Abstract
Phagocytic and pathogen sensing receptors are responsible for particle uptake and inflammation. It is unclear how these receptors' systems influence each other's function to shape an innate response. The class-A scavenger receptors SR-A (scavenger receptor A) and MARCO (macrophage receptor with collagenous structure) are 2 well-characterized phagocytic receptors that are unable to initiate inflammatory responses by themselves, yet are implicated in the pathogenesis of various inflammatory disorders. However, the mechanism for such an apparent discrepancy is still unclear. We utilized SR-A−/−, MARCO−/−, and SR-A−/−-MARCO−/− mice, along with microbe-derived, environmental, and synthetic polyanions to assess the inflammatory responses following combinatorial ligation of SR-A/MARCO and selected Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain (NOD)–like receptors (NLRs) by their shared ligands. In addition to ligating SR-A and MARCO, these agonists also selectively activated the cell-surface sensor TLR4, endosomal TLR3, and the cytosolic NOD2 and NALP3 (NACHT domain–, leucine-rich repeat–, and pyrin domain–containing protein 3). We show that, following recognition of common ligands, SR-A and MARCO attenuate TLR4-mediated responses while enhancing responses by the intracellular TLR3, NOD2, and NALP3. We conclude that SR-A/MARCO-mediated rapid ligand internalization prevented sensing by surface TLRs while increasing ligand availability in intracellular compartments, thus allowing sensing and robust responses by intracellular sensors.
Introduction
Innate immune recognition of microbes by pattern recognition receptors (PRRs) initiates 2 distinct effector responses, phagocytosis and inflammation, to promote pathogen clearance, killing, tissue repair, and induction of adaptive immunity.1 Phagocytic PRRs, such as scavenger receptors, and pathogen sensors, such as Toll-like receptors (TLRs) and NOD-like receptors (NLRs), mediate uptake and initiate inflammation, respectively.2,3 Although much has been learned about these 2 processes separately, knowledge is still limited on how these processes are coordinated during the host response. Functional collaboration between innate receptor families must exist to coordinate uptake and inflammation; breakdown in such receptor collaboration can lead to defective pathogen clearance and inflammatory pathologies.4
The scavenger receptor A (SR-A) and macrophage receptor with collagenous structure (MARCO) are 2 well-characterized, nonopsonic phagocytic receptors on myeloid cells.5,6 Due to their shared multidomain structure and their ability to bind modified low-density lipoprotein and other polyanionic ligands (a common feature of all scavenger receptors), they are defined as members of the class-A scavenger receptor family. Both molecules are multiligand, multifunctional receptors7 ; share large numbers of overlapping but distinct polyanionic ligands; and carry out common functions such as cellular adhesion,8 migration,9 phagocytosis, and antigen presentation.10 Both receptors have been shown to bind a range of isolated microbial components11-14 and intact Gram-positive and Gram-negative organisms,15,16 and their nonredundant role in host defense has been established in a variety of bacterial infection models.17-20
Although SR-A and MARCO recognize a range of highly inflammatory ligands, these receptors by themselves are unable to initiate inflammatory signaling, which is usually triggered by sensing PRRs that share overlapping ligand repertoires. However, SR-A and MARCO have been implicated in the pathogenesis of a range of chronic and acute inflammatory pathologies, including atherosclerosis, Alzheimer disease, granuloma formation, and septic shock.21 Studies utilizing both microbial and sterile inflammatory stimuli and distinct models of inflammation have suggested both proinflammatory and anti-inflammatory roles; however, the mechanisms for such apparent discrepancies are unknown. Therefore, how SR-A and MARCO modulate inflammatory responses without being directly involved in signaling is not only poorly understood, but often contradictory.
We hypothesized that when a common inflammatory ligand is recognized by both SR-A/MARCO and another sensing PRR, SR-A and MARCO modulate the function of the sensor, altering the inflammatory outcome. Direct functional collaboration between several scavenger receptors and TLRs on the cell surface has been implicated in combinatorial recognition and an appropriate tailored response.22,23 Pathogen sensing occurs in both cell surface and intracellular compartments, PRRs can be dynamically shuttled in various subcellular compartments, and a specific subcellular location determines distinct signaling responses of PRRs.24 In addition, SR-A and MARCO ligands range from highly inflammatory bacterial lipopolysaccharides (LPS) to potent anti-inflammatory ligands such as apoptotic cells.11,12,25 Furthermore, there are differences between SR-A and MARCO in structure, splice variants, ligand recognition, tissue distribution, and regulation in response to inflammatory stimuli.5,6 Therefore, we reasoned that the combination of these distinct features of SR-A/MARCO and other pathogen sensors determines selectivity, specificity, and a repertoire of innate immune recognition and inflammatory responses.
To test this, we systematically assessed the innate immune responses following ligation of SR-A/MARCO and selected cell-surface (TLR4), endosomal (TLR3), and cytosolic (nucleotide-binding oligomerization domain 2 [NOD2] and NACHT domain–, leucine-rich repeat–, and pyrin domain–containing protein 3 [NALP3]) sensors by their shared ligands. We used SR-A−/−, MARCO−/−, and SR-A/MARCO double knockout (DKO) mice, along with a range of microbe-derived, environmental, and synthetic polyanions, which, in addition to ligating SR-A and MARCO, also selectively activated specific pathogen sensors. To co-ligate SR-A/MARCO and TLR4, we utilized a clinically important Gram-negative human pathogen, Neisseria meningitidis (NM). SR-A and MARCO are 2 major nonopsonic phagocytic receptors for NM, but such recognition is independent of neisserial LPS.26,27 In contrast, NM-mediated cytokine responses depend on TLR4-mediated recognition of NM LPS and do not require SR-A/MARCO-mediated NM uptake.26 We utilized polyinosinic acid (polyI) a synthetic polyanionic ligand for SR-A/MARCO that also stimulates intracellular TLR3 to release inflammatory cytokines.28 Similarly, the bacterial cell wall product muramyl dipeptide (MDP) stimulated the cytosolic sensor NOD2, and silica, asbestos, and a combination of NM and extracellular ATP stimulated NALP3 inflammasomes and engaged SR-A and MARCO.29,30
We show that SR-A/MARCO competes with TLR4 for ligand recognition, limiting its inflammatory response, but strikingly enhances TLR3-, NOD2-, and NALP3-mediated inflammatory responses. We conclude that rapid ligand internalization via SR-A/MARCO prevents sustained sensing by surface TLRs. However, such internalization also increases ligand availability in intracellular compartments, allowing sustained sensing and robust responses by distinct intracellular pathogen sensors.
Methods
Mouse strains
SR-A−/−, MARCO−/−, and SR-A−/−-MARCO−/− DKO mouse strains of the C57BL/6J background were kindly provided by the original investigators.17,31 Animals were housed in specific pathogen-free conditions, and all procedures involving animals were conducted according to the requirements of the United Kingdom Home Office Animals Scientific Procedures Act, 1986.
Bacterial culture and fluorescent labeling
The encapsulated serogroup B strains of pathogenic NM strain MC58 were cultured in an atmosphere of 5% CO2 overnight at 37°C on brain-heart infusion medium (Oxoid), supplemented with Levinthal reagent (10% vol/vol) and solidified with agar (1% wt/vol). For fluorescent labeling, NM was harvested and resuspended in 70% ethanol overnight at 4°C and labeled with RdGnX (RdGnX-NM) from Molecular Probes according to the manufacturer's instructions.
Isolation and culture of primary Mϕ
Biogel-elicited peritoneal Mϕ (Bg-PM) were prepared by intraperitoneal injection of 1 mL of polyacrylamide gel P-100 (Bio-Rad) beads at a 2% wt/vol suspension in endotoxin-free phosphate-buffered saline (PBS).27 Four days after injection, cells were harvested by peritoneal lavage with ice-cold PBS and plated on bacteriological plastic dishes in a defined serum-free medium, OPTIMEM-1, supplemented with 500 IU/mL of penicillin-streptomycin and 2mM glutamine. After 3-4 hours, plates were washed 3 times to remove nonadherent cells and Biogel beads. After washing, the purity of Mϕ in the adherent monolayer was > 98%.
In vitro stimulation of Mϕ to activate TLR and NLR pathways
To activate TLR3 and TLR4 pathways, Mϕ were stimulated with different concentrations of polyI (100 μg/mL) and 20 multiplicity of infectious units (MOI) of inactivated NM, respectively. To activate NOD2, cells were first primed with 20 ng/mL of recombinant murine interferon-γ (Mϕ priming is needed to up-regulate NOD2 signaling components in vitro) and then stimulated with 50 μg/mL of MDP. NALP3 inflammasomes were activated by stimulating Mϕ for 6 hours with either 100 μg/mL of polyI or 20 MOI of inactivated NM, followed by a short pulse (≤ 5 minutes) of 1mM ATP; 6 hours after the ATP pulse, supernatants were collected and interleukin-1β (IL1β) secretion was analyzed by enzyme-linked immunosorbent assay (ELISA). In some cases, ATP pulsing was replaced by overnight stimulation with 250 μg/mL of silica (MIN-U-SIL 5; US Silica Company) or 250 μg/mL of asbestos to activate NALP3 inflammasomes, and IL1β secretion was measured.
Sterile peritonitis model
To induce sterile peritonitis, mice were injected intraperitoneally with 1 × 105 colony-forming units (CFU) of ethanol-inactivated NM or 100 μg of purified MDP suspended in 200 μL of PBS. Mice in the control group were injected with the same volume of PBS alone. Two hours after injection, mice were killed and inflammatory cellular infiltrate was collected by peritoneal lavage with 1 mL of ice-cold PBS. Care was taken to avoid erythrocyte contamination. Cells were separated by centrifugation, and leukocyte populations were analyzed by multicolor flow cytometry using well-characterized cell-type–specific markers. Cytokine levels in lavage fluid were measured by ELISA. To assess NALP3-mediated IL1β secretion, 200 μL of ATP (1mM, pH 7.4) was injected 2 hours after the first NM injection, and peritoneal lavage was performed after another 2 hours following the method of Griffiths et al with modifications.32
Cytokine and chemokine analysis
Levels of IL6 and tumor necrosis factor-α (TNF-α; BD Biosciences), macrophage inflammatory protein 2 (MIP2) and keratinocyte-derived cytokine (KC; R&D Systems), and IL1β (eBioscience) were determined by ELISA according to the manufacturers' instructions. To ensure that differences in cell plating did not contribute to differences in results, the number of cells plated in parallel wells was determined by incubation with carboxyfluorescein diacetate succinimidyl ester from Molecular Probes using a plate reader. No significant cell loss was observed in any genotype (data not shown).
Real-time PCR
Mϕ were stimulated with 20 NM/Mϕ or 100 ng/mL of LPS for 3 hours, RNA was extracted with TRI-reagent, treated with RNase-free DNAse I, and purified with an RNeasy kit (QIAGEN). Two micrograms of RNA were reverse transcribed using the Omniscript RT kit (QIAGEN) with a random hexamer primer. Resulting cDNA was analyzed by real-time polymerase chain reaction (PCR). Each reaction was prepared with .25× SYBR Green I nucleic acid stain (Invitrogen), 10nM fluorescein isothiocyanate (FITC; Bio-Rad), 50mM KCl, 20mM Tris-HCl (pH 8.3), 25mM MgCl2, 100 μg/mL of bovine serum albumin (BSA), and 4μM deoxynucleotide triphosphate (Denville Scientific) with 1.25 U Choice-Taq (Denville Scientific). Real-time PCR amplification conditions were 95°C (3 minutes) and 45 cycles of 95°C (15 seconds), 60°C (30 seconds), and 72°C (30 seconds). The following IL1β primers were used: forward, 5′ACGGACCCCAAAAGATGAAG3′, reverse, 5′TTCTCCACAGCCACAATGAG3′.
Flow cytometry to assay surface antigen expression and association of bacteria
Multicolor flow cytometry was performed on peritoneal exudate cells to analyze the expression of various surface antigens. Antibody staining was performed according to conventional protocols at 4°C in the presence of 2mM NaN3. Cells were incubated at 4°C for 30 minutes in FACS blocking buffer. Directly conjugated primary antibodies were added and incubated for another 1 hour. Cells were washed 3 times with FACS wash buffer (5% heat-inactivated rabbit serum, 0.5% BSA, 5mM EDTA [ethylenediaminetetraacetic acid], and 10 μg/mL of 2.4G2 antibody) and fixed with 2% paraformaldehyde. Fixed cells were analyzed on an FACSCalibur (BD Biosciences) using FlowJo software Version 3 (TreeStar). To assess in vivo bacterial clearance, mice were injected intraperitoneally with 106 CFU of fluorescently labeled inactivated NM, peritoneal exudate cells were isolated and fixed, and total NM uptake was measured by flow cytometry.
ELISA overlay assays
OPTI-EIA plates (96-well; Corning) were coated overnight with candidate ligands (10 μg/mL unless stated otherwise) at 4°C in replicates of 10. The wells were blocked with 10% LPS-free BSA before incubation with postnuclear supernatant prepared from wild-type (WT) or SR-A−/− bone marrow–derived macrophages (BMMϕ) for 2 hours in the presence of 5mM EDTA. Five wells of each candidate were incubated with WT or SR-A−/− lysate. The wells were washed with PBS containing 0.1% Tween 20, and incubated at room temperature for 2 hours with 10 μg/mL of anti–SR-A antibody (2F8). Binding of the SR-A antibody was detected by incubating the wells with a horseradish peroxidase–conjugated anti–rat antibody (BioLegend) and visualized using TMB reagent (BD Bioscience) according to the manufacturer's instructions. The average of 5 replicates for each condition was plotted and results are representative of at least 3 independent experiments. The statistical significance of results was determined using the paired Student t test and significance tested at the 95% confidence level (P ≤ .05).33 The MARCO overlay ELISA assay was optimized by adjusting the above protocol as follows: ELISA plates were coated with potential ligands and 5% milk was used as a blocking agent. Ligands were incubated with recombinant extracellular MARCO (sMARCO) at a final concentration of 3 μg/mL. sMARCO was produced in 293/EBNA cells and purified using a His tag as described in Sankala et al.12 In parallel, we incubated the ligands with another unrelated His-tagged Mϕ surface protein, EMR1 (sEMR1), to control for nonspecific binding. sMARCO and sEMR1 were detected using the monoclonal antibodies ED31 for MARCO and BC9 for EMR1; primary antibodies were detected with horseradish peroxidase–conjugated secondary antibodies. During optimization of this protocol, appropriate isotype-matched control antibodies for ED31 and BC9 were included. Because no nonspecific binding was observed for ED31 or BC9, we excluded these isotype controls for subsequent assays.
Results
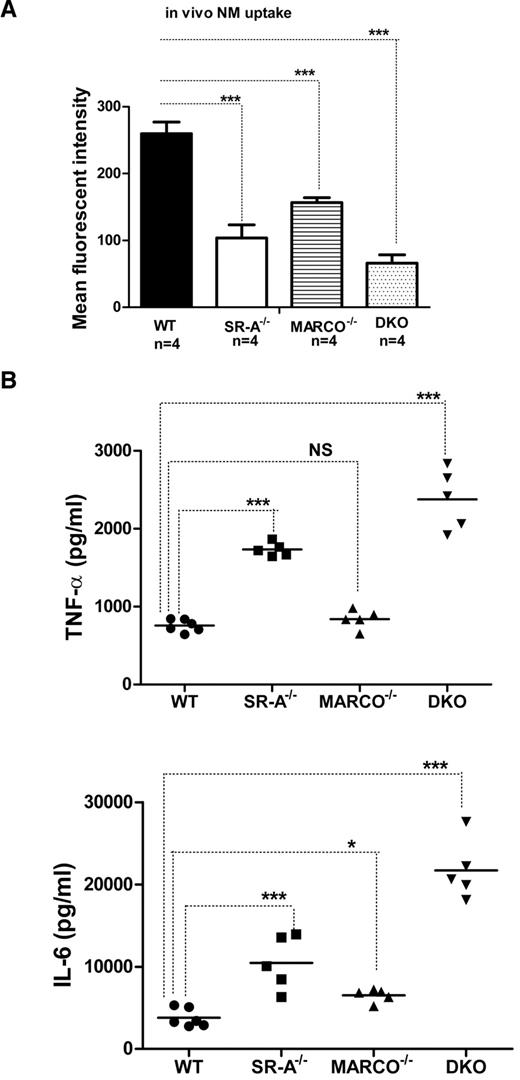
Lack of SR-A and MARCO induces an exaggerated cytokine response during sterile peritonitis in vivo
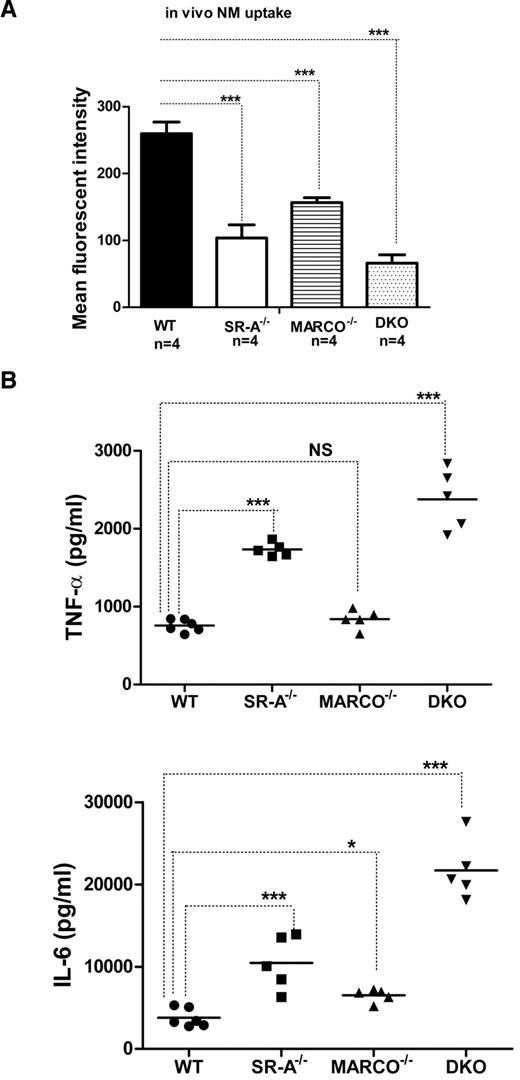
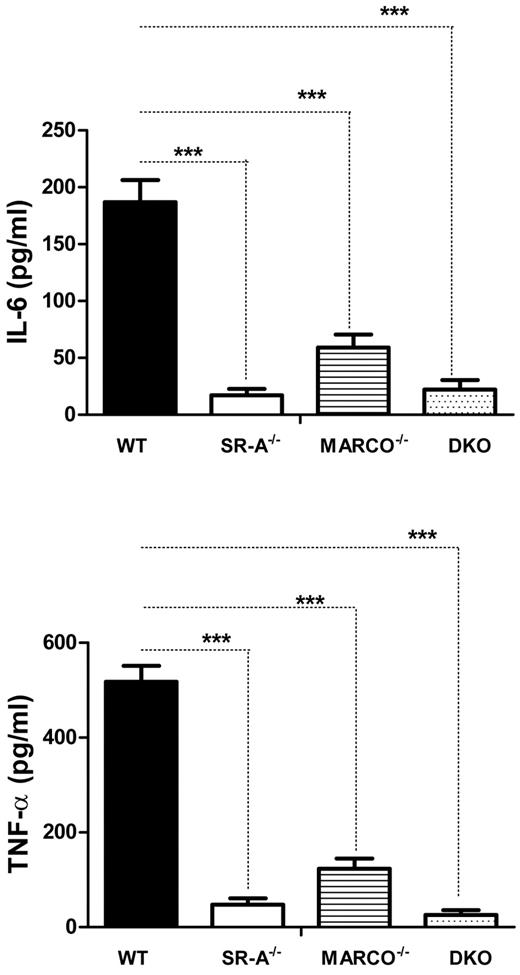
SR-A and MARCO are 2 major nonopsonic uptake receptors for the Gram-negative pathogen NM, while TLR4 is essential for NM-induced inflammatory responses.26,27 We previously reported that although Bg-PM and resident peritoneal macrophages (RPMs) from SR-A−/− and DKO mice are significantly deficient in NM uptake, they showed a modest increase in TNF-α secretion.27 In the present study, we set out to compare how SR-A and MARCO contribute to NM clearance and modulate early cytokine responses in vivo. WT, SR-A−/−, MARCO−/−, and DKO mice were injected intraperitoneally with fluorescently labeled NM, peritoneal exudate cells were isolated after 45 minutes, and NM uptake was measured by flow cytometry. Our results showed that NM uptake was reduced in all 3 knockout strains, indicating important roles for these 2 receptors in bacterial clearance in vivo (Figure 1A). Levels of the proinflammatory cytokines TNF-α and IL6 were also compared in peritoneal lavage fluid 2 hours after intraperitoneal challenge with 1 × 105 CFU of ethanol-inactivated NM (Figure 1B). Induction of cytokine was highest in DKO mice compared with WT mice, followed by SR-A−/− mice, but MARCO−/− mice showed only a modest increase over the WT strain, indicating that both SR-A and MARCO mediated clearance of NM partially limited TLR4-NM–mediated cytokine response by efficient ligand scavenging, with a predominant contribution of SR-A. These findings are consistent with a previous report that SR-A mice show enhanced bacterial load and produce strikingly high levels of proinflammatory cytokines during NM infection.20 The use of ethanol-fixed organisms in the present study ruled out any potential immune modulation by replicating pathogens while retaining the SR-A/MARCO and TLR4 ligand activity.
SR-A and MARCO enhance bacterial clearance in vivo and limit NM-TLR4–mediated inflammatory cytokine responses. (A) SR-A−/−, MARCO−/−, and DKO mice show defective bacterial clearance in vivo. 106 CFU of RdGNX-labeled NM was injected into the peritoneal cavity of WT, SR-A−/−, MARCO−/−, and DKO mice. After 45 minutes, peritoneal exudate cells were harvested by lavage and bacterial uptake was analyzed by flow cytometry. (B) SR-A−/−, MARCO−/−, and DKO mice showed enhanced cytokine production following intraperitoneal injection of 105 CFU of inactivated NM. Secretion of TNF-α (top panel) and IL6 (bottom panel) was measured by ELISA in peritoneal lavage fluid 2 hours after NM injection. Data presented are representative of 3 independent experiments. One-way ANOVA with Dunnett multiple comparison test was used to assess statistical significance. *P ≤ .05; ***P < .001
SR-A and MARCO enhance bacterial clearance in vivo and limit NM-TLR4–mediated inflammatory cytokine responses. (A) SR-A−/−, MARCO−/−, and DKO mice show defective bacterial clearance in vivo. 106 CFU of RdGNX-labeled NM was injected into the peritoneal cavity of WT, SR-A−/−, MARCO−/−, and DKO mice. After 45 minutes, peritoneal exudate cells were harvested by lavage and bacterial uptake was analyzed by flow cytometry. (B) SR-A−/−, MARCO−/−, and DKO mice showed enhanced cytokine production following intraperitoneal injection of 105 CFU of inactivated NM. Secretion of TNF-α (top panel) and IL6 (bottom panel) was measured by ELISA in peritoneal lavage fluid 2 hours after NM injection. Data presented are representative of 3 independent experiments. One-way ANOVA with Dunnett multiple comparison test was used to assess statistical significance. *P ≤ .05; ***P < .001
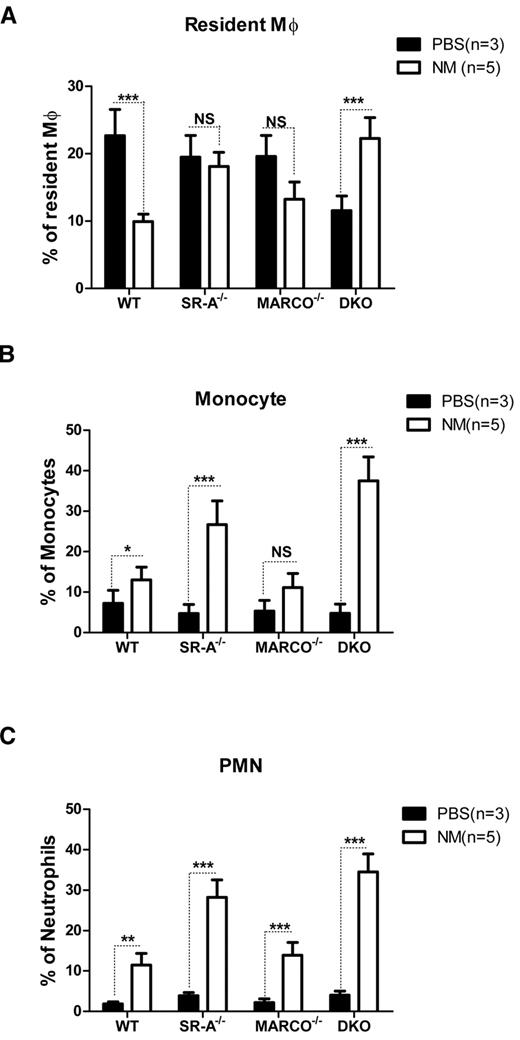
SR-A and MARCO differentially regulate trafficking of inflammatory cells during sterile peritonitis
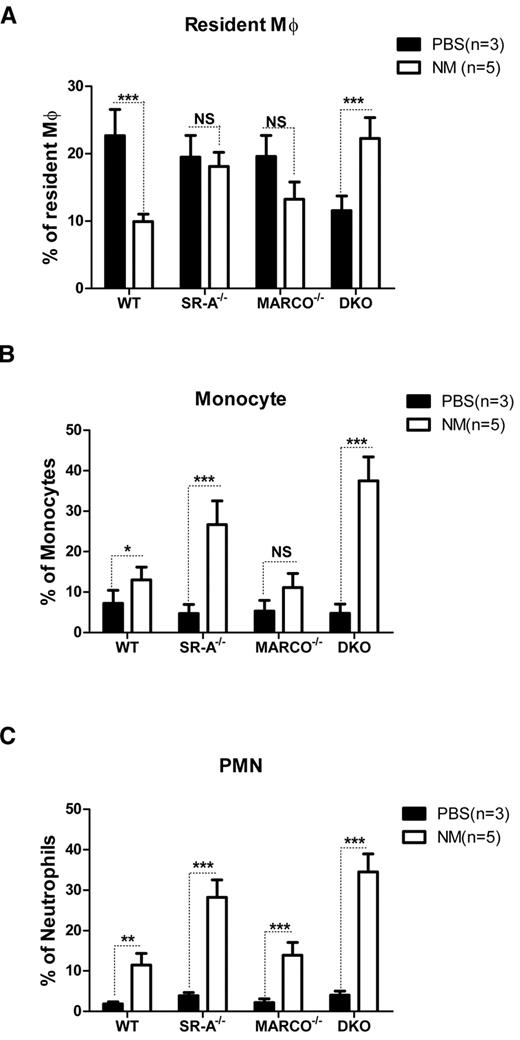
In addition to inflammatory cytokine release, recruitment of various leukocyte populations is another hallmark of inflammation. To determine how SR-A and MARCO deficiency modulates leukocyte trafficking during the early phase of acute peritonitis, the composition of various leukocyte populations was analyzed by flow cytometry 2 hours after intraperitoneal injection of NM or control PBS. Results showed similar numbers of F4/80hiCD11bhiGR-1−ve RPMs in PBS-injected SR-A−/−, MARCO−/−, and WT mice, but fewer macrophages in DKO mice. The number of these cells strikingly decreased in WT mice following NM injection. Although less strikingly, MARCO−/− mice showed a similar drop in CD11bhiF4/80hi cell number following NM challenge; however, this drop was not observed in SR-A−/− mice, and DKO mice showed even smaller increases in CD11bhiF4/80hi cell numbers following NM challenge (Figure 2A). It is important to note that all of the CD11bhiF4/80hi cells in the NM-treated groups were not necessarily RPMs; hyperactivated monocytes acquiring the F4/80hi phenotype may have contributed to this pool in SR-A−/− and DKO mice. In contrast to resident Mϕ, F4/80lowCD11b+veGR-1med inflammatory monocytes (Figure 2B) and CD11bhiGR-1hiF4/80−ve neutrophil (Figure 2C) populations were recruited in all mouse strains following NM challenge, but induced recruitment was strikingly higher in SR-A−/− and DKO mice compared with the WT and MARCO−/− strain. We conclude that SR-A and MARCO limit the excessive recruitment of inflammatory monocytes and neutrophils during peritonitis.
SR-A and MARCO regulate leukocyte trafficking during NM-induced peritonitis. WT, SR-A−/−, MARCO−/−, and DKO mice were injected intraperitoneally with 1 × 105 CFU of inactivated NM or equal volumes of PBS. After 2 hours, the percentage of resident macrophages (A), recruited monocytes (B), and neutrophils (C) from total leukocytes were determined in peritoneal exudates by flow cytometry and compared among strains. No significant difference in the number of monocytes and neutrophils was observed among the mouse strains in the vehicle-injected groups. Similarly, no significant difference was observed in the number of resident macrophages in PBS-treated SR-A and MARCO mice compared with WT mice, but PBS-treated DKO mice showed significantly fewer resident macrophages (P ≤ .01). Comparisons between PBS-treated (n ≥ 3) and NM-treated (n = 5) groups are presented and are representative of 3 independent experiments. The 2-way ANOVA with Bonferroni posttest was used to assess statistical significance. *P ≤ .05; **P ≤ .01; ***P ≤ .001
SR-A and MARCO regulate leukocyte trafficking during NM-induced peritonitis. WT, SR-A−/−, MARCO−/−, and DKO mice were injected intraperitoneally with 1 × 105 CFU of inactivated NM or equal volumes of PBS. After 2 hours, the percentage of resident macrophages (A), recruited monocytes (B), and neutrophils (C) from total leukocytes were determined in peritoneal exudates by flow cytometry and compared among strains. No significant difference in the number of monocytes and neutrophils was observed among the mouse strains in the vehicle-injected groups. Similarly, no significant difference was observed in the number of resident macrophages in PBS-treated SR-A and MARCO mice compared with WT mice, but PBS-treated DKO mice showed significantly fewer resident macrophages (P ≤ .01). Comparisons between PBS-treated (n ≥ 3) and NM-treated (n = 5) groups are presented and are representative of 3 independent experiments. The 2-way ANOVA with Bonferroni posttest was used to assess statistical significance. *P ≤ .05; **P ≤ .01; ***P ≤ .001
SR-A and MARCO limit inflammatory CXC chemokine production in vitro and in vivo
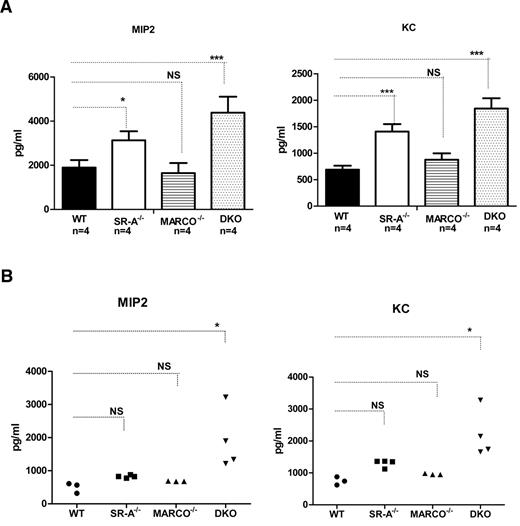
Previously, SR-A has been shown to limit neutrophil chemoattractant MIP2 (CXCL1) and KC (CXCL2) production during sterile peritonitis.34 Because we observed enhanced inflammatory cell recruitment in SR-A−/− and DKO mice, we set out to compare the contributions of SR-A and MARCO in NM-mediated inflammatory chemokine production. Bg-PM from WT, SR-A−/−, MARCO−/−, and DKO mice were stimulated with NM, and MIP2 and KC levels in the culture supernatant were measured. No chemokine secretion was observed in untreated controls of either mouse strain (data not shown), but SR-A−/− and DKO Mϕ produced significantly higher levels of MIP2 and KC after NM stimulation (Figure 3A). Similar to in vitro experiments, DKO mice produced significantly higher levels of MIP2 and KC in vivo after intraperitoneal NM challenge (Figure 3B). However, enhanced chemokine response was less prominent in SR-A−/− and MARCO−/− mice in vivo (Figure 3B). These data indicate that SR-A and, to some extent, MARCO regulate inflammatory chemokine production in response to NM challenge, possibly by rapid NM clearance.
SR-A and MARCO regulate NM-mediated inflammatory chemokine secretion. (A) After NM stimulation, Bg-PM from SR-A−/−, MARCO−/−, and DKO mice (all n = 4) produced enhanced levels of the inflammatory chemokines MIP2 (left panel) and KC (right panel) compared with WT cells. Mouse strains were stimulated with 20 MOI of ethanol-inactivated NM for 16 hours. Levels of MIP2 and KC were measured by ELISA. No chemokine was detected in the untreated groups (data not shown). (B) Enhanced levels of MIP2 (left panel) and KC (right panel) were produced in the peritoneal cavity of SR-A−/−, MARCO, and DKO mice injected intraperitoneally with 105 CFU of inactivated NM or an equal volume of PBS. After 2 hours, animals were killed and peritoneal lavage performed using 1 mL of PBS. Levels of MIP2 and KC were measured by ELISA. No chemokine was detected in vehicle-injected controls in any mouse strain (data not shown). Statistical significance was assessed by 2-way ANOVA with Bonferroni posttest. One-way ANOVA with Dunnett multiple comparison test was used to assess statistical significance. *P ≤ .05; ***P < .001.
SR-A and MARCO regulate NM-mediated inflammatory chemokine secretion. (A) After NM stimulation, Bg-PM from SR-A−/−, MARCO−/−, and DKO mice (all n = 4) produced enhanced levels of the inflammatory chemokines MIP2 (left panel) and KC (right panel) compared with WT cells. Mouse strains were stimulated with 20 MOI of ethanol-inactivated NM for 16 hours. Levels of MIP2 and KC were measured by ELISA. No chemokine was detected in the untreated groups (data not shown). (B) Enhanced levels of MIP2 (left panel) and KC (right panel) were produced in the peritoneal cavity of SR-A−/−, MARCO, and DKO mice injected intraperitoneally with 105 CFU of inactivated NM or an equal volume of PBS. After 2 hours, animals were killed and peritoneal lavage performed using 1 mL of PBS. Levels of MIP2 and KC were measured by ELISA. No chemokine was detected in vehicle-injected controls in any mouse strain (data not shown). Statistical significance was assessed by 2-way ANOVA with Bonferroni posttest. One-way ANOVA with Dunnett multiple comparison test was used to assess statistical significance. *P ≤ .05; ***P < .001.
PolyI- and TLR3-mediated cytokine response requires SR-A and MARCO
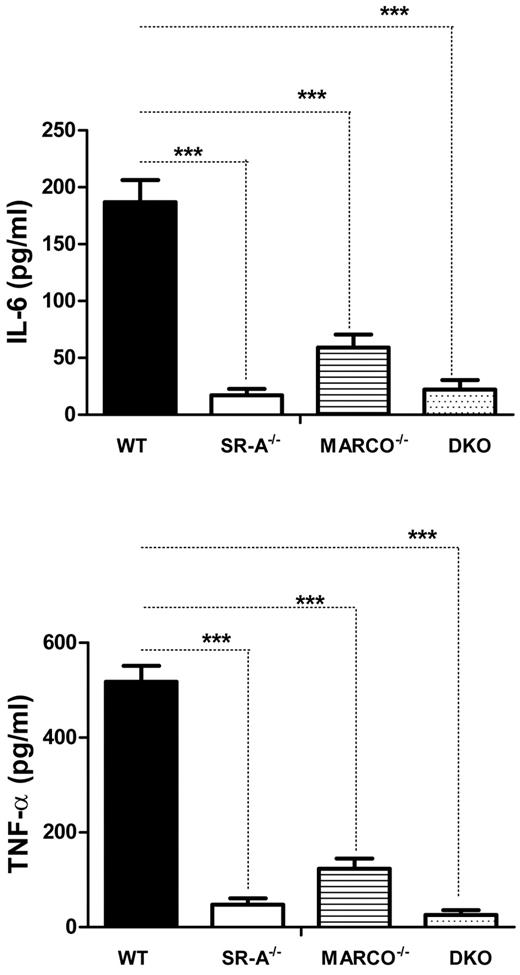
TLR4 recognizes NM at the cell surface without requiring internalization,26 signaling through the adaptor protein MyD88 (myeloid differentiation factor 88) to induce an innate immune response, and we show that SR-A and MARCO regulate such responses. To investigate how SR-A and MARCO modulate the inflammatory response by intracellular TLR3, which signals independently of MyD88, we utilized polyI, a synthetic polyanionic ligand for SR-A, MARCO, and intracellular TLR3.28 WT, SR-A−/−, MARCO−/−, and DKO macrophages were stimulated with polyI, and secretion of cytokines was measured. Results showed that polyI induced TNF-α and IL6 in WT Mϕ as expected, but cytokine secretion was almost completely abolished in SR-A−/− and DKO macrophages. MARCO−/− macrophages also showed a significant decrease in cytokine response following polyI stimulation (Figure 4). In contrast, polyC (a control nonligand for SR-A and MARCO) did not induce any cytokine secretion in either mouse strain (not shown). This finding indicates that SR-A-mediated and, to a lesser extent, MARCO-mediated internalization of polyI enhances TLR3-mediated inflammatory cytokine secretion.
SR-A and MARCO promote TLR3 activation by ligand delivery. Bg-PM from SR-A, MARCO, and DKO mice show reduced cytokine production following polyI stimulation compared with WT control. TNF-α and IL6 concentrations were measured in supernatants following 16 hours of stimulation of Bg-PM with 100 μg/mL of polyI. Two-way ANOVA with the Bonferroni posttest was used to assess statistical significance. ***P < .001
SR-A and MARCO promote TLR3 activation by ligand delivery. Bg-PM from SR-A, MARCO, and DKO mice show reduced cytokine production following polyI stimulation compared with WT control. TNF-α and IL6 concentrations were measured in supernatants following 16 hours of stimulation of Bg-PM with 100 μg/mL of polyI. Two-way ANOVA with the Bonferroni posttest was used to assess statistical significance. ***P < .001
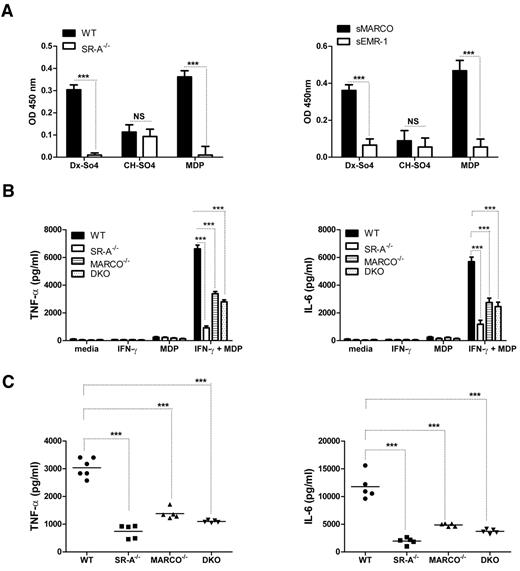
SR-A and MARCO are involved in MDP delivery and are essential for NOD2-mediated cytokine response
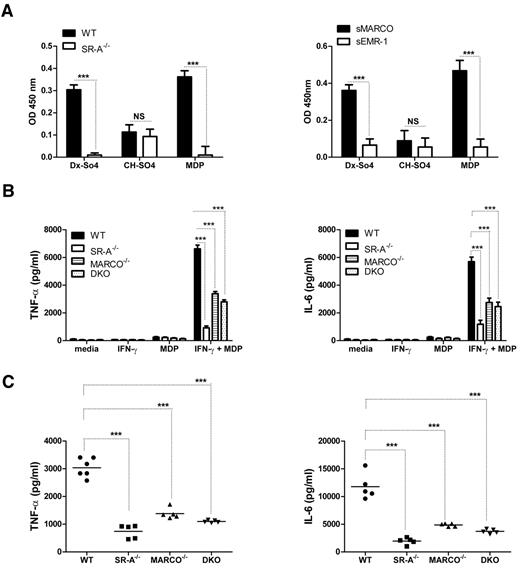
Because SR-A and MARCO regulated the NM-mediated inflammatory response of cell surface–expressed TLR4, but enhanced the response of endosomal TLR3, we investigated how SR-A and MARCO modulated responses of other classes of intracellular pathogen sensors such as NLRs, which include NOD1/2 and NALPs. Unlike TLRs, NOD2 is a cytosolic sensor for the bacterial cell wall component MDP, but also activates nuclear factor-κB signaling through the specific adaptor RIP2 (receptor interacting protein 2).29 To determine whether SR-A and MARCO can recognize the NOD2 ligand MDP, we utilized a novel solid-phase assay for ligand detection using solubilized or recombinant receptor and known ligands as a control. Results confirmed that both SR-A and MARCO can recognize MDP (Figure 5A). To determine the role of SR-A and MARCO in MDP-NOD2–mediated inflammatory responses, TNF-α and IL6 levels were compared among WT and SR-A−/−, MARCO−/−, and DKO Mϕ following MDP stimulation, in vitro (Figure 5B), and in the peritoneal lavage fluid of these mice 2 hours after intraperitoneal MDP injection (Figure 5C). Results confirmed that SR-A−/−, MARCO−/−, and DKO mice and isolated Mϕ from these animals secreted significantly fewer inflammatory cytokines, indicating that SR-A and MARCO are required for an MDP response.
SR-A and MARCO bind MDP and enhance NOD2 response. (A) SR-A (left panel) and MARCO (right panel) bind to various polyanionic ligands. A known ligand (dextran sulfate, Dx-SO4) and a non-ligand (chondroitin sulfate, Ch-SO4) for SR-A and MARCO were used to coat wells of a 96-well plate, along with MDP. SR-A–specific ligand activity was determined by overlaying postnuclear cell lysates from WT and SR-A−/− BMMϕ and detected with specific anti–SR-A monoclonal antibody 2F8. Similarly, recombinant sMARCO and specific anti–MARCO monoclonal antibody ED31 were utilized to identify MARCO specific binding. (B) After MDP stimulation, SR-A−/−, MARCO−/−, and DKO Mϕ produced decreased levels of TNF-α (left panel) and IL6 (right panel) compared with WT cells. IFN-γ–primed Bg-PM from each mouse strain were stimulated with 100 μg/mL of MDP for 16 hours. Levels of TNF-α and IL6 were measured by ELISA. (C) Decreased levels of TNF-α (left panel) and IL6 (right panel) were produced in the peritoneal cavity of SR-A−/−, MARCO, and DKO mice injected intraperitoneally with 100 μg of MDP or an equal volume of PBS. After 2 hours, animals were killed and peritoneal lavage performed using 1 mL of PBS. Levels of TNF-α and IL6 were measured by ELISA. Statistical significance for panels A and B was assessed by 2-way ANOVA with the Bonferroni posttest. One-way ANOVA with Dunnett multiple comparison test was used to assess statistical significance for panel C. ***P < .001
SR-A and MARCO bind MDP and enhance NOD2 response. (A) SR-A (left panel) and MARCO (right panel) bind to various polyanionic ligands. A known ligand (dextran sulfate, Dx-SO4) and a non-ligand (chondroitin sulfate, Ch-SO4) for SR-A and MARCO were used to coat wells of a 96-well plate, along with MDP. SR-A–specific ligand activity was determined by overlaying postnuclear cell lysates from WT and SR-A−/− BMMϕ and detected with specific anti–SR-A monoclonal antibody 2F8. Similarly, recombinant sMARCO and specific anti–MARCO monoclonal antibody ED31 were utilized to identify MARCO specific binding. (B) After MDP stimulation, SR-A−/−, MARCO−/−, and DKO Mϕ produced decreased levels of TNF-α (left panel) and IL6 (right panel) compared with WT cells. IFN-γ–primed Bg-PM from each mouse strain were stimulated with 100 μg/mL of MDP for 16 hours. Levels of TNF-α and IL6 were measured by ELISA. (C) Decreased levels of TNF-α (left panel) and IL6 (right panel) were produced in the peritoneal cavity of SR-A−/−, MARCO, and DKO mice injected intraperitoneally with 100 μg of MDP or an equal volume of PBS. After 2 hours, animals were killed and peritoneal lavage performed using 1 mL of PBS. Levels of TNF-α and IL6 were measured by ELISA. Statistical significance for panels A and B was assessed by 2-way ANOVA with the Bonferroni posttest. One-way ANOVA with Dunnett multiple comparison test was used to assess statistical significance for panel C. ***P < .001
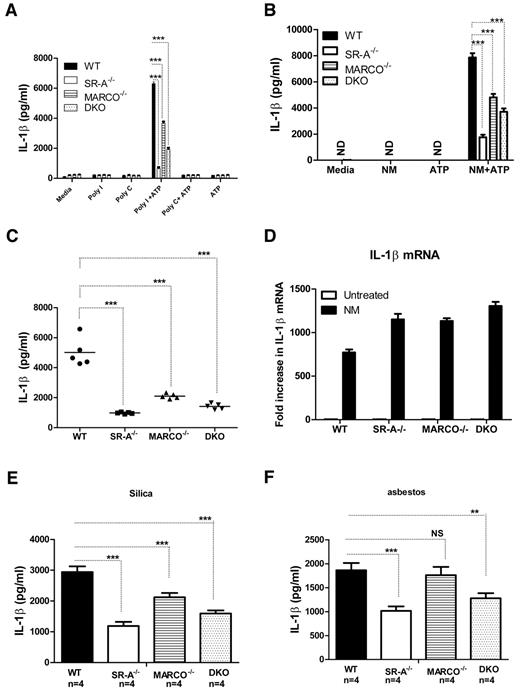
SR-A/MARCO-mediated ligand delivery is essential for NALP3-mediated IL1β secretion
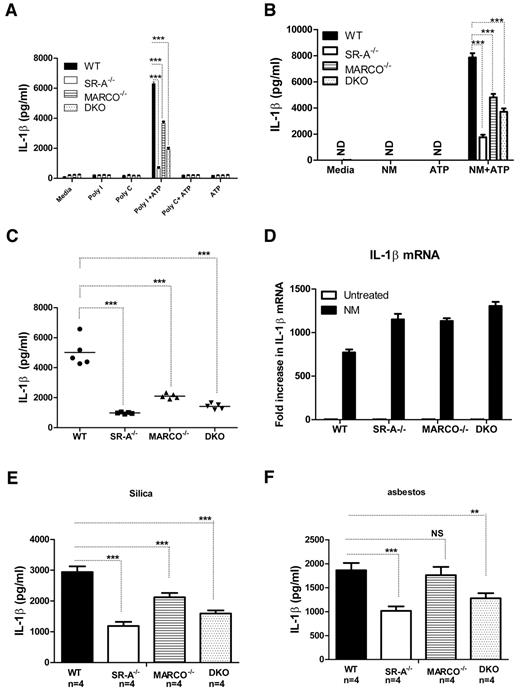
Stimulation of NALP3 inflammasomes does not activate nuclear factor-κB, but activates caspase-1 cleavage. Caspase-1 cleaves pro-IL1β to mature IL1β.35 We investigated whether SR-A and MARCO are able to modulate NALP3-mediated IL1β secretion. WT, SR-A−/−, MARCO−/−, and DKO Mϕ were stimulated with polyI, polyC (Figure 6A), or inactivated NM (Figure 6B) for 6 hours to induce pro-IL1β, followed by a short pulse of ATP. After 6 hours of further incubation, supernatants were harvested and the secretion of IL1β was analyzed with ELISA. The SR-A−/−, MARCO−/−, and DKO Mϕ produced significantly less IL1β compared with WT Mϕ. Furthermore, we detected significantly less IL1β in the peritoneal lavage fluid of SR-A−/−, MARCO−/−, and DKO mice after sequential intraperitoneal injection with NM and ATP (Figure 6C), indicating that SR-A and MARCO are required for efficient IL1β production following NALP3 activation both in vitro and in vivo. To exclude the possibility that the reduced IL1β secretion observed in SR-A−/−, MARCO−/−, and DKO mice was not due to a defect in TLR-mediated IL1β mRNA induction, we compared IL1β mRNA levels by quantitative PCR in Bg-PM after NM treatment. No IL1β mRNA was detected in untreated groups, but all 3 KO strains consistently expressed more IL1β mRNA after NM treatment (Figure 6D), indicating that defects in IL1β secretion were not due to defective IL1β induction, but rather to defective NALP3-mediated cleavage of pro-IL1β to mature IL1β. To further assess the involvement of SR-A and MARCO in NALP3-mediated sterile inflammation, we stimulated NM-primed Bg-PM with silica crystals or asbestos, 2 well-characterized activators of NALP3 inflammasomes, as well as selective ligands for SR-A and MARCO, and measured IL1β levels in the supernatant. Results showed that SR-A−/− and DKO macrophages produced significantly reduced IL1β after silica or asbestos stimulation (Figure 6E), indicating that SR-A- and MARCO-mediated internalization of these environmental particles enhances NALP3-mediated IL1β secretion.
SR-A/MARCO-mediated ligand recognition promotes NALP3-mediated IL1β secretion. WT, SR-A−/−, MARCO−/−, and DKO Mϕ (all n = 4) were stimulated with 100 μg/mL of polyI, polyC (A), or 20 MOI of inactivated NM (B) for 6 hours and then pulsed for 5 minutes with 1mM ATP. After a further 6 hours, supernatants were harvested and the levels of IL1β were measured by ELISA. (C) WT, SR-A−/−, MARCO−/−, and DKO mice were challenged intraperitoneally with 105 CFU of inactivated NM, followed by a short ATP pulse 2 hours later. Levels of IL1β in peritoneal lavage fluid were measured by ELISA and compared among strains. (D) -Fold changes in IL1β mRNA expression are shown after stimulating Bg-PM from WT, SR-A−/− and MARCO−/−, and DKO mice with 20 MOI of NM for 4 hours. Each treatment condition was analyzed in triplicate and data are representative of 3 independent experiments. (E) Bg-PM from WT, SR-A, and MARCO, DKO mice were primed with 20 MOI of inactivated NM, washed to remove excess bacteria, and then further stimulated overnight with either 250μg/mL of silica or asbestos. IL1β levels were measured by ELISA. NM, silica, and asbestos alone did not induce any secreted IL1β (data not shown). Each treatment condition was measured in triplicate and data presented are representative of 3 independent experiments. Statistical significance for panels A, B, and E was assessed by 2-way ANOVA with Bonferroni posttest. One-way ANOVA with Dunnett multiple comparison test was used to assess statistical significance for panel C. *P ≤ .05; ***P < .001.
SR-A/MARCO-mediated ligand recognition promotes NALP3-mediated IL1β secretion. WT, SR-A−/−, MARCO−/−, and DKO Mϕ (all n = 4) were stimulated with 100 μg/mL of polyI, polyC (A), or 20 MOI of inactivated NM (B) for 6 hours and then pulsed for 5 minutes with 1mM ATP. After a further 6 hours, supernatants were harvested and the levels of IL1β were measured by ELISA. (C) WT, SR-A−/−, MARCO−/−, and DKO mice were challenged intraperitoneally with 105 CFU of inactivated NM, followed by a short ATP pulse 2 hours later. Levels of IL1β in peritoneal lavage fluid were measured by ELISA and compared among strains. (D) -Fold changes in IL1β mRNA expression are shown after stimulating Bg-PM from WT, SR-A−/− and MARCO−/−, and DKO mice with 20 MOI of NM for 4 hours. Each treatment condition was analyzed in triplicate and data are representative of 3 independent experiments. (E) Bg-PM from WT, SR-A, and MARCO, DKO mice were primed with 20 MOI of inactivated NM, washed to remove excess bacteria, and then further stimulated overnight with either 250μg/mL of silica or asbestos. IL1β levels were measured by ELISA. NM, silica, and asbestos alone did not induce any secreted IL1β (data not shown). Each treatment condition was measured in triplicate and data presented are representative of 3 independent experiments. Statistical significance for panels A, B, and E was assessed by 2-way ANOVA with Bonferroni posttest. One-way ANOVA with Dunnett multiple comparison test was used to assess statistical significance for panel C. *P ≤ .05; ***P < .001.
Discussion
We examined how SR-A/MARCO influenced inflammatory responses by selected cell surface and intracellular PRRs when co-ligated by their shared ligands. We report that SR-A and MARCO attenuated plasma membrane TLR4-mediated responses by competing for ligand recognition and limiting ligand availability on the cell surface for sustained sensing, while strikingly increasing responses by the intracellular pathogen sensors TLR-3, NOD2, and NALP3 by enhanced ligand delivery.
Innate recognition of Gram-negative organisms by SR-A and MARCO is complex. Both receptors can interact with the major Gram-negative cell wall component LPS,11,12 but such interaction is not essential for SR-A/MARCO-mediated uptake of Gram-negative organisms.26,27 Similarly, TLR4 rather than SR-A and MARCO is required for LPS-mediated inflammatory responses. While LPS-independent ligands have been implicated in SR-A/MARCO-mediated uptake, the significance of SR-A/MARCO-mediated LPS recognition is unclear. Hampton et al proposed that SR-A binds and detoxifies LPS, limiting its biological response.11 Bacille Calmette-Guérin–primed SR-A−/− animals have been shown to be more susceptible to septic shock following additional LPS challenge, suggesting a regulatory role for SR-A in limiting LPS responses.36 Infection of SR-A−/− mice with a pathogenic strain of NM was associated with strikingly high proinflammatory cytokine secretion and septic shock in addition to defective bacterial clearance.20 Our current finding that SR-A−/−, MARCO−/−, and DKO mice showed defective bacterial clearance but produced higher levels of inflammatory cytokines after intraperitoneal NM challenge is consistent with most previous reports11,36 ; although enhancement of LPS responses has also been reported.37
Apart from the secretion of bioactive mediators, another hallmark of inflammation is the infiltration of specific leukocyte populations into affected tissue. This process is regulated by a series of complex changes in damaged tissue, vascular endothelia, and migrating leukocytes. SR-A and MARCO play important roles in various aspects of this process, including cell adhesion,8 shape change,38 cytoskeletal rearrangement,39 and chemokine secretion by tissue-resident macrophages.34 We assessed the contribution of SR-A and MARCO to the trafficking of leukocytes following intraperitoneal injection of NM. Our observation that PBS-injected DKO mice showed fewer numbers of RPMs is consistent with a previous report that DKO mice have reduced numbers of selected tissue-resident Mϕ.31 Following NM challenge, the number of CD11bhiF4/80hi resident type macrophages in WT mice decreased significantly, possibly due to emigration into lymphatics. This decrease was not observed in SR-A−/− mice, and DKO mice even showed increased numbers of CD11bhiF4/80hi cells (Figure 2A). This could have been due to defective emigration of SR-A−/− and DKO RPMs from the peritoneal cavity, but also to higher expression of F4/80 on DKO monocytes after a primed or hyperresponsive state. The enhanced PMN and monocyte recruitment in SR-A−/− and DKO mice compared with WT mice (Figure 2A-B) was possibly due to enhanced inflammatory chemokine secretion by these animals, and is consistent with a previous report that SR-A−/− mice produced higher levels of CXC chemokines during inflammation.34 We conclude that SR-A and MARCO orchestrate leukocyte trafficking by regulating chemokine secretion. In addition, SR-A and MARCO may also regulate the emigration of RPMs from the peritoneal cavity. Accumulation of CD11bhi F4/80hi cells in SR-A−/− and DKO mice after NM challenge could be due to the failure of RPMs to emigrate from the peritoneal cavity due to defects in cellular adhesion. Alternatively, recruited monocytes from DKO mice may express higher levels of F4/80 and contribute to the CD11bhiF4/80hi population. SR-A and MARCO down-regulated surface-expressed TLR4 responses by clearance of ligand, but enhanced inflammatory responses initiated by intracellular TLRs7/8 and 9.13,14 Because all of these TLRs utilize the common signaling adaptor MyD88, we tested how SR-A/MARCO regulate inflammatory responses by the MyD88-independent endosomal TLR3. Our observation that SR-A/MARCO-mediated ligand recognition also enhanced a TLR3 response established that SR-A and MARCO enhance both MyD88-dependent and MyD88-independent responses of intracellular TLRs.
We extended these observations by dissecting the relationship between SR-A/MARCO and other classes of intracellular pathogen sensors such as NOD2 and NALP3. NOD2 recognizes the bacterial cell wall product MDP, and signaling through the adaptor protein RIP2 activates NF-kB to induce an inflammatory response.29 MDP is a minimal subcomponent of peptidoglycan, a complex carbohydrate found in almost all bacterial cell walls.40 Although peptidoglycan is an excellent ligand for several SRs, including SR-A and MARCO, it has not been determined whether these receptors recognize MDP. Using a solid-phase assay, we showed that SR-A and MARCO also recognize MDP (Figure 5A). Recently, Nunez et al reported that the general SR inhibitor polyI did not block endocytosis of FITC-labeled MDP, concluding that SRs are unable to recognize MDP.41 A possible explanation is that because FITC is larger than MDP, it might have masked MDP, making it inaccessible to SRs. Enhanced SR-mediated delivery of MDP into Mϕ by chemical conjugation of polyanions has been shown to increase its biological response, inducing high levels of inflammatory mediators and enhanced macrophage tumoricidal activity.42,43 We have shown that SR-A and MARCO are specific receptors for MDP and are required for the NOD2-mediated cellular response (Figure 5). The role of NOD2 in neisserial disease is relatively underinvestigated compared with TLRs. However, it has been reported that NOD2 plays an important role in inflammatory responses by microglial cells during neisserial invasion in the central nervous system.44 Usually, MDP is released in the cytosol following lysosomal degradation of bacterial cell walls. Free serum MDP is rare even during other fulminant bacteremia, but NM constantly sheds outer membrane vesicles by membrane blebbing, which contains muropeptides. Moreover, antibiotics are regularly used to treat meningococcal bacteremia, further releasing large amounts of cell wall degradation products capable of activating NOD1/2. SR-A/MARCO-mediated direct recognition of MDP could be particularly relevant in these situations.
The secretion of IL1β requires 2 steps, TLR-mediated induction of pro-IL1β, followed by active caspase-1–mediated cleavage of pro-IL1β into mature IL1β. Cleavage of pro-caspase-1 into active caspase-1 by activation of the NALP3 inflammasome itself requires combined stimulation from a microbial agonist and extracellular ATP, a process that is independent of MyD88. Because NM by itself induced enhanced IL1β mRNA in SR-A−/−, MARCO−/−, and DKO mice, it is likely that reduced IL1β secretion in SR-A, MARCO, and DKO Mϕ is due to reduced NALP3 activation. Although NOD2/NALP3-mediated cytokine secretion was reduced in both SR-A−/− and DKO macrophages compared with WT cells, DKO macrophages consistently produced more cytokine than SR-A−/− macrophages. This apparent discrepancy is most likely due to compensatory up-regulation of other scavenger receptors in DKO macrophages. How NALP3 inflammasomes are activated is unclear, but NM internalization might be important for this process. Although the role of NALP3 in NM infection has not been directly tested, it has been shown that lipooligosaccharides from the closely related Neisseria gonorrheae activates NALP3 inflammasomes by a cathepsin-B–mediated pathway.45 It is known that meningococcal and gonorrheal lipooligosaccharides are structurally and functionally conserved, so it is likely that meningococci are capable of activating NALP3 by a similar mechanism. Many bacteria secrete extracellular ATP, which has been shown to modulate host immunity.46 In addition, inflammasome-activating signals can also come from host cells, and ligation of different PRRs often triggers the release of extracellular ATP from host monocytes, which, by acting through an autocrine/paracrine loop, can activate NALP3.47 Interactions between SR and inflammasomes could be relevant beyond pathogen-mediated acute septicemia because, as we have shown, SR-A and MARCO are required for optimal inflammatory responses to environmental pollutants such as silica and asbestos, known activators of NALP3,48-50 as well as ligands for SR-A and MARCO.21
On the basis of our present study and previous reports from our group, we propose a model in which SR-A and MARCO modulate inflammatory responses via cell-surface TLRs by at least 2 distinct mechanisms. SR-A and MARCO by their scavenging function rapidly remove LPS and Gram-negative organisms and limit TLR4 ligand availability on the cell surface, thus attenuating TLR4 responses. In contrast, SR-A/MARCO bind the complex mycobacterial lipid trehalose dimycolate at the cell surface, acting as a coreceptor to present it to TLR2, enhancing its response.23 Similar functional collaboration between TLRs and other members of the SR family has been reported by others.22 While SR-A and MARCO can differentially enhance or limit surface TLR2 and TLR4 responses, the paradigm emerges that SR-A and MARCO amplify responses of both intracellular TLRs and NLRs, possibly increasing ligand availability in intracellular compartments. Intracellular TLRs are recruited into endosomes to recognize ligands, but how cytosolic sensors access ligands from endosomal compartments is still unclear. Passive leakage, specific transport through membrane channels, and cathepsin B–mediated destabilization of phagolysomes have been proposed as potential mechanisms.50
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr R. Nussenblatt, National Institutes of Health, Bethesda, MD, for support.
S.M. was supported by the E. P. Abraham Trust, Oxford, and is presently funded by a Wellcome Trust Flexible Travel Award Fellowship. A.V. was supported by the Fondation pour la Recherche Medicale. S.G. was supported by a program grant from the Medical Research Council, United Kingdom.
Authorship
Contribution: S.M. and S.G. designed the research; S.M. and A.V. performed experiments; Y.C., B.L., and K.T. provided new reagents and knockout mouse strains and interpreted data; S.M., S.G., and A.V. analyzed data; and S.M. and S.G. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Siamon Gordon, Sir William Dunn School of Pathology, South Parks Rd, University of Oxford, OX1 3RE, United Kingdom; e-mail: siamon.gordon@path.ox.ac.uk.