 488 antibody (Invitrogen) for 15 minutes at room temperature. After washing, cells were resuspended in ACD/PBS buffer containing 0.025% Triton-X 100 and propidium iodide, and read on a flow cytometer within 30 minutes.
Quantitative RT-PCR
cDNA was made from BSA-enriched megakaryocytes by reverse transcription according to the manufacturer's protocol (Superscript II; Invitrogen). Quantitative RT-PCR was performed using the SYBR Green method (Applied Biosystems). β-actin served as an internal control, and gene expression was normalized first to β-actin (ΔCT) and second to the receptor for thrombopoietin (TPO-R; ΔΔCT). The arbitrary unit was calculated based on the following formula: 2−ΔΔCT × 100 000. The following primer sets were used: β-actin, (forward) GGTCATCACTATTGGCAACG, (reverse) ACGGATGTCAACGTCACACT; TPO-R, (forward) ACTGCTGCTAAAGTGGCAATTTC, (reverse) GACCCGGTGTAGGTCTGGAA; CD41, (forward) ACAAGCCTATCTGCCACA, (reverse) CGACACGTTGAGGCTTAG; PF4, (forward) CGGTTCCCCAGCTCATAGC, (reverse) GCCGGTCCAGGCAAATTT; GATA-1, (forward) AAAGATGGAATCCAGACGAGG, (reverse) GTCAAGGCTATTCTGTGTACC; GPIbα, (forward) CCTGGAAGAAGCTCTGTTCCTCC, (reverse) CATTGGTCTGCAGGCTCGTC; GPIbβ, (forward) TGACCGGCAACAACCTGAC, (reverse) CAGCAGAGTAGACCGGGTG; and GPIX, (forward) TGCGACCACAGATACTCAGG, (reverse) ACTGAACGCAGGCTATTGTTG.
Histology and immunofluorescence microscopy of bone/megakaryocytes
Femur bones were cleaned and fixed in 4% paraformaldehyde in 0.1M PBS, pH 7.4, decalcified in EDTA-based decalcification buffer, and placed in 30% sucrose/PBS before embedding in optimal cutting temperature freezing medium. Next, 0.7-μM sections were cut using a cryostat and slides were stained with H&E for histology. The number of megakaryocytes per field was determined by manually counting in longitudinal bone sections under 200× magnification. Alternatively, for immunofluorescence, slides were refixed in ice-cold acetone or with 2% paraformaldehyde and permeabilized with ice-cold methanol. Background staining was blocked with 10% BSA/FACS buffer and FcR blocking antibody. Primary and secondary antibody was diluted in 5% BSA/FACS buffer. Slides were washed in PBS and counterstained with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI), mounted with anti-fade mounting medium, and immunofluorescence was visualized with a Leica Axioplan 2 microscope at a 200× magnification.
Protein extraction, immunoprecipitation, and Western blot analysis
Protein extraction, immunoprecipitation, and Western blot analysis were carried out as described previously.8 Briefly, IB9-IX cells were harvested by trypsin-EDTA or 5μM EDTA and exposed to dithiobis(succinimidyl propionate) (Thermo Scientific), for 30 minutes at room temperature, washed in PBS, and lysed on ice in radioimmunoprecipitation assay lysis buffer in the presence of a protease inhibitor cocktail (Sigma-Aldrich). Nuclear-free protein lysate was quantified by Bradford assay, and an equal amount of lysate was incubated with GPIb-IX complex antibodies or isotype control. GPIbα was immunoprecipitated using a goat anti–human antibody (C-20; Santa Cruz Biotechnology) that recognizes the C-terminus of GPIbα and protein-G agarose beads (Genescript). GPIbβ was immunoprecipitated with a mouse anti–human antibody (Gi27; Serotec) and protein-G agarose beads. GPIX-FLAG was immunoprecipitated using anti-FLAG antibody (BioM2; Sigma-Aldrich) and NeutrAvidin beads (Thermo Scientific).
GST pull-down assay
Full-length human gpIX cDNA was subcloned into pGEX-pMagEmcs vector. GST fusion proteins were isolated on glutathione-Sepharose 4B beads (Amersham Biosciences). Purified mouse gp96/grp94 was incubated with GST alone or with GST-hgpIX in the presence of 20mM HEPES, pH 7.2, 50mM KCl, 5mM MgCl2, 20mM Na2MO4, 0.5% NP40, and 1mM ATP, followed by incubation with glutathione-Sepharose 4B beads at 4°C overnight, and then washed 3 times, boiled in Laemmli buffer, and resolved by SDS-PAGE.
shRNA knockdown of gp96/grp94, induction of ER stress, and inhibition of degradation
shRNA oligonucleotides (21-bp) corresponding to different positions along the DNA sequence of gp96/grp94 were cloned into pLL3.7 vector. 293T cells were cotransfected with shRNA, Δ8.9, and VSV-G plasmids at a ratio of 2:2:1. Forty-eight hours later, lentivirus was collected and cells were transduced by spin infection at 1800g for 1.5 hours at 32°C. gp96/grp94 knockdown in Chinese hamster ovary (CHO) cells was achieved by 2-3 rounds of shRNA (against position 119 of gp96/grp94) transduction on consecutive days. On day 8, knockdown efficiency was determined by intracellular stain or Western blot for gp96/grp94 using 9G-10 antibody. Induction of ER stress was achieved by 18-24 hours of treatment with 5 μg/mL tunicamycin, 5μM thapsigargin, 10 μg/mL of antimycin C, or by 4-6 hours of treatment with 10 μg/mL of brefeldin-A. To examine degradation of the gpIb-IX complex, empty vector (EV), gp96/grp94 shRNA (119) Ib9-IX cells, or thapsigargin-stressed Ib9-IX cells were treated for 6 hours with various inhibitors, 100μM kifunensine, or 100μM chloroquine, followed by immunoblot analysis.
Statistical analysis
The Student t test was used for statistical analysis. P < .05 was considered significant.
Results
gp96/grp94 deletion in the hematopoietic system causes macrothrombocytopenia but does not affect megakaryopoiesis
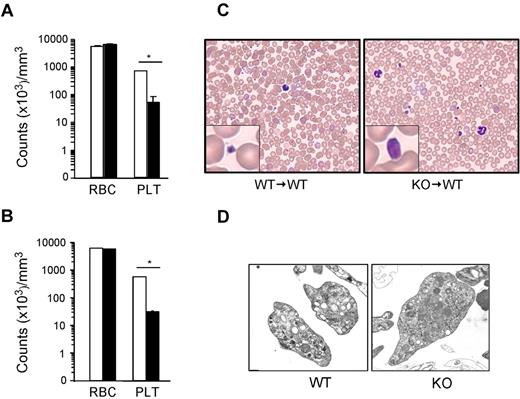
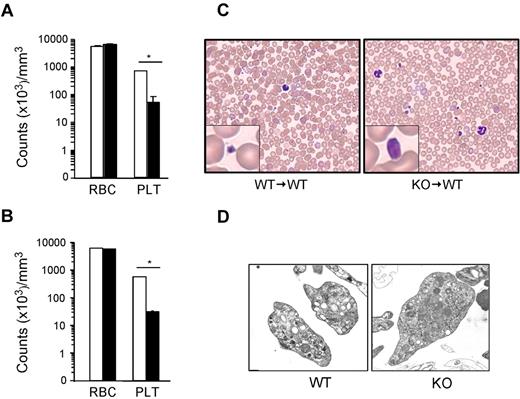
gp96/grp94 has been shown to be an essential chaperone for both the TLR and integrin families. Recently, we described a novel tamoxifen-inducible gp96/grp94 KO mouse and unveiled the central role of gp96/grp94 in early lymphopoiesis, but not in myelopoiesis.10 CBC analysis post tamoxifen-induced deletion (PTD) of gp96/grp94 not only revealed leukocytosis and granulocytosis, but also considerable thrombocytopenia (more than a 13-fold reduction). To understand the etiology of thrombocytopenia, we performed experiments using inducible gp96/grp94 KO mice and WT recipient mice reconstituted with KO bone marrow (KO → WT). CBC was performed before and after 2 weeks of tamoxifen injections. As expected, KO mice had significant thrombocytopenia 14 days PTD (an ∼19-fold reduction of peripheral blood platelets), and this effect was intrinsic to the hematopoietic system because deletion of gp96/grp94 in KO → WT bone marrow chimeric mice yielded identical results (Figure 1A-B). However, erythropoiesis was normal. Strikingly, peripheral blood smears revealed the presence of giant platelets in KO and KO → WT mice (Figure 1C). KO platelets were distinctly larger than WT platelets, occasionally approaching the size of mature red blood cells (Figure 1C). There was no evidence of platelet aggregation on the smear. Abundant granules were visible in both WT and KO platelets using ultrastructural analysis (Figure 1D). Therefore, gp96/grp94 deletion in the hematopoietic system results in profound macrothrombocytopenia.
Loss of gp96/grp94 results in macrothrombocytopenia. CBC analysis on WT (□, n = 5) and KO (■, n = 5) mice (A) or WT → WT (□, n = 4) and KO → WT (■, n = 5) bone marrow chimeras (B) 14 days PTD. RBC indicates red blood cells; PLT, platelets. *P < .05. (C) Representative Hem-3 staining of peripheral blood smears. (D) Ultrastructure transmission electron microscopy of WT and KO platelets. Magnifications are WT, 15 000×; and KO, 10 000×.
Loss of gp96/grp94 results in macrothrombocytopenia. CBC analysis on WT (□, n = 5) and KO (■, n = 5) mice (A) or WT → WT (□, n = 4) and KO → WT (■, n = 5) bone marrow chimeras (B) 14 days PTD. RBC indicates red blood cells; PLT, platelets. *P < .05. (C) Representative Hem-3 staining of peripheral blood smears. (D) Ultrastructure transmission electron microscopy of WT and KO platelets. Magnifications are WT, 15 000×; and KO, 10 000×.
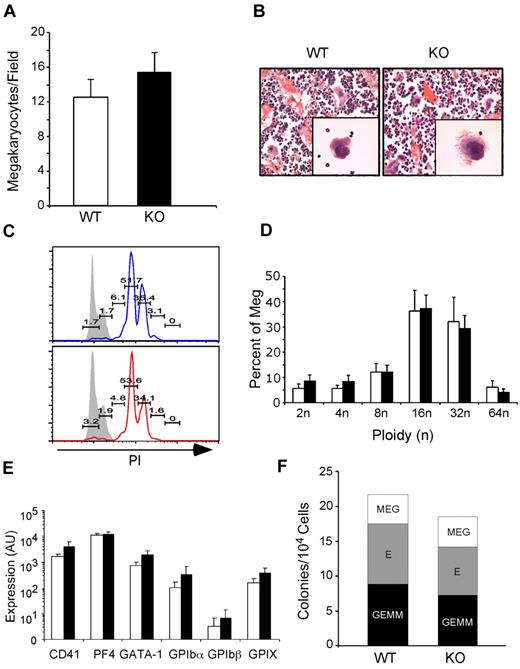
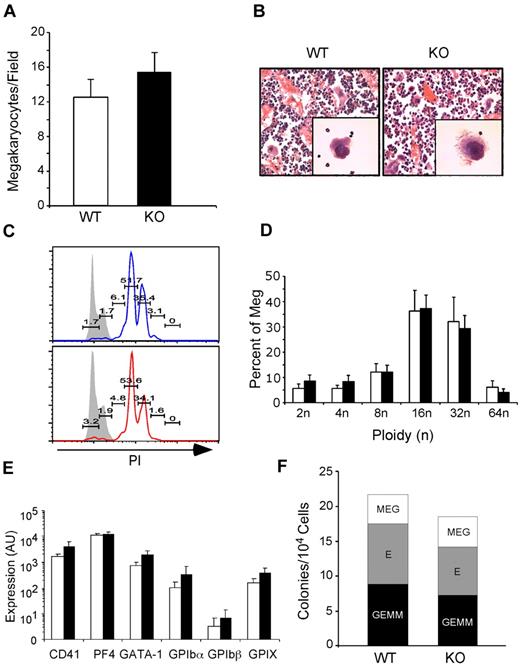
Thrombopoiesis is the process by which platelets are produced from megakaryocytes in the bone marrow. To determine whether macrothrombocytopenia in KO mice was due to inefficient production of megakaryocytes, we examined bone sections from KO mice for the presence of megakaryocytes. At 1 month PTD, KO → WT mice had normal if not slightly increased megakaryocyte numbers with typical multilobulated nuclei and morphology (Figure 2A-B). Megakaryocytes were also present in nonchimeric KO mice 14 days PTD. Moreover, ploidy analysis showed no difference between WT and KO megakaryocytes (Figure 2C-D). Using quantitative PCR analysis, we also found no difference between WT and KO megakaryocytes in the expression of developmentally important megakaryocyte-specific genes, including CD41, PF4, GATA-1, and the platelet GPIb-IX complex (Figure 2E). Finally, culturing KO bone marrow progenitors in a complete methylcellulose CFU assay demonstrated normal differentiation of granulocyte-erythroid-monocyte-megakaryocyte, erythroid, and megakaryocyte colonies (Figure 2F). gp96/grp94 ablation does not lead to reduced production of megakaryocytes, and therefore macrothrombocytopenia in KO mice must be due to a qualitative defect with megakaryocytes.
gp96/grp94 deletion does not negatively affect megakaryopoiesis. (A) Number of megakaryocytes per field (200× magnification) in bone sections of pooled WT and WT → WT (□, n = 4) and KO and KO → WT (■, n = 4) mice 2-4 weeks PTD. No significant differences were observed in the number of megakaryocytes in nonchimeric and chimeric mice (P > .05). (B) Morphologic analysis of WT and KO bone marrow megakaryocytes by light microscopy after H&E staining and the ultrastructure of the demarcation membrane of megakaryocytes by transmission electron microscopy (magnification, 12 000×). (C) Propidium iodide (PI) ploidy analysis of bone marrow CD41+FSChiSSChi megakaryocytes from WT → WT (top) and KO → WT mice (bottom). Numbers represent percentage of PI+ cells indicated over the total population of megakaryocytes. Gray histograms represent control staining of lymphocyte gaiting. (D) Bar graph of megakaryocyte ploidy analysis of 5 WT → WT (□) and 5 KO → WT mice (■). No statistical difference was found between WT and KO mice. (E) BSA gradient–enriched megakaryocytes from WT (n = 3) and KO (n = 3) mice were analyzed for expression of the indicated genes relative to TPO-R expression. TPO-R expression was similar between WT and KO mice. (F) Number of granulocyte-erythroid-monocyte-megakaryocyte (GEMM), erythroid (E), and megakaryocyte (MEG) colonies per 104 cells from WT → WT (n = 2) and KO → WT (n = 2) mice 2 months PTD.
gp96/grp94 deletion does not negatively affect megakaryopoiesis. (A) Number of megakaryocytes per field (200× magnification) in bone sections of pooled WT and WT → WT (□, n = 4) and KO and KO → WT (■, n = 4) mice 2-4 weeks PTD. No significant differences were observed in the number of megakaryocytes in nonchimeric and chimeric mice (P > .05). (B) Morphologic analysis of WT and KO bone marrow megakaryocytes by light microscopy after H&E staining and the ultrastructure of the demarcation membrane of megakaryocytes by transmission electron microscopy (magnification, 12 000×). (C) Propidium iodide (PI) ploidy analysis of bone marrow CD41+FSChiSSChi megakaryocytes from WT → WT (top) and KO → WT mice (bottom). Numbers represent percentage of PI+ cells indicated over the total population of megakaryocytes. Gray histograms represent control staining of lymphocyte gaiting. (D) Bar graph of megakaryocyte ploidy analysis of 5 WT → WT (□) and 5 KO → WT mice (■). No statistical difference was found between WT and KO mice. (E) BSA gradient–enriched megakaryocytes from WT (n = 3) and KO (n = 3) mice were analyzed for expression of the indicated genes relative to TPO-R expression. TPO-R expression was similar between WT and KO mice. (F) Number of granulocyte-erythroid-monocyte-megakaryocyte (GEMM), erythroid (E), and megakaryocyte (MEG) colonies per 104 cells from WT → WT (n = 2) and KO → WT (n = 2) mice 2 months PTD.
gp96/grp94 KO mice have increased bleeding time and defective platelet function
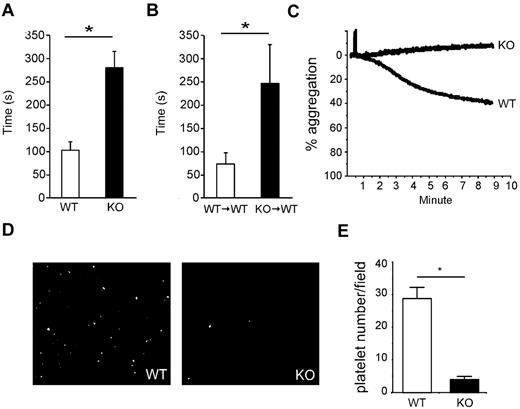
We measured bleeding time to determine the impact of gp96/grp94 loss on the platelet function. WT or WT → WT mice had a bleeding time of < 2 minutes (Figure 3A-B). In comparison, all KO and KO → WT mice had significantly prolonged bleeding times that averaged > 4 minutes (bleeding had to be experimentally stopped at 5 minutes with intervention to prevent death) (Figure 3A-B). In some of the KO and KO → WT mice, hemostasis could not be achieved and these mice had to be euthanized.
Increased bleeding time and failure of platelets to respond to VWF in gp96/grp94 KO mice. Bleeding time analysis of WT (□, n = 5) and KO (■, n = 5) mice (A) and WT → WT (□, n = 8) and KO → WT (■, n = 9) mice (B) 14 days PTD. *P < .05. (C) Aggregation of pooled purified WT (n = 10) and KO (n = 13) platelets 2 months PTD in response to VWF/botrocetin. (D) Representative adherence to immobilized VWF under shear stress of fluorescently labeled platelets from WT (n = 10) and KO (n = 13) mice 2 months PTD. (E) Average number of adhered platelets of WT and KO origin shown in panel D in 10 randomly selected fields. *P < .01.
Increased bleeding time and failure of platelets to respond to VWF in gp96/grp94 KO mice. Bleeding time analysis of WT (□, n = 5) and KO (■, n = 5) mice (A) and WT → WT (□, n = 8) and KO → WT (■, n = 9) mice (B) 14 days PTD. *P < .05. (C) Aggregation of pooled purified WT (n = 10) and KO (n = 13) platelets 2 months PTD in response to VWF/botrocetin. (D) Representative adherence to immobilized VWF under shear stress of fluorescently labeled platelets from WT (n = 10) and KO (n = 13) mice 2 months PTD. (E) Average number of adhered platelets of WT and KO origin shown in panel D in 10 randomly selected fields. *P < .01.
Macrothrombocytopenia can be associated with acquired disorders such as idiopathic thrombocytopenia and myelodysplastic syndrome.26 The inherited disorders include problems caused by mutations in nonmuscle myosin heavy chain-A (eg, May-Hegglin anomaly, Sebastian syndrome, Fechtner syndrome, and Epstein syndrome) and GPIb-IX-V complex (eg, Bernard-Soulier syndrome, Mediterranean macrothrombocytopenia/Bernard-Soulier syndrome carrier, and DiGeorge/velocardiofacial syndrome). In addition, mutation of the transcription factor GATA-1 can also lead to macrothrombocytopenia and several other defects in hematopoiesis.27 Given that gp96/grp94 is an ER chaperone, gp96/grp94 KO is unlikely to affect the expression of the cytoplasmic nonmuscle myosin heavy chain-A or transcription factors.
We tested the hypothesis that the loss of the GPIb-IX complex in KO mice could serve as the molecular basis of macrothrombocytopenia. We performed the standard platelet functional assay in response to VWF/botrocetin, and found that KO platelets completely failed to aggregate in response to VWF/botrocetin treatment (Figure 3C). In addition, we assessed the ability of KO platelets to stably adhere to VWF under shear stress, a model that mimics platelet adhesion in vivo and, again, KO platelets were significantly impaired in their ability to stably adhere to VWF (Figure 3D-E). In the present study, gp96/grp94 deficiency led to the appearance of multiple characteristic features of human BSS, including macrothrombocytopenia, giant platelets, prolonged bleeding time, and a failure to aggregate in response to VWF.
gp96/grp94 deletion is correlated with the loss of GPIbα on platelets and megakaryocytes
To determine the molecular basis of BSS-like diseases in gp96/grp94 KO mice, we examined the expression of GPIbα (CD42b) on platelets and megakaryocytes as identified by the platelet-specific integrin CD41 (integrin αIIb chain). We found that KO platelets began losing surface GPIbα expression on day 7 PTD, and this loss was nearly complete on day 12 (Figure 4A top). Loss of GPIbα expression closely mirrored the kinetics of loss of the gp96/grp94 protein, which has a half-life of approximately 3-4 days. The effect of the loss of gp96/grp94 on platelet GPIbα expression was also both hematopoietic and cell intrinsic (Figure 4B). Accordingly, the loss of GPIbα expression in KO platelets was concomitant with an approximately 20-fold increase in platelet size (Figure 4A-B bottom). Immunofluorescence of bone sections from KO mice revealed a similar kinetic loss of GPIbα expression on megakaryocytes after tamoxifen treatment, and was correlated with the loss of gp96/grp94 expression (Figure 4C). CD41 was reduced slightly but was never absent from gp96/grp94 KO cells. Enrichment of WT and KO megakaryocytes using a BSA discontinuous gradient followed by Western blot for gp96/grp94 and GPIbα further confirmed the absolute requirement of gp96/grp94 for the expression of GPIbα (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). We conclude that gp96/grp94 is necessary for the cell-surface expression of GPIbα on platelets and megakaryocytes.
Cell-intrinsic loss of GPIbα expression on platelets is correlated with the loss of gp96/grp94 and GPIbα expression on megakaryocytes. (A) Top: Flow cytometry of GPIbα expression on platelets (CD41+Ter119−) from a WT mouse and a KO mouse at the indicated days PTD. The Y-axis is the cell count. The gray histogram represents background staining with isotype-matched antibody. Bottom: Flow cytometry of forward scatter profile (FSC) on platelets (CD41+Ter119−) at the indicated time points PTD. Data are representative of at least 3 independent experiments. Numbers indicate mean fluorescence intensity for GPIbα and mean size for FSC. (B) Top: Flow cytometry of CD41+ expression on Ter119-gated platelets from WT → WT (blue), KO → WT (red), and mixed (WT, blue; KO, red) bone marrow chimeras 1 month PTD. Bottom: GPIbα on WT → WT (blue), KO → WT (red), and mixed (WT, blue; KO, red) bone marrow chimeras from the above gates. Data represent 2 independent experiments with multiple mice. (C) Top: Immunofluorescence of GPIbα expression on megakaryocytes (CD41+) of bone sections from WT and KO mice at the indicated time points PTD. Bottom: Immunofluorescence of gp96/grp94 expression on megakaryocytes (CD41+) of bone sections from WT and KO mice at the indicated time points PTD. Slides were counterstained with DAPI. Data are representative of at least 3 independent experiments.
Cell-intrinsic loss of GPIbα expression on platelets is correlated with the loss of gp96/grp94 and GPIbα expression on megakaryocytes. (A) Top: Flow cytometry of GPIbα expression on platelets (CD41+Ter119−) from a WT mouse and a KO mouse at the indicated days PTD. The Y-axis is the cell count. The gray histogram represents background staining with isotype-matched antibody. Bottom: Flow cytometry of forward scatter profile (FSC) on platelets (CD41+Ter119−) at the indicated time points PTD. Data are representative of at least 3 independent experiments. Numbers indicate mean fluorescence intensity for GPIbα and mean size for FSC. (B) Top: Flow cytometry of CD41+ expression on Ter119-gated platelets from WT → WT (blue), KO → WT (red), and mixed (WT, blue; KO, red) bone marrow chimeras 1 month PTD. Bottom: GPIbα on WT → WT (blue), KO → WT (red), and mixed (WT, blue; KO, red) bone marrow chimeras from the above gates. Data represent 2 independent experiments with multiple mice. (C) Top: Immunofluorescence of GPIbα expression on megakaryocytes (CD41+) of bone sections from WT and KO mice at the indicated time points PTD. Bottom: Immunofluorescence of gp96/grp94 expression on megakaryocytes (CD41+) of bone sections from WT and KO mice at the indicated time points PTD. Slides were counterstained with DAPI. Data are representative of at least 3 independent experiments.
gp96/grp94 stabilizes the GPIb-IX complex
To elucidate the underlying molecular mechanism of gp96/grp94 in the folding of the GPIb-IX complex, we used a well-established CHO cell line that stably expresses the GPIbα, GPIbβ, and GPIX of the GPIb-IX complex (CHO-Ib9).28 We also enforced the expression of GPIX tagged with the FLAG epitope at the C-terminus in CHO-Ib9 cells (Ib9-IX).
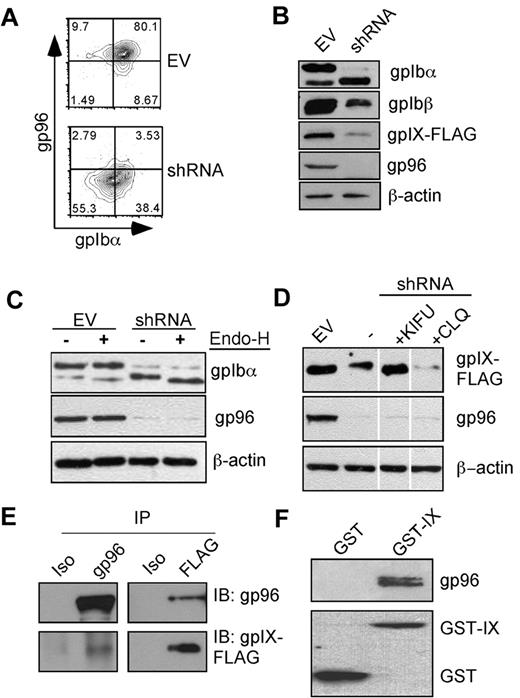
We performed shRNA knockdown experiments to examine the effect of gp96/grp94 ablation on the fate of the GPIb-IX complex in IB9-IX cells. We used the pLL3.7 lentiviral vector29 to express gp96/grp94 shRNA along with the enhanced green fluorescent protein in a bicistronic fashion. Eight days after infection with gp96/grp94 shRNA, but not with an EV, we observed the knock-down of gp96/grp94 intracellularly and the concurrent reduction of GPIbα on the cell surface (Figure 5A).
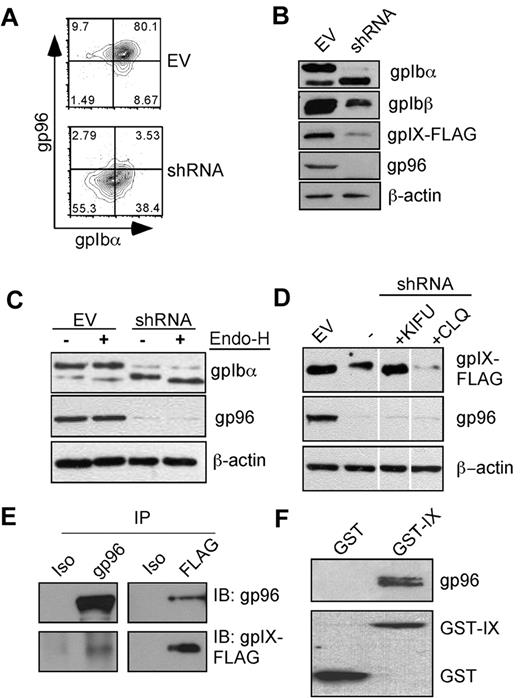
gp96/grp94 chaperones the GPIb-IX complex. (A) Knockdown of gp96/grp94 results in concomitant loss of GPIbα surface expression. Flow cytometric staining was performed for surface GPIbα (AK2), followed by intracellular staining for gp96/grp94 in Ib9 cells. Data are representative of 5 independent experiments. (B) gp96/grp94 is important for the stable expression of multiple subunits of the GPIb-IX complex. Individual components of the complex were blotted from the total cell lysate of Ib9 cells transduced with either EV or gp96/grp94 shRNA lentivector. (C) Differential glycosylation of GPIbα in the absence of gp96/grp94 as shown by Endo-H treatment and Western blot for GPIbα in EV and gp96/grp94 shRNA Ib9-IX cells. (D) The GPIX subunit is degraded in the absence of gp96/grp94. Western blot analysis for GPIX subunit expression in EV and gp96/grp94 shRNA-treated Ib9-IX cells and in gp96/grp94 shRNA Ib9-IX cells after treatment with the inhibitors kifunensine (KIFU) and chloroquine (CLQ). Vertical lines have been inserted to indicate a repositioned gel lane. Data are representative of 2 independent experiments. (E) Coimmunoprecipitation of GPIX and gp96/grp94 in CHO-Ib9/IX cells (Ib9/IX). GPIX-FLAG or gp96/grp94 proteins were immunoprecipitated, followed by immunoblotting. Isotype control antibodies are included. (F) Recombinant GST-gpIX or GST was incubated with mouse gp96/grp94, followed by GST pull-down and immunoblot for gp96/grp94 and GST, respectively.
gp96/grp94 chaperones the GPIb-IX complex. (A) Knockdown of gp96/grp94 results in concomitant loss of GPIbα surface expression. Flow cytometric staining was performed for surface GPIbα (AK2), followed by intracellular staining for gp96/grp94 in Ib9 cells. Data are representative of 5 independent experiments. (B) gp96/grp94 is important for the stable expression of multiple subunits of the GPIb-IX complex. Individual components of the complex were blotted from the total cell lysate of Ib9 cells transduced with either EV or gp96/grp94 shRNA lentivector. (C) Differential glycosylation of GPIbα in the absence of gp96/grp94 as shown by Endo-H treatment and Western blot for GPIbα in EV and gp96/grp94 shRNA Ib9-IX cells. (D) The GPIX subunit is degraded in the absence of gp96/grp94. Western blot analysis for GPIX subunit expression in EV and gp96/grp94 shRNA-treated Ib9-IX cells and in gp96/grp94 shRNA Ib9-IX cells after treatment with the inhibitors kifunensine (KIFU) and chloroquine (CLQ). Vertical lines have been inserted to indicate a repositioned gel lane. Data are representative of 2 independent experiments. (E) Coimmunoprecipitation of GPIX and gp96/grp94 in CHO-Ib9/IX cells (Ib9/IX). GPIX-FLAG or gp96/grp94 proteins were immunoprecipitated, followed by immunoblotting. Isotype control antibodies are included. (F) Recombinant GST-gpIX or GST was incubated with mouse gp96/grp94, followed by GST pull-down and immunoblot for gp96/grp94 and GST, respectively.
Terminally misfolded GPs are rapidly degraded to prevent the induction of the UPR. This is most often accomplished by trimming of the N-glycan by the ER mannosidase I and retrotranslocation of the misfolded and deglycosylated protein back into the cytosol for degradation, a process known as ER-associated degradation (ERAD).23 Consistent with a role for gp96/grp94 in the steady-state folding of the GPIb-IX complex, knockdown of gp96/grp94 resulted in the reduction of the intracellular expression of all subunits of the GPIb-IX complex, particularly GPIX (Figure 5B).
GPs acquire endoglycosidase-H (Endo-H) resistance after exiting the ER due to modification of N-glycans in the Golgi compartment.30 Proteins that are misfolded or incompletely assembled are not licensed to leave the ER through a process known as ER quality control, and thus remain Endo-H sensitive.23 GPIbα is known to undergo both N- and O-linked glycosylation,22 the former occurring in the ER and the latter in the Golgi. Immature GPIbα has a mass of ∼ 90 kDa; however, extensive O-linked glycosylation results in a significant increase in GPIbα mass to ∼130 kDa. N-linked glycosylation does not contribute significantly to the increase in GPIbα mass. Strikingly, after gp96/grp94 knockdown, GPIbα was found to be mostly immature and Endo-H sensitive (Figure 5C), suggesting its retention in the ER. This was in stark contrast to EV cells, which had mostly mature and Endo-H–resistant GPIbα.
To confirm whether GPIX reduction in the absence of gp96/grp94 was due to increased degradation of GPIX, we treated cells with inhibitors of the ERAD pathway, and found that GPIX expression could be rescued by treatment of cells with an inhibitor of ER mannosidase I, kifunensine, but not with a lysosomal inhibitor, chloroquine (Figure 5D). Because glycan trimming is the first critical step for ERAD of GPs, these data are consistent with the classic concept of ERAD-mediated degradation of GPIX in the absence of gp96/grp94.
We hypothesized that gp96/grp94 chaperones the GPIb-IX complex by binding directly to the luminal side of one or more subunits of the complex. We immunoprecipitated each individual component of the GPIb-IX complex using antibodies directed against the cytoplasmic tails, and examined its association with gp96/grp94. We found that gp96/grp94 could be coprecipitated with the GPIX subunit (Figure 5E), but not with GPIbα or GPIbβ (supplemental Figure 2). By ectopically expressing GPIX or its ectodomain without the other 2 subunits in a murine pre-B-cell line, we also observed complex formation between gp96/grp94 and GPIX (supplemental Figure 3). Furthermore, an in vitro experiment demonstrated that purified gp96/grp94 bound directly to N-terminal GST-tagged gpIX, but not to GST itself (Figure 5F). These data suggest that the association between gp96/grp94 and free GPIX is the critical step in the assembly of the GPIb-IX complex, which is consistent with the known importance of GPIX in the subsequent GPIb-IX complex formation.31,32
The GPIb-IX complex is sensitive to general ER stress
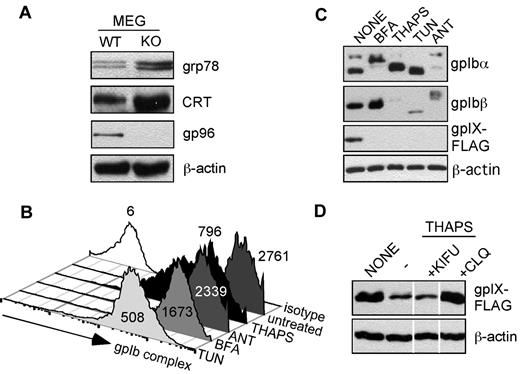
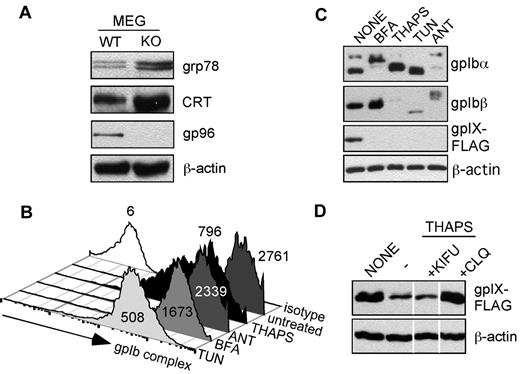
We next examined the impact of gp96/grp94 loss on the general UPR in megakaryocytes and Ib9-IX cells. We found that the UPR is strongly induced, as indicated by a 2- to 4-fold up-regulation in ER HSPs, grp78 (Bip), and calreticulin (Figure 6A). To investigate the possibility that the ensuing ER stress downstream of loss of gp96/grp94 could explain the loss of the GPIb-IX complex and the BSS-like phenotype in gp96/grp94 KO mice, we treated WT IB9-IX cells with a panel of known ER stressors: brefeldin A, thapsigargin, tunicamycin, and antimycin A. We found that virtually all treatments resulted in the reduction of the cell-surface expression of the GPIb-IX complex, with tunicamycin and thapsigargin resulting in an ∼ 3- to 5-fold reduction in surface GPIbα expression (Figure 6B). This result was not surprising, given that the induction of ER stress and UPR are known to arrest nonessential protein synthesis/folding. Therefore, the residual surface GPIbα staining may reflect preexisting GPIb-IX complexes that would be resistant to ER stressors, because the total protein expression level of GPIb-IX complex was significantly reduced (Figure 6C). Similar to the gp96/grp94 shRNA treatment, the GPIX subunit is the most labile molecule in response to all ER stressors (Figure 6C). Therefore, we sought to determine the mechanism of GPIX degradation in response to general ER stress. Thapsigargin was chosen as the ER stressor because, unlike tunicamycin, which inhibits N-linked glycosylation, thapsigargin treatment does not directly affect GPIX glycosylation. Interestingly, GPIX expression on thapsigargin-treated cells could be rescued by treatment with the lysosomal inhibitor chloroquine, but not with kifunensine (Figure 6D). Therefore, the mechanism of GPIX degradation differs with regard to its dependence on gp96/grp94 for steady-state folding and general ER stress. In gp96/grp94 KO cells, GPIX degradation occurs through a classic ERAD pathway that is dependent upon the trimming of N-glycans, whereas in thapsigargin-stressed WT cells, GPIX degradation requires the post-ER acidic compartment. Therefore, we suggest that the loss of gp96/grp94 itself, but not the induction of general ER stress, is responsible for the failure of proper folding and assembly of the GPIb-IX complex in gp96/grp94KO mice. We conclude that the GPIb-IX complex is critically dependent on the integrity of gp96/grp94.
GPIb-IX complex is sensitive to ER stress. (A) Western blot for indicated proteins in primary megakaryocytes from WT and KO mice. (B) Effect of ER stress on the expression level of cell-surface GPIb-IX complex. Flow cytometry for GPIbα (AK2) expression after Ib9-IX cells were treated with the ER stressors thapsigargin (THAPS), antimycin A (ANT), brefeldin A (BFA), or tunicamycin (TUN) or left untreated. Isotype staining is shown. Numbers represents mean fluorescence intensity. Data are representative of 2 independent experiments. (C) Western blot for GPIbα, GPIbβ, and GPIX expression in Ib9-IX cells after indicated treatments. (D) GPIX is degraded in response to ER stress. Western blot analysis for GPIX subunit expression in thapsigargin-exposed Ib9-IX cells after cotreatment with kifunensine (KIFU) and chloroquine (CLQ). Vertical lines have been inserted to indicate a repositioned gel lane. Data are representative of 2 independent experiments.
GPIb-IX complex is sensitive to ER stress. (A) Western blot for indicated proteins in primary megakaryocytes from WT and KO mice. (B) Effect of ER stress on the expression level of cell-surface GPIb-IX complex. Flow cytometry for GPIbα (AK2) expression after Ib9-IX cells were treated with the ER stressors thapsigargin (THAPS), antimycin A (ANT), brefeldin A (BFA), or tunicamycin (TUN) or left untreated. Isotype staining is shown. Numbers represents mean fluorescence intensity. Data are representative of 2 independent experiments. (C) Western blot for GPIbα, GPIbβ, and GPIX expression in Ib9-IX cells after indicated treatments. (D) GPIX is degraded in response to ER stress. Western blot analysis for GPIX subunit expression in thapsigargin-exposed Ib9-IX cells after cotreatment with kifunensine (KIFU) and chloroquine (CLQ). Vertical lines have been inserted to indicate a repositioned gel lane. Data are representative of 2 independent experiments.
Discussion
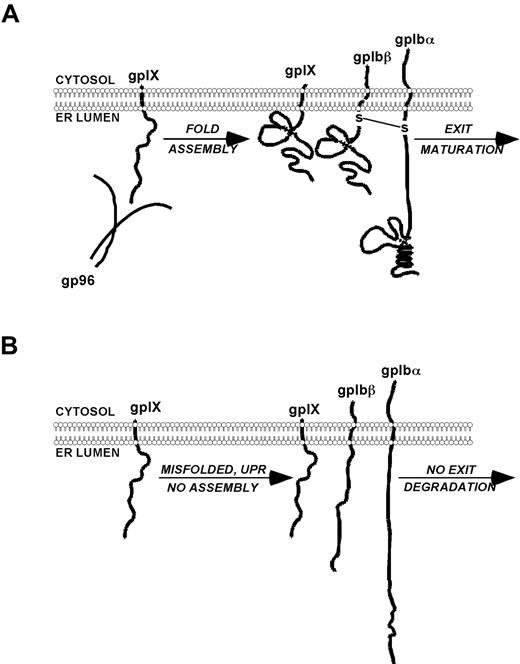
In the present study, we uncovered a novel role of a major ER luminal HSP in the folding of the GPIb-IX complex, a receptor for the platelet VWF. Genetic KO of gp96/grp94 in mice led to a complete loss of the mature GPIb-IX complex in a cell-intrinsic fashion, and also resulted in macrothrombocytopenia and prolonged bleeding time consistent with human BSS. Furthermore, we demonstrated that gp96/grp94 interacted with GPIX, but not with other subunits of the GPIb-IX complex. In the absence of gp96/grp94, GPIX became unstable and was catabolized via the classic ERAD pathway. These results suggest a model in which gp96/grp94 chaperones GPIX selectively, enabling the complex assembly with GPIbα and GPIbβ, and that in its absence, intracellular retention and degradation of the unassembled subunits occur (Figure 7). Our study has established for the first time the importance of gp96/grp94 as an essential molecular chaperone for GPIX that is required for the assembly of the platelet GPIb-IX complex.
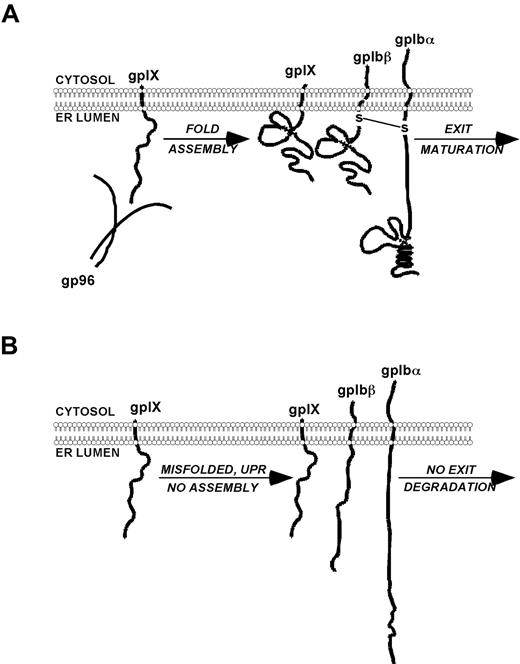
Proposed model of the role of gp96/grp94 in the chaperone and assembly of the GPIb-IX complex. (A) gp96/grp94 binds to GPIX and promotes the normal maturation of the GPIb-IX complex, resulting in normal-sized platelets and homeostasis. (B) In the absence of gp96/grp94, GPIX and consequently GPIbβ and GPIbα are misfolded, retained in the ER, or degraded, resulting in a BSS-like disorder.
Proposed model of the role of gp96/grp94 in the chaperone and assembly of the GPIb-IX complex. (A) gp96/grp94 binds to GPIX and promotes the normal maturation of the GPIb-IX complex, resulting in normal-sized platelets and homeostasis. (B) In the absence of gp96/grp94, GPIX and consequently GPIbβ and GPIbα are misfolded, retained in the ER, or degraded, resulting in a BSS-like disorder.
gp96/grp94 is the most abundant chaperone in the ER lumen and is one of the key downstream chaperones in ER quality control, acting by mediating the UPR.33 gp96/grp94 is also critical for folding members of both TLR and integrin families,7,,–10 even though there is no apparent sequence/structural homology between the 2. One hypothesis is that gp96/grp94 is involved in the coordinated multimerization of selective protein substrates through binding to and stabilizing protein-folding intermediates, thereby preventing inappropriate protein-protein interactions and aggregations.
The GPIb complex is one such multisubunit complex that likely requires the help of ER chaperones such as gp96/grp94 to orchestrate its proper folding, assembly, and thus normal maturation and transport. Unfortunately, study of the folding of the GPIb complex in BSS has been limited to the role of each of its constituents, GPIbα, GPIbβ, and GPIX (GPV plays little role in the assembly of the GPIb complex).34 Mutations in GPIbα, GPIbβ, and GPIX result in an incomplete or misfolded GPIb complex in humans that leads to BSS. The assembly of the GPIb complex takes place in the ER, and occurs immediately after the synthesis of its constituent polypeptides.22 The newly assembled GPIb complex takes only approximately 160 minutes to reach the cell surface, undergoing numerous posttranslational modifications as a part of its normal maturation.22 Efficient transport of the ligand-binding subunit of the complex, GPIbα, to the plasma membrane requires both GPIbβ and GPIX, which direct GPIbα away from lysosomes and therefore protect it from proteolysis, essentially acting as chaperones for GPIbα. Mutations in GPIbα that prevent the normal maturation and assembly of the GPIb complex do not necessarily result in GPIbα degradation.35 However, mutations in either GPIbβ or GPIX result in the instability of GPIbα and the dissociation of the GPIb complex.36,37 Therefore, the interaction between GPIX and GPIbβ subunits is required for the normal expression of the GPIb complex.31 Consistent with a role for gp96/grp94 in folding the gpIb complex, surface and intracellular expression of GPIbα was markedly reduced in KO megakaryocytes in the present study. In vitro knockdown of gp96/grp94 by shRNA led to the decreased expression of GPIX and GPIbβ, the intracellular retention of GPIbα, and decreased assembly with GPIbβ.
There was a slight reduction of cell-surface CD41 (αIIb) on KO megakaryocytes and platelets. However, as reported previously, unlike other bona fide client proteins such as β2 integrin, the surface expression of this integrin is never lost in the absence of gp96/grp94.10 However, the subtle reduction of CD41 in gp96/grp94 KO cells may also contribute to the observed abnormality of KO megakaryocytes and platelets. Given that gp96/grp94 KO is incompatible with life in mice because of early embryonic lethality,38 it is unlikely that the null mutation of gp96/grp94 contributes to the congenital BSS. However, the results of our study clearly illustrate that perturbation of the protein quality-control system in the ER could contribute to platelet disorders. This notion is reminiscent of the recent discovery of the chaperone disorder causing combined factor V and factor VIII deficiency.39 Indeed, treatment of cells expressing the GPIb complex with known chemical inducers of ER stress has revealed rather remarkable sensitivity of the GPIb complex to ER stress. This suggests that gp96/grp94, and possibly other ER chaperones, may participate in the biogenesis of the GPIb complex. The aberrant expression of gp96/grp94 may also cause acquired BSS. Intriguingly, BSS-like hematologic indicators such as giant platelets and reduction of surface GPIbα expression have been reported in subsets of patients with myelodysplasia.40,41
In conclusion, our study has established gp96/grp94 as a molecular chaperone for gpIX and the gpIb complex. We have provided yet another striking example of the defect of protein quality control in causing a unique genetic disorder. Further study of the mechanism of gp96/grp94 in the assembly of the GPIb complex will be useful in uncovering the biology of gp96/grp94 chaperone machinery and in contributing to our understanding of the roles of protein quality control in the regulation of platelet development and function.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the laboratory staff of Dr Leonardo Aguila at the University of Connecticut Health Center (Farmington, CT) for help with the methylcellulose CFU assay; Drs Pramod Srivastava, Kang Liu, Leo Lefrancois, Harry Drabkin, Joachim Deeg, and Lynn Puddington for their helpful input; and Ms Samantha Cronin for her secretarial assistance.
This work was supported by National Institutes of Health grants AI070603, AI077283, and HL100556 (to Z.L.) and T32AI070603 (to M.S.).
National Institutes of Health
Authorship
Contribution: M. S. and Z. L. conceived the idea, designed the research, and wrote the manuscript; S.W., F. H., and B.L. assisted in the binding analysis between gp96/grp94 and GPIX; A.S. and X.D. performed the platelet function study; M.S. performed the rest of the experiments; and all authors including R.B. contributed to the analysis of the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation of M.S. is Department of Immunobiology, Yale University School of Medicine, New Haven, CT.
Correspondence: Zihai Li, Department of Microbiology & Immunology, Medical University of South Carolina (MUSC), HO-612B, Charleston, SC 29425; e-mail: zihai@musc.edu.