Abstract
Epithin/PRSS14, a type II transmembrane serine protease, is involved in normal epithelial development and tumor progression. Here we report, as an interacting substrate of epithin, a receptor tyrosine kinase Tie2 that is well known for important roles in the vessel stability. Epithin interacts with and degrades the Tie2 extracellular portion that contains the ligand-binding domain. Epithin is located in the neighbor of Tie2-expressing vessels in normal tissue. Furthermore, epithin can cleave and degrade Tie2 not only in the same cell but also from neighboring cells nearby, resulting in the degradation of the Tie2 ectodomain. The remaining Tie2 fragment was highly phosphorylated and was able to recruit a downstream effector, phosphatidylinositol 3-kinase. Knocking down epithin expression using short hairpin RNA in thymoma cell severely impaired the migration through endothelial cells that show the actin rearrangement during the process. The diminution of epithin protein expression in 4T1 breast cancer cells caused the significant decrease in the number of transendothelial migrating cells in vitro as well as in those of metastasizing tumor nodules in vivo, Therefore, we propose that epithin, which regulates endothelial Tie2 functions, plays a critical role in the fine tuning of transendothelial migration for normal and cancer cells.
Introduction
Epithin (also known as PRSS14, matriptase, MT-SP1, and ST14), a member of the type II transmembrane serine protease family,1,2 was cloned from mouse thymic epithelial cells3 and human epithelial cancer cells.4,5 As commonly observed in the type II transmembrane serine protease family member,1 epithin consists of a relatively short cytoplasmic domain, a single transmembrane domain, a stem region, and a serine protease domain in the extracellular part. In the stem region, there are 2 CUB and 4 LDLRA domains that are thought to regulate the serine protease activity or to interact with other proteins.1,6
Initially, expression of epithin is described mostly in normal and malignant epithelial tissues.3,7 The normal functions of epithin were derived from expression studies in targeted mouse models. These studies revealed severe defects leading to perinatal death; defects found were in hair, and in thymocyte survival,8 the epidermal barrier function,9,10 and neural tube closure.11,12 In addition, functionally defective mutations in the loci of mouse and human result in severe skin disease, ichthyosis.13,14 In contrast, its overexpression in the skin of transgenic mice resulted in spontaneous tumorigenesis and potentiated carcinogen-induced tumor formation analogous to sustained phorbol 12-myristate 13-acetate (PMA) exposure.15 Moreover, a mouse model injected with gastric cancer cells showed that activated epithin promotes metastasis.16 In another study, the down-regulation of epithin resulted in suppression of the invasiveness and tumorigenic capacity of the ovarian cancer cells.17 Along with many earlier reports on the strong association between the overexpression of epithin and human epithelial cancers, there is little doubt that epithin plays the important roles during all stages of epithelial tumors.15,18-20
Epithin degrades extracellular matrix proteins, including fibronectin, laminin, gelatin, and collagen type IV, and cleaves many substrates, such as the protease-activated receptor-2, pro-urokinase plasminogen activator,21 and pro-hepatocyte growth factor22 at specific sites.19 A database search based on target amino acid sequences successfully identified specific epithin substrates. However, the presence of multiple protein interacting domains suggests that more epithin substrates are present and they can be screened by searching for the interacting proteins. Previously, we showed that PMA, which activates protein kinase C, can induce the translocation of epithin to the areas of cell-cell contact while it is dispersed throughout the plasma membrane in the absence of stimuli.23 We also showed that the translocation of epithin is followed by its activation and ectodomain shedding.23 Based on these findings, we made an attempt to screen the novel binding substrates of epithin after inducing its translocation and activation with PMA.
Tie2, an endothelial cell specific receptor tyrosine kinase, is well known for the roles in vessel remodeling and stabilization, inflammation, and endothelial permeability.24 Its functions are known to be modulated by specific ligands, the angiopoietin family.24,25 The previous paradigm for the regulation of Tie2 is that angiopoietin-1 (Ang1) binding to Tie2 induces tyrosine phosphorylation and evokes signaling events that induce vessel stability by maintaining interactions between endothelial cells, periendothelial support cells, and the matrix.26,27 In general, the signaling through Tie2 appears to depend on the balance between agonistic Ang1 and antagonistic Ang2. However, these effects are somewhat more complex and context-dependent.24,25 In addition, activated Tie2 transduces signals to different types of pathways depending on the presence or absence of cell-cell contacts.28,29
The present study shows that Tie2 is a substrate that binds to the LDLRA domains of epithin and that this binding is necessary for the proteolysis. It also demonstrates that epithin can degrade Tie2 in cis as well as trans and that epithin-mediated degradation of Tie2 induces Tie2 downstream signaling and participates in transendothelial cell migration.
Methods
Coimmunoprecipitation
The 427.1.86 cells on a 100-mm dish were transfected with Tie2 or Tie2 mutant cDNA. Twenty-four hours after transfection, cells were maintained without serum for 12 hours, treated with 10mM PMA and/or 10mM leupeptin for various times, and lysed in 50mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 1% NP-40, 150mM NaCl, 1mM ethylenediaminetetraacetic acid with protease inhibitors. The lysates were subjected to immunoprecipiation as described previously.23
Coculture
For the first coculture experiments (see Figure 5), MS1 cells and 427.1.86 or 427 Epi-KD cells were detached, and the same numbers of each cell types were mixed and plated. In the second coculture experiment, MS1 cells grown on a gelatin-coated coverslip were transferred into a culture dish containing 427.1.86 or 427 Epi-KD cells. Eighteen hours after each coculture, the cells were lysed and analyzed by Western blotting.
Transendothelial migration assay
MS1 cells were cultured on gelatin-coated coverslips in 12-well plates to form a confluent monolayer. GFP-427 and GFP-427-Epi-KD cells detached using Cellstripper (Cellgro) were added to the MS1 monolayer. After 8 hours, cells were stained with rhodamine phalloidin and observed with a fluorescence microscope. Detailed protocol is available in supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
In vivo metastasis assay
A total of 5 × 105 4T1-derived clones in phosphate-buffered saline were injected into tail vein in Balb/c mice. Mice were killed after 14 days, and Indian ink in phosphate-buffered saline was injected into trachea to stain lungs. The stained lungs were dissected and washed with phosphate-buffered saline mildly before taking images and counting nodules. All animal studies were carried out under a protocol approved by Inha University.
Results
PMA induces a transient interaction between epithin and Tie2
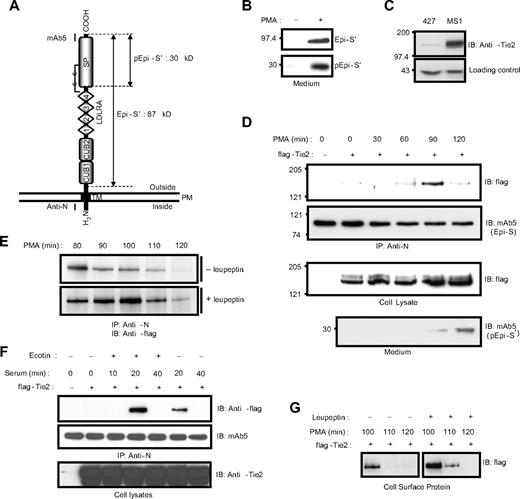
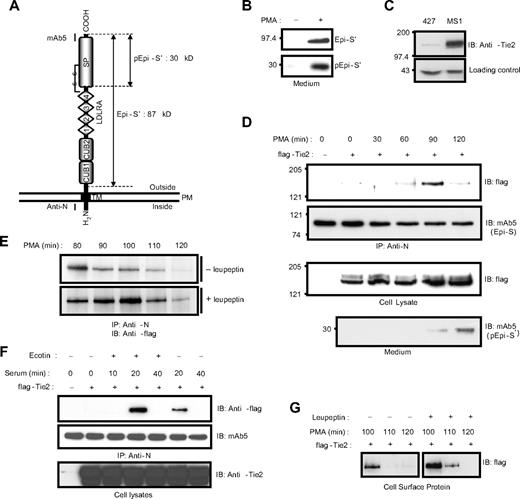
Previously, we showed that PMA induces the translocation of epithin to the cell-cell contact area, where epithin is released.23 Here, we also observed that the PMA-induced translocation of epithin to the cell-cell contact area is coupled with the activation of epithin. As shown in Figure 1A and B, a 30-kDa fragment of the epithin protease domain resulting from the cleavage for activation is apparent in the medium after the PMA treatment (Figure 1B). Therefore, we hypothesized that the translocation of epithin is directly linked to the proteolytic function of epithin on the membrane. To test this hypothesis, we induced the translocation and screened epithin-binding proteins in the thymic epithelial cell line, 427.1.86 cells, which endogenously express epithin. We treated 427.1.86 cells with 10μM PMA for 1 hour, in which condition the translocation of epithin was fully induced, but only approximately 5% of epithin was shed23 and then performed a large-scale immunoprecipitation. After analyzing the coprecipitated proteins with sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by silver-staining and mass spectrometry, we found a receptor tyrosine kinase, Tie2, as a relevant candidate (supplemental Figure 1).
Epithin interacts with Tie2. (A) Domain structure of epithin. The estimated molecular weights of a fragment produced by shedding, Epi-S′ (87 kDa), and a fragment produced by activation, pEpi-S′ (30 kDa), are designated. Regions that are recognized by the anti-N antibody and mAb5 are marked. SP indicates serine protease; TM, transmembrane region; and PM, plasma membrane. (B) The 427.1.86 cells were treated with or without 10μM PMA for 90 minutes. Proteins in the media were precipitated and analyzed by Western blotting with mAb5. (Top panel) The released epithin (Epi-S′). (Bottom panel) The activated epithin (pEpi-S′) after activation site cleavage and reduction of the disulfide bond shown in panel A. (C) The level of endogenous Tie2 in 427.1.86 cells and MS1 cells was analyzed by Western blotting using anti-Tie2 antibody (top panel). A nonspecific band is shown below as a loading control. (D) The 427.1.86 cells were transfected with flag-tagged Tie2. Twenty-four hours after the transfection, cells were deprived of serum for 12 hours and treated with 10μM PMA for the indicated time. Epithin in cell lysates was precipitated by anti-N antibody, and the precipitates were analyzed by Western blotting using anti-flag antibody to detect Tie2 (top panel) and mAb5 for epithin (top middle panel). The Tie2 levels in the cell lysates were analyzed by Western blotting using anti-flag antibody (bottom middle panel). To access amounts of the activated epithin, media from cells in each condition were collected, precipitated, and analyzed by Western blotting using mAb5 (bottom panel). (E) The 427.1.86 cells transfected with flag-tagged Tie2 were treated with PMA alone (top panel) or with both PMA and 20μM leupeptin (bottom panel) for the indicated time, and epithin and Tie2 interaction was analyzed as described in panel D. (F) The 427.1.86 cells transfected with flag-tagged Tie2 were deprived of serum for 12 hours and treated with 10% serum for the indicated time in the presence or absence of 500nM ecotin, a serine protease inhibitor, as indicated. After epithin was immunoprecipitated, the precipitates were analyzed by Western blotting using anti-flag antibody (top panel) and mAb5 (middle panel). The cell lysates were analyzed with Western blotting using anti-Tie2 antibody (bottom panel). (G) The 427.1.86 cells transfected with flag-tagged Tie2 were treated with PMA alone (left) or with PMA and leupeptin (right), and the cell surface proteins were then biotinylated. The resulting biotinylated proteins were pulled down by streptavidin beads and analyzed by Western blot using anti-flag antibody.
Epithin interacts with Tie2. (A) Domain structure of epithin. The estimated molecular weights of a fragment produced by shedding, Epi-S′ (87 kDa), and a fragment produced by activation, pEpi-S′ (30 kDa), are designated. Regions that are recognized by the anti-N antibody and mAb5 are marked. SP indicates serine protease; TM, transmembrane region; and PM, plasma membrane. (B) The 427.1.86 cells were treated with or without 10μM PMA for 90 minutes. Proteins in the media were precipitated and analyzed by Western blotting with mAb5. (Top panel) The released epithin (Epi-S′). (Bottom panel) The activated epithin (pEpi-S′) after activation site cleavage and reduction of the disulfide bond shown in panel A. (C) The level of endogenous Tie2 in 427.1.86 cells and MS1 cells was analyzed by Western blotting using anti-Tie2 antibody (top panel). A nonspecific band is shown below as a loading control. (D) The 427.1.86 cells were transfected with flag-tagged Tie2. Twenty-four hours after the transfection, cells were deprived of serum for 12 hours and treated with 10μM PMA for the indicated time. Epithin in cell lysates was precipitated by anti-N antibody, and the precipitates were analyzed by Western blotting using anti-flag antibody to detect Tie2 (top panel) and mAb5 for epithin (top middle panel). The Tie2 levels in the cell lysates were analyzed by Western blotting using anti-flag antibody (bottom middle panel). To access amounts of the activated epithin, media from cells in each condition were collected, precipitated, and analyzed by Western blotting using mAb5 (bottom panel). (E) The 427.1.86 cells transfected with flag-tagged Tie2 were treated with PMA alone (top panel) or with both PMA and 20μM leupeptin (bottom panel) for the indicated time, and epithin and Tie2 interaction was analyzed as described in panel D. (F) The 427.1.86 cells transfected with flag-tagged Tie2 were deprived of serum for 12 hours and treated with 10% serum for the indicated time in the presence or absence of 500nM ecotin, a serine protease inhibitor, as indicated. After epithin was immunoprecipitated, the precipitates were analyzed by Western blotting using anti-flag antibody (top panel) and mAb5 (middle panel). The cell lysates were analyzed with Western blotting using anti-Tie2 antibody (bottom panel). (G) The 427.1.86 cells transfected with flag-tagged Tie2 were treated with PMA alone (left) or with PMA and leupeptin (right), and the cell surface proteins were then biotinylated. The resulting biotinylated proteins were pulled down by streptavidin beads and analyzed by Western blot using anti-flag antibody.
As Tie2 is initially known to express mainly in endothelial cells, the 427.1.86 cells of epithelial nature were tested as to whether they express Tie2. Western blot analysis showed that Tie2 expression was clearly observed in the 427.1.86 cells (Figure 1C), although its expression level is relatively low compared with that in mouse pancreatic endothelial cells, MS1. Because of its low expression level, the expression of Tie2 was enhanced by transiently transfecting an N-terminal flag-tagged Tie2 construct into the 427.1.86 cells to verify the interaction between epithin and Tie2. As shown in Figure 1D, the interaction determined by immunoprecipitation was not clearly observed in the absence of stimuli (Figure 1D top panel). However, when the cells were stimulated with PMA, the interaction was observed at 90 minutes after the stimulation, verifying the interaction between epithin and Tie2 in a PMA-dependent manner. Surprisingly, the interaction was no longer sustained after the 90-minute peak (Figure 1D top panel). Moreover, the disappearance of Tie2 in the epithin immunoprecipitates was correlated with the appearance of activated epithin in the medium (Figure 1D bottom panel). Therefore, it was hypothesized that Tie2 becomes degraded after the interaction. To test this, cells were treated with both PMA and a serine protease inhibitor, leupeptin, and the interaction was examined. The serine protease inhibitor treatment not only enhanced but also sustained the interaction for an additional 20 minutes (Figure 1E), suggesting that serine protease activity is indeed involved in the disappearance of Tie2 in the epithin immunoprecipitates. The transient interaction and the stabilizing effect of the serine protease inhibitor were also observed in cells stimulated with serum (Figure 1F), which is also known to induce the translocation, activation, and shedding of epithin.30 In addition, cell surface biotinylation experiments showed that Tie2 on surfaces of the 427.1.86 cells is reduced when treated with PMA; leupeptin stabilizes this with similar kinetics (Figure 1G), as observed in the immunoprecipitation experiments. From these observations, it was concluded that Tie2 on the cell surface is degraded shortly after interacting with epithin because of the serine protease activity.
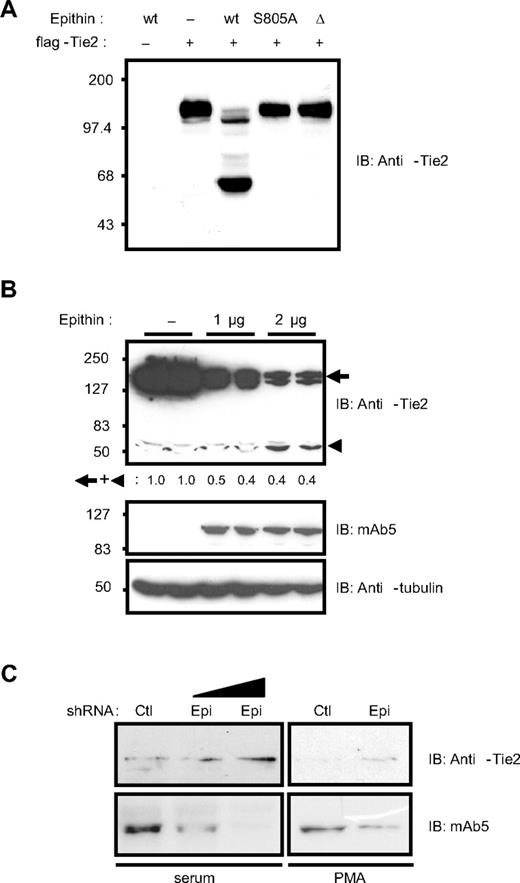
Epithin regulates the Tie2 protein level by proteolytic cleavage
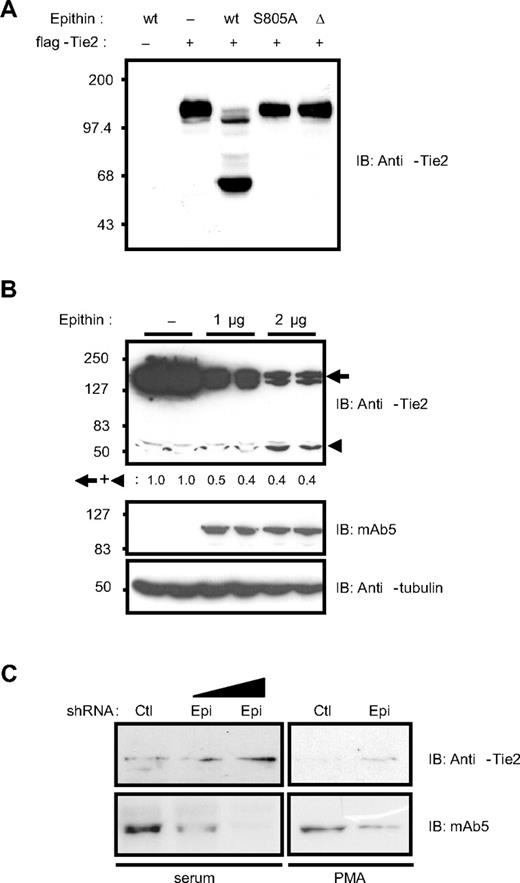
The data presented in Figure 1 strongly suggest that epithin is the candidate serine protease responsible for the degradation of Tie2. Therefore, the ability of epithin to degrade Tie2 was tested in both gain- and loss-of-function experiments. First, wild-type epithin, activity-dead mutant (epithinS805A),4,31 or activation-defective isoform (epithinΔ)32 was cotransfected with Tie2 into HEK293T cells in which both epithin and Tie2 expression was not observed. In the cells cotransfected with wild-type epithin and Tie2, the intact Tie2 band was significantly reduced; instead, smaller fragments including a major approximately 60-kDa Tie2 fragment was detected by anti-Tie2 antibody raised against its cytoplasmic tail (Figure 2A). However, the Tie2 protein in the cells transfected with epithinS805A or epithinΔ remained intact, showing that Tie2 can be truncated by epithin and that the cleavage requires the protease activity of epithin. The Western blot analysis using anti-flag antibody did not detect the corresponding N-terminal fragments (∼ 80 kDa) resulting from the cleavage either in the cell lysates or in the media, suggesting that there may be more than one cleavage leading to the degradation of the extracellular domain of Tie2 (data not shown). Whether ectopically expressed epithin could degrade endogenously expressed Tie2 in endothelial cells was also investigated. As shown in Figure 2B, when epithin is transfected into an endothelial cell line, MS1, which endogenously expresses Tie2 but not epithin, the disappearance of intact Tie2 as well as the appearance of approximately 60 kDa truncated product was observed (Figure 2B). Quantification of the intensities of the remaining Tie2 bands showed that the amount of truncated Tie2 product is much less than the reduced amount of intact Tie2, suggesting that the Tie2 was further degraded after epithin-mediated cleavage. Finally, when the epithin expression was knocked down by transiently transfecting short hairpin RNA (shRNA) into 427.1.86 cells expressing both epithin and Tie2 endogenously, Tie2 was stabilized in the presence of either serum or PMA stimulation (Figure 2C). Taken together, these data show that epithin can negatively regulate the Tie2 protein level via proteolytic cleavage and degradation.
Epithin can degrade Tie2. (A) Wild-type epithin (wt), epithin protease activity dead mutant (S805A), or epithin protease dead isoform (Δ) was cotransfected with flag-Tie2 into HEK293T cells as indicated. Twenty-four hours after transfection, the cells were analyzed by Western blotting using anti-Tie2 antibody (against its cytoplasmic tail). (B) MS1 cells were transfected with the indicated amount of epithin (duplicated), and the protein level of endogenous Tie2 was analyzed by Western blotting using anti-Tie2 antibody (top). The expression levels of epithin (middle) and tubulin (bottom) as a loading control are also shown. The sum of the band intensities of intact Tie2 and truncated Tie2 are shown as relative values. (C) pSUPER-control or pSUPER-epi was transfected into 427.1.86 cells. Twenty-four hours after the transfection, cells were lysed immediately (left) or deprived of serum for 12 hours and treated with PMA for 90 minutes before lysis (right). In each condition, the levels of endogenous Tie2 and epithin were analyzed by Western blotting with anti-Tie2 antibody or mAb5, respectively.
Epithin can degrade Tie2. (A) Wild-type epithin (wt), epithin protease activity dead mutant (S805A), or epithin protease dead isoform (Δ) was cotransfected with flag-Tie2 into HEK293T cells as indicated. Twenty-four hours after transfection, the cells were analyzed by Western blotting using anti-Tie2 antibody (against its cytoplasmic tail). (B) MS1 cells were transfected with the indicated amount of epithin (duplicated), and the protein level of endogenous Tie2 was analyzed by Western blotting using anti-Tie2 antibody (top). The expression levels of epithin (middle) and tubulin (bottom) as a loading control are also shown. The sum of the band intensities of intact Tie2 and truncated Tie2 are shown as relative values. (C) pSUPER-control or pSUPER-epi was transfected into 427.1.86 cells. Twenty-four hours after the transfection, cells were lysed immediately (left) or deprived of serum for 12 hours and treated with PMA for 90 minutes before lysis (right). In each condition, the levels of endogenous Tie2 and epithin were analyzed by Western blotting with anti-Tie2 antibody or mAb5, respectively.
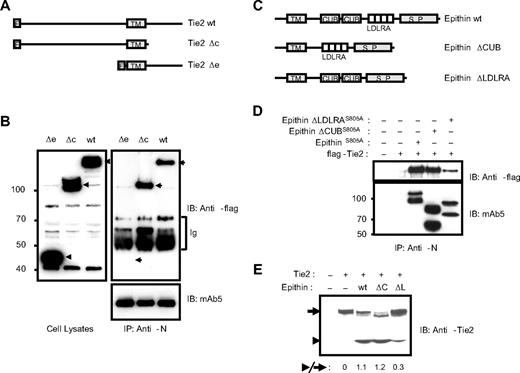
The interaction via LDLRA domain regulates the proteolysis
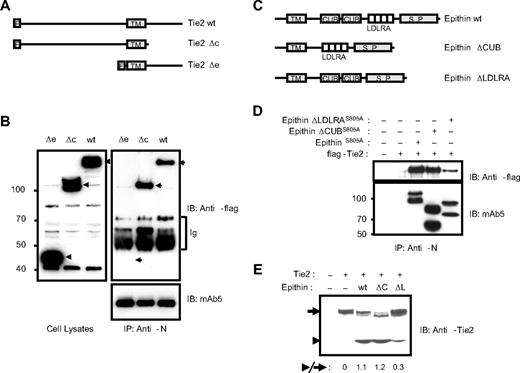
Next, the domains among the cytoplasmic, CUB, and LDLRA domains in epithin responsible for the interaction were determined. As the signal sequence for targeting type II transmembrane protein, epithin, to the membrane may be in the cytoplasmic domain and its deletion would cause problems in the topology of epithin, the cytoplasmic domain-deleted Tie2 mutant, Tie2Δc (Figure 3A), was used to test the importance of the cytoplasmic domain in the interaction. Tie2Δc as well as Tie2Δe, the extracellular domain-deleted Tie2 mutant (Figure 3A), were transfected into the 427.1.86 cells, and the cells were stimulated with PMA to induce the interaction, as in the experiment in Figure 1D. Tie2Δc was coimmunoprecipitated with epithin efficiently as wild-type Tie2 was, although Tie2Δe was not (Figure 3B), indicating that the cytoplasmic domain is not required for the interaction. Next, the effect of CUB and LDLRA domains was tested by assessing the interaction of the CUB domain-deleted epithin mutant, epithinΔCUB, and LDLRA domain-deleted epithin mutant, epithinΔLDLRA, with Tie2 (Figure 3C). Because epithin protease activity decreases the interaction by degrading Tie2, the protease-inactive S805A epithin mutants were used to stabilize the interaction of epithin with Tie2. Each protease-inactive epithin construct was transfected with Tie2 into HEK293T cells, and the interaction was analyzed by immunoprecipitation. In this experiment, the LDLRA domain-deleted mutant, epithinΔLDLRAS805A, showed reduced interaction with Tie2, whereas the CUB domain-deleted mutants did not show a defect in the interaction (Figure 3D), indicating that the LDLRA domains contribute to the interaction with Tie2. Interestingly, the deletion of the LDLRA domains also showed a defect in the Tie2 cleavage. When the LDLRA domain-deleted wild-type epithin (bearing protease activity) was tested for Tie2 cleavage, the deletion caused severe impairment compared with the wild-type and CUB domain-deleted epithin (Figure 3E), although the degree of activation of epithinΔLDLRA was shown to be equal to those of wild-type and epithinΔCUB.6 Therefore, the inefficient cleavage induced by epithinΔLDLRA suggests that the interaction is a prerequisite for the cleavage.
Epithin interacts with Tie2 through its LDLRA domain. (A) Schematic diagram showing Tie2 mutants. wt indicates wild-type; Δc, cytoplasmic domain deleted; Δe, extracellular domain deleted; F, flag tag; and TM, transmembrane region. (B) Wild-type Tie2 and its mutants were transfected into 427.1.86 cells, deprived of serum for 12 hours, and treated with PMA for 90 minutes. The cell lysates (left) and epithin immunoprecipitates using anti-N antibody from the cell lysates (right) were analyzed by Western blotting using anti-flag antibody. The amounts of precipitated epithin were also verified with mAb5. Ig indicates immunoglobulins. (C) Schematic diagram showing epithin deletion mutants. S.P. indicates serine protease domain. (D) EpithinS805A or each deletion mutant bearing the S805A mutation was cotransfected with flag-tagged Tie2 into HEK293T cells. The cell lysates were immunoprecipitated with anti-N antibody and analyzed by Western blotting using anti-flag antibody (top panel) and mAb5 (bottom panel). (E) Flag-tagged Tie2 was cotransfected with wild-type epithin or deletion mutants (ΔC, epithinΔCUB; ΔL, epithinΔLDLRA) into HEK293T cells. The Tie-2 level in each cell lysates was analyzed by Western blotting using anti-Tie2 antibody. The ratios of the truncated Tie2 (arrowhead) to the intact Tie2 (arrow) are indicated.
Epithin interacts with Tie2 through its LDLRA domain. (A) Schematic diagram showing Tie2 mutants. wt indicates wild-type; Δc, cytoplasmic domain deleted; Δe, extracellular domain deleted; F, flag tag; and TM, transmembrane region. (B) Wild-type Tie2 and its mutants were transfected into 427.1.86 cells, deprived of serum for 12 hours, and treated with PMA for 90 minutes. The cell lysates (left) and epithin immunoprecipitates using anti-N antibody from the cell lysates (right) were analyzed by Western blotting using anti-flag antibody. The amounts of precipitated epithin were also verified with mAb5. Ig indicates immunoglobulins. (C) Schematic diagram showing epithin deletion mutants. S.P. indicates serine protease domain. (D) EpithinS805A or each deletion mutant bearing the S805A mutation was cotransfected with flag-tagged Tie2 into HEK293T cells. The cell lysates were immunoprecipitated with anti-N antibody and analyzed by Western blotting using anti-flag antibody (top panel) and mAb5 (bottom panel). (E) Flag-tagged Tie2 was cotransfected with wild-type epithin or deletion mutants (ΔC, epithinΔCUB; ΔL, epithinΔLDLRA) into HEK293T cells. The Tie-2 level in each cell lysates was analyzed by Western blotting using anti-Tie2 antibody. The ratios of the truncated Tie2 (arrowhead) to the intact Tie2 (arrow) are indicated.
Epithin can degrade Tie2 of the neighboring cells in a trans manner
Although Tie2 expression was observed in the thymic epithelial 427.1.86 cells, Tie2 is usually found in endothelial cells and epithin is found in epithelial cells. Therefore, we investigated the expression of epithin in normal thymus (supplemental Figure 2; supplemental Video 1). The immunohistochemistry with specific antibody showed the vascular pattern, and there is significant overlap between epithin expression and vessels of thymus (supplemental Figure 2Biii,vi). These suggest that epithin-expressing cells are located close to the vessel endothelium. In more careful examination with Z sections, epithin expression resided in the surrounding cells of, and in direct contact with, endothelial cells (supplemental Video 1). The cells expressing epithin are probably not a type of pericyte/mural cells because α-smooth muscle actin did not stain the cells expressing epithin (supplemental Figure 2C). This close proximity suggests a possible interplay between them. Therefore, we hypothesized that Tie2 in the vessels can be degraded by epithin in trans.
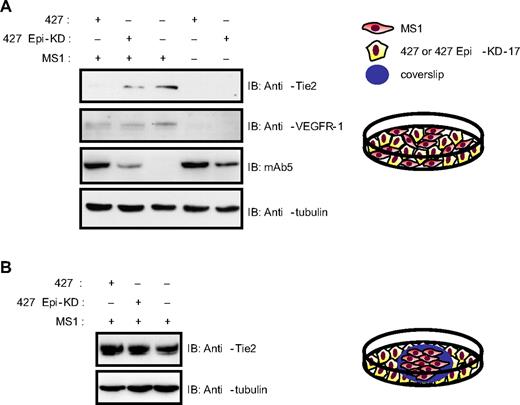
To test this idea in vitro, MS1 cells were mixed with the 427.1.86 cells, and this mixture was plated on the same dish to determine whether epithin expressed in the 427.1.86 cells can degrade Tie2 expressed in MS1 cells (Figure 4A diagram). As a control, 427 Epi-KD cells, stably expressing shRNA for epithin, were used instead of 427.1.86 cells. The results showed that the coculture of MS1 cells with 427.1.86 cells induced a significant decrease in the Tie2 level (Figure 4A lane 1) compared with the coculture with 427 Epi-KD cells (Figure 4A lane 2). VEGFR-1, another receptor expressed in endothelial cell surfaces, was not decreased (Figure 4A lanes 1 and 2), suggesting that the degradation activity of epithin is specific to Tie2. As epithin is also released into the medium in a soluble form (Figure 1B), it was also determined whether the released epithin can degrade Tie2 on endothelial cells using another type of coculture in which the 2 cell types are in separate areas but share the same medium (Figure 4B diagram). Interestingly, Tie2 was not degraded by sharing the same medium, with no cell-cell contacts (Figure 4B). These results suggest that the Tie2 degradation is mediated by the membrane-bound form of epithin but not by the released form. Taken together, these data clearly show that epithin can degrade Tie2 not only in cis but also in trans, although not in a paracrine fashion.
Epithin can degrade Tie2 in a trans fashion. (A) For the coculture of 427.1.86 or 427 Epi-KD cells with MS1 cells (lanes 1 and 2), 2 types of cells were mixed together and cocultured. Eighteen hours after plating, the cells were lysed and analyzed by Western blotting for Tie2, VEGFR-1, epithin, and tubulin. (B) MS1 cells on a gelatin-coated coverslip were transferred into a culture dish containing 427.1.86 or 427 Epi-KD cells and were cocultured for 18 hours. MS1 cells were lysed and analyzed by Western blotting for Tie2 and tubulin. Schematic drawings of each coculture experiment are shown.
Epithin can degrade Tie2 in a trans fashion. (A) For the coculture of 427.1.86 or 427 Epi-KD cells with MS1 cells (lanes 1 and 2), 2 types of cells were mixed together and cocultured. Eighteen hours after plating, the cells were lysed and analyzed by Western blotting for Tie2, VEGFR-1, epithin, and tubulin. (B) MS1 cells on a gelatin-coated coverslip were transferred into a culture dish containing 427.1.86 or 427 Epi-KD cells and were cocultured for 18 hours. MS1 cells were lysed and analyzed by Western blotting for Tie2 and tubulin. Schematic drawings of each coculture experiment are shown.
Epithin-induced Tie2 cleavage generates Tie2 signaling
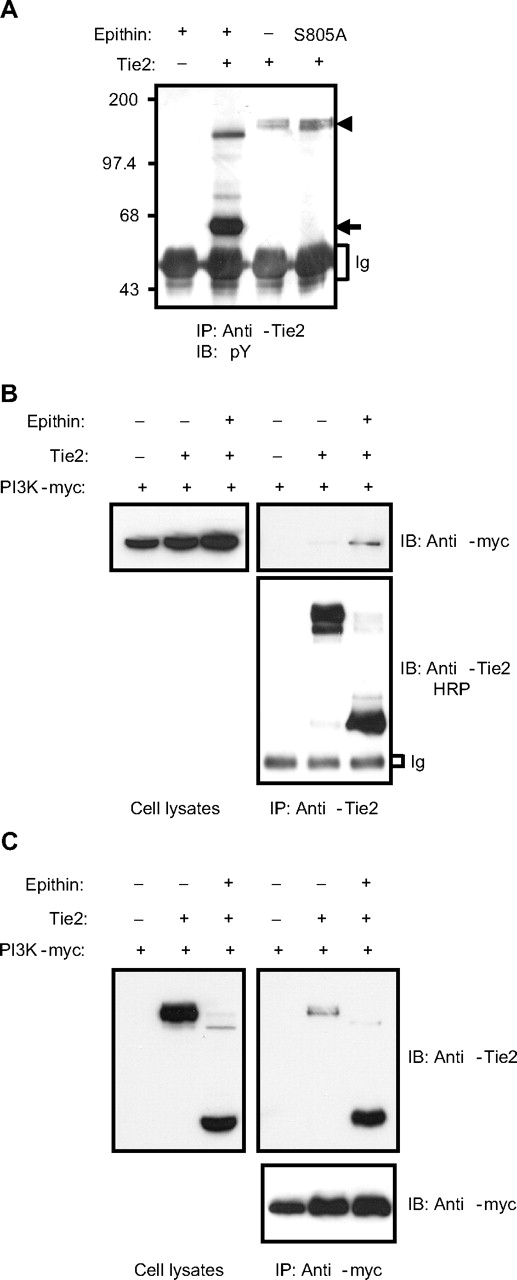
As commonly observed in receptor tyrosine kinases, the occupancy of the ligand-binding site of Tie2 by its ligand, Ang1, induces oligomerization of Tie2 and phosphorylation of tyrosine residues in its cytoplasmic domain. This recruits signaling molecules, such as Dok-R and phosphatidylinositol 3-kinase (PI3K) and transduces signaling.33 Given that the truncated Tie2 may not bind its ligand as most of its extracellular domain is missing (Figure 2A-B), it was expected that no Tie2 signaling would occur when the cleavage occurs. However, very interestingly, the epithin-mediated cleavage induced the tyrosine phosphorylation of Tie2 in a ligand-independent manner. When the truncated Tie2 observed in the epithin-transfected cells was immunoprecipitated and compared with intact Tie2 in terms of the degree of tyrosine phosphorylation using a phosphotyrosine specific antibody, the truncated Tie2 was found to be heavily phosphorylated (Figure 5A). The truncated Tie2 and its increased phosphorylation level were also observed in primary endothelial cells (human umbilical vein endothelial cells; supplemental Figure 3). We also observed enhanced phosphorylation in a genetically truncated Tie2 construct, which has similar size to the cleaved Tie2 (supplemental Figure 4), suggesting that the presence of the extracellular domain of Tie2 somehow blocks the spontaneous phosphorylation. Finally, we observed that the p85 subunit of PI3K interacts much more efficiently with the truncated Tie2 than intact Tie2 (Figure 5B-C), suggesting that the phosphorylation of truncated Tie2 induces downstream Tie2 signaling.
Epithin-induced Tie2 cleavage generates Tie2 signaling. (A) Tie2 was transfected with or without epithin into HEK293T cells. Twenty-four hours after transfection, cells were lysed and immunoprecipitated by anti-Tie2 antibody and blotted with antiphosphotyrosine antibody. The sizes of the intact and truncated Tie2 are indicated with arrowheads and arrows, respectively. (B) HEK293T cells were cotransfected with epithin, Tie2, and myc-tagged p85 subunit of PI3K as indicated. Twenty-four hours after transfection, cells were lysed and immunoprecipitated with anti-Tie2 antibody. The p85 subunit and the Tie2 in the precipitates were analyzed by Western blotting using anti-myc antibody and horseradish peroxidase (HRP)-conjugated anti-Tie2 antibody, respectively (right). The expression level of the p85 subunit in the cell lysates is shown (left). (C) HEK293T cells were transfected and lysed as in panel B, and the lysates were immunoprecipitated by anti-myc antibody. Tie2 and p85 subunits in the precipitated (right) and Tie2 expression (left) were analyzed by Western blotting.
Epithin-induced Tie2 cleavage generates Tie2 signaling. (A) Tie2 was transfected with or without epithin into HEK293T cells. Twenty-four hours after transfection, cells were lysed and immunoprecipitated by anti-Tie2 antibody and blotted with antiphosphotyrosine antibody. The sizes of the intact and truncated Tie2 are indicated with arrowheads and arrows, respectively. (B) HEK293T cells were cotransfected with epithin, Tie2, and myc-tagged p85 subunit of PI3K as indicated. Twenty-four hours after transfection, cells were lysed and immunoprecipitated with anti-Tie2 antibody. The p85 subunit and the Tie2 in the precipitates were analyzed by Western blotting using anti-myc antibody and horseradish peroxidase (HRP)-conjugated anti-Tie2 antibody, respectively (right). The expression level of the p85 subunit in the cell lysates is shown (left). (C) HEK293T cells were transfected and lysed as in panel B, and the lysates were immunoprecipitated by anti-myc antibody. Tie2 and p85 subunits in the precipitated (right) and Tie2 expression (left) were analyzed by Western blotting.
Epithin enhances transendothelial migration
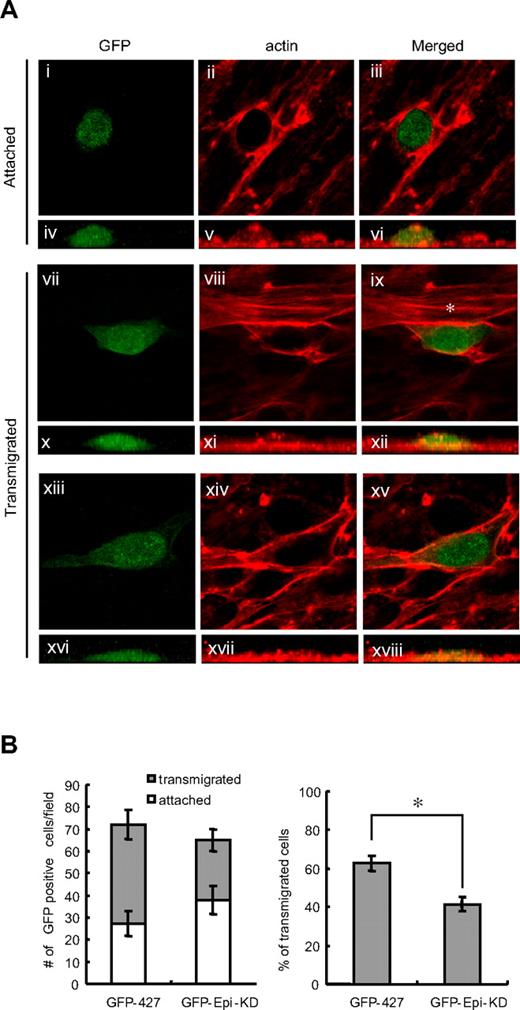
From the accumulated results, we hypothesized that epithin may have a role in transendothelial migration by degrading Tie2-dependent endothelial cell-cell contacts28,29 and by modulating Tie2 signaling in endothelial cells. To test this idea, the transendothelial migration of the 427.1.86 cells was compared with that of epithin knockdown 427 Epi-KD cells using in vitro transendothelial migration assay.34 For this experiment, GFP-427 and GFP-427 Epi-KD cells, which stably express green fluorescent protein (GFP), were used to distinguish these cells from endothelial cells. The GFP-427 and GFP-427 Epi-KD cells were seeded on a confluent monolayer of endothelial cells, MS1. After 8 hours of incubation, the cells were fixed, stained for actin filaments, and observed under the confocal microscope with Z sections. It was found that the round-shaped cells shown in the horizontal image (Figure 6Aiii) showed a hemisphere attached on top of the MS1 cells in the vertically reconstituted image (Figure 6Avi), whereas spindle-shaped or polygonal-shaped cells (Figure 6Aix,xv) were spread and located between the MS1 cells (Figures 6Axii,xviii), as described elsewhere.34
Epithin enhances transendothelial migration. (A) GFP-427 or GFP-427 Epi-KD cells were seeded on a confluent monolayer of MS1 cells. After incubation for 8 hours, the cells were fixed and stained for F-actin with rhodamine-conjugated phalloidin. Based on their morphology visualized by GFP and F-actin, round-shaped cells were counted as attached cells and spindle- or polygonal-shaped cells were counted as transmigrated cells. The representative images of attached (i-iii) and transmigrated (vii-ix,xiii-xv) cells in a sequence of the event are shown. Green represents the GFP signal (i,iv,vii,x,xiii,xvi); and red, the F-actin staining (ii,v,viii,xi,xiv,xvii) in the merged image (iii,vi,ix,xii,xv,xviii). Vertical images (iv-vi,x-xii,xvi-xviii) are also shown to visualize the height of the GFP-positive cells. (ix) *Actin stress fiber. (B) The mean values of the number of attached cells or transmigrated cells per field from 3 independent experiments are shown as bar graphs (left). The percentage of the number of transmigrated cells to the total number of transmigrated and attached cells is shown in each cell type (right). Error bars represent the SD. *P < .001.
Epithin enhances transendothelial migration. (A) GFP-427 or GFP-427 Epi-KD cells were seeded on a confluent monolayer of MS1 cells. After incubation for 8 hours, the cells were fixed and stained for F-actin with rhodamine-conjugated phalloidin. Based on their morphology visualized by GFP and F-actin, round-shaped cells were counted as attached cells and spindle- or polygonal-shaped cells were counted as transmigrated cells. The representative images of attached (i-iii) and transmigrated (vii-ix,xiii-xv) cells in a sequence of the event are shown. Green represents the GFP signal (i,iv,vii,x,xiii,xvi); and red, the F-actin staining (ii,v,viii,xi,xiv,xvii) in the merged image (iii,vi,ix,xii,xv,xviii). Vertical images (iv-vi,x-xii,xvi-xviii) are also shown to visualize the height of the GFP-positive cells. (ix) *Actin stress fiber. (B) The mean values of the number of attached cells or transmigrated cells per field from 3 independent experiments are shown as bar graphs (left). The percentage of the number of transmigrated cells to the total number of transmigrated and attached cells is shown in each cell type (right). Error bars represent the SD. *P < .001.
To quantify the transmigrated cells, the spindle-shaped or polygonal-shaped GFP-positive cells were scored as transmigrated cells. It was found that 63% of the GFP-427 cells attached initially to MS1 were transmigrated but that epithin knockdown significantly decreased the percentage of transmigrated cells to 42% (Figure 6B right), although there were no significant differences in the initial attachments of GFP-427 and GFP-427 Epi-KD cells (Figure 6B left). Interestingly, well-organized actin stress fiber formation was dominantly observed in endothelial cells neighboring the transmigrating cells (Figure 6Aix) when the GFP-427 cells were not completely spread out on the bottom surface (Figure 6Axii). However, actin stress fibers were not observed in those endothelial cells when the epithelial cells were completely spread and located between the endothelial cells (Figure 6Axv,xviii). Therefore, these data suggest that epithin is involved in the transendothelial migration and that the actin fibers are reorganized in the endothelial cells by the invasion of epithelial cells during transmigration.
Epithin enhances cancer cell metastasis
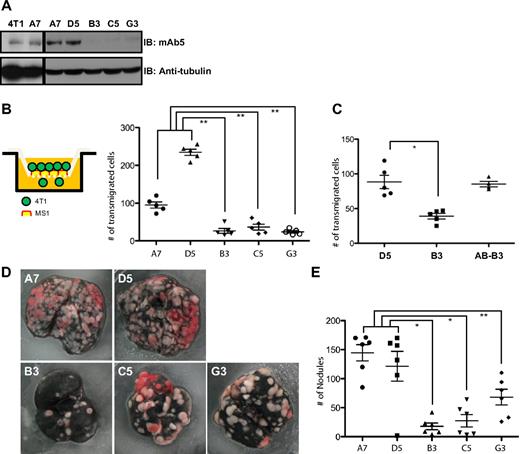
Because the transendothelial migration is an essential step for metastasis, we validate our results further by analyzing cancer cell metastasis in more physiologic conditions. For this analysis, we used a mouse mammary tumor cell line, 4T1, which is well known for efficient metastasis to the lung.35 As often observed in metastatic cancer cells, epithin expression was well detected in the 4T1 cell line (Figure 7A, lane 1). These cells were infected with lentivirus particles encoding shRNA for epithin or control shRNA. As the infection efficiency was not 100%, we further cloned those cells to generate 2 clones, A7 and D5, from control shRNA-infected cells, and 3 clones, B3, C5, and G3, from epithin shRNA-infected cells (Figure 7A). When their abilities of transendothelial migration were tested in the Boyden chamber covered with confluent monolayer of mouse endothelial cells, MS1, the epithin knockdown clones (B3, C5, and G3) showed significant defects (Figure 7B), confirming the important role of epithin in transendothelial migration. This effect was also shown with thymoma line 427.1.86 and 427 Epi-KD (supplemental Figure 5). The defect in transendothelial migration was reversed by reconstitution with human ortholog of epithin, matriptase (Figure 7C). Similar to 427.1.86 cells, 4T1 cells can also degrade Tie2 expressed in MS1 cells when cocultured (supplemental Figure 6). In addition, down-regulation of Tie2 by shRNA also increased transendothelial migration of 4T1 cells (supplemental Figure 7), suggesting that Tie2 has an endothelial barrier function. Therefore, although epithin may play multiple roles during transendothelial migration, these data strongly suggest that the epithin-induced degradation of Tie2 is critical in transendothelial migration by reducing the barrier function of Tie2.
Knockdown of epithin reduces metastatic ability of 4T1 cells. (A) The levels of epithin in 4T1 cell line (4T1) and its derivatives (A7, D5, B3, C5, and G3) were measured by Western blot with antiepithin mAb5 antibody. An equal amount of protein loading was confirmed by antitubulin blot (not shown). (B-C) The number of transmigrated 4T1 cells through MS1-confluent Boyden chamber was counted and plotted. Each dot in the graph represents the number of transmigrated cells in a microscopic field. The add-back clone AB-B3 was generated by stably transfecting pcDNA3.1/Matriptase (human ortholog of mouse epithin) into B3. (D) Each of 4T1 clones was injected into tail veins of 6 Balb/c mice. After 2 weeks, mice were killed and their lungs were stained with Indian ink. Representative lung images of each group are shown. (E) The numbers of tumor nodules on the lungs were counted and plotted. (B-E) Data are mean plus or minus SE. *P < .001. **P < .01.
Knockdown of epithin reduces metastatic ability of 4T1 cells. (A) The levels of epithin in 4T1 cell line (4T1) and its derivatives (A7, D5, B3, C5, and G3) were measured by Western blot with antiepithin mAb5 antibody. An equal amount of protein loading was confirmed by antitubulin blot (not shown). (B-C) The number of transmigrated 4T1 cells through MS1-confluent Boyden chamber was counted and plotted. Each dot in the graph represents the number of transmigrated cells in a microscopic field. The add-back clone AB-B3 was generated by stably transfecting pcDNA3.1/Matriptase (human ortholog of mouse epithin) into B3. (D) Each of 4T1 clones was injected into tail veins of 6 Balb/c mice. After 2 weeks, mice were killed and their lungs were stained with Indian ink. Representative lung images of each group are shown. (E) The numbers of tumor nodules on the lungs were counted and plotted. (B-E) Data are mean plus or minus SE. *P < .001. **P < .01.
Next, we injected those clones into the tail vein of syngenic Balb/c mice to test their metastatic transendothelial migration. After 2 weeks, mice were killed and their lungs were counterstained with Indian ink to visualize tumor nodules as white cell masses. As shown in Figure 7D, almost the entire surface was covered with tumor cells in the lungs from mice that were injected with epithin-expressing A7 and D5 cells, whereas the areas covered with tumor cells were evidently reduced in the lungs from mice injected with B3, C5, and G3 (Figure 7D). In addition, the numbers of tumor nodules are much less in the mice injected with these epithin knockdown cells (Figure 7E). Furthermore, the more tumor nodules were developed, the less body weights were shown in 4T1-injected mice (supplemental Figure 8). These data clearly show that epithin is required for efficient cancer cell metastasis.
Discussion
LDLRA domains provide the specific binding sites for Tie2
As other trypsin-like serine proteases, the target amino acid sequences that epithin can cleave are not highly stringent.21 Therefore, to prevent improper proteolysis, the protease should have a mechanism that ensures the action of the protease only on its specific substrates. In this regard, the nonprotease domains found in many type II transmembrane proteases1 can make an important contribution to their substrate specificity. The LDLRA domain is commonly found in type II membrane serine proteases.1 Although the LDLRA domains of epithin have been considered to function in the protein-protein interactions, Tie2 is the first binding partner identified for this domain. As shown in Figure 3, deletion of the LDLRA domains generated a severe defect not only in the interaction but also in its ability to degrade Tie2. This result strongly suggests that the relatively broad sequence specificity of this protease can be compensated by the specific protein-protein interaction that is mediated by the domains present outside of the protease domain. The Tie2 interaction through the LDLRA domain may also play a role in the activation process of epithin. As suggested earlier,6 the LDLRA domains can play a dual role in epithin activation: they serve as a binding domain for activators and function as an autoinhibitory domain. Thus, it is possible that Tie2 binding to epithin induces changes in the protein structure to release the autoinhibitory LDLRA domain, leading to its activation and robust proteolytic cleavage of Tie2, as shown in the overexpression system (Figure 2A).
Proteolytic modulation of Tie2 activity by epithin
It is a novel finding that Tie2 can be modulated by the protease activity of type II transmembrane serine protease, epithin. Under conditions in which epithin in the 427.1.86 thymic epithelial cell is translocated to the cell-cell contact area, epithin interaction with Tie2 is induced (Figure 1). After the interaction, the extracellular domain of Tie2 was degraded, resulting in elimination of the ligand-binding site. Therefore, the cells become insensitive to the ligands, Ang1/Ang2. Consistently, Ang1-induced phosphorylation of Akt, a well-known downstream effector of Tie2 signaling, was significantly reduced in epithin-transfected endothelial cells (data not shown). Therefore, epithin can inhibit ligand-dependent Tie2 activation by the proteolytic cleavage of the ectodomain. In addition, the generation of excess amount of soluble Tie2 ectodomain further inhibits Tie2 activation by sequestering their ligands. However, epithin-mediated degradation also induces the tyrosine phosphorylation of the truncated Tie2 in a ligand-independent manner (Figure 5A). Moreover, the phosphorylated Tie2 fragment can recruit a downstream effector, the p85 subunit of PI3K (Figure 5B-C). Thus, the truncated Tie2 can no longer be regulated by angiopoietins but can nonetheless elicit its own signaling. This new type of modulation yields different outcomes in Tie2 signaling compared with those by Ang1/Ang2. Ang1/Ang2 is secreted as a soluble ligand. Hence, its effects can potentially target the cells in a wider range and/or at a longer distance when it is used as a paracrine ligand. In contrast, epithin degrades Tie2 only in the same cell (Figures 1,Figure 2–3) or in neighboring cells in close proximity (Figure 4) through the specific molecular interaction (Figures 1,3). Therefore, it is more likely that the localized regulation of Tie2 by epithin is useful when modulation of Tie2 in a confined region is required. For example, during transendothelial migration, endothelial cell junctions physically stabilized by the Ang1 and Tie2 complex28 should be disrupted only in the region where the transmigrating cell is attached, whereas other junctions remain intact.
Interestingly, a soluble Tie2, part of the extracellular domain including the ligand binding site, was detected in the conditioned media of PMA-stimulated endothelial cells and in human sera36 ; in addition, an elevated level of soluble Tie2 was noticed in the sera of patients with recurrent cancer or with the development of metastasis.37 These observations suggest the presence of alternative pathways for the proteolytic modulation of Tie2.
Then, how can the cleavage of Tie2 by epithin activate Tie? We found that a genetically truncated Tie2 as well as the cleaved Tie2 by epithin are phosphorylated much more than wild-type Tie2 (supplemental Figure 4). More interestingly, we found that the membrane proximal region of Tie2 cytoplasmic domain contains a possible coiled-coil region (supplemental Figure 9). Because coiled-coil regions in many proteins are known to mediate oligomerization, these results suggest that the transmembrane and intracellular domain of Tie2 may have a tendency of oligomerization through the coiled-coil region and the extracellular domain of Tie2 may inhibit the spontaneous oligomerization. Therefore, it may be possible that the epithin-mediated degradation of the extracellular domain abolishes the inhibitory effect resulting in oligomerization and cross-phosphorylation of Tie2. Interestingly, another receptor tyrosine kinase, ErbB2, is also known to exhibit increased kinase activity and signaling when extracellular domain of the receptor is truncated.38 Therefore, this type of regulation for receptor tyrosine kinase can be more generalized.
Roles of epithin-mediated Tie2 regulation in transendothelial migration
It is shown that: epithin-expressing cells are located near Tie2-expressing vessels (supplemental Figure 2), epithin can degrade Tie2 in neighboring cells in a trans manner (Figure 4), epithin can induce Tie2 signaling by proteolytic cleavage (Figure 5), and epithin enhances transendothelial migration as well as metastasis (Figures 6, 7). Based on these data, we propose 3 possible mechanisms by which epithin can facilitate metastasis. First, when cancer cells approach the endothelial cells of the vessel, epithin expressed in extravasating cancer cells interacts with and degrades Tie2 in the Tie2/Ang1 complex that holds the endothelial cells together at the cell-cell junction,28,29 thus transiently loosening the cell-cell junctions. Second, the proteolytic cleavage of Tie2 induces Tie2 signaling through recruiting PI3K (Figure 5), leading to actin cytoskeletal rearrangement and retraction of the endothelial cells,28,29 which can further facilitate epithin-expressing cell pass through the gaps between endothelial cells. Indeed, during transendothelial migration, actin stress fibers have often been observed in endothelial cells,39 and the formation of actin stress fibers facilitates transendothelial migration by inducing endothelial cell retraction. Consistently, stress fiber formation was observed in the endothelial cells in contact with the epithin-expressing transmigrating cells (Figure 6A). PI3K signaling is well known to mediate actin remodeling.40 This stimulates the formation of actin stress fibers through the activation of RhoA.41 Therefore, PI3K signaling evoked by truncated Tie2 (Figure 5) may be involved in the formation of actin stress fibers in endothelial cells (Figure 6A), leading to endothelial cell retraction. Third, when epithin-expressing cells escape the vasculature, the matrix-degrading activity of epithin19 can generate a pathway for the cells to invade further into the layers of the extracellular matrix. In these ways, we propose, epithin can participate in transendothelial migration as well as in metastasis.
Epithin-mediated Tie2 regulation in other biologic situations
Tie2 signaling is known to play important roles in the regulation of the vascular function by stabilizing vascular structures,42 inhibiting apoptosis of endothelial cells,43,44 or by promoting endothelial cell migration and angiogenesis.27,45 Therefore, the expression of epithin in endothelial cells may regulate these functions of Tie2 by degrading it. Indeed, epithin expression is induced during the formation of capillary-like structures in vitro, suggesting a potential role of epithin in the vascular function and angiogenesis.46 We found evidence that epithin induces migration of endothelial cells, vessel morphogenesis, and other angiogenic processes.47 Therefore, it would be of high interest to find out if epithin knockout mice8 show any vessel phenotype and/or insufficient Tie2 signaling. Indeed, the reddish appearance of the knockout mice may implicate some vascular defects, which are consistent with insufficient Tie-2 signaling. In addition, in our preliminary studies, keratin 5 promoter-driven epithin overexpressing mouse-K5 matriptase shows enhanced angiogenesis and leaky ear vessels when irritated with mustard oil (data not shown).
Tie2 signaling is also known to maintain hematopoietic stem cells in the bone marrow niche.48 Ang1 produced by osteoblasts in the bone marrow activates Tie2 on hematopoietic stem cells, resulting in tight adhesion of the hematopoietic stem cells to the niche. The absence of the Ang1/Tie2 interaction appears to induce both the exit of those hematopoietic stem cells from the niche and their differentiation.48 In addition, several reports show that epithin is expressed in the hematopoietic lineage cells of monocytes, macrophages, and mast cells.49-51 These results raise the intriguing possibility that epithin may also be involved in the regulation of the exit of hematopoietic stem cells from their niche by proteolytic cleavage. Indeed, we found that transendothelial cell migration of activated macrophages were dependent on epithin (data not shown). The specific roles of epithin in hematopoiesis and homeostasis remain to be studied further.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Research Program for New Drug Target Discovery (2009-0083366) grant (M.K.) and by the Brain Research Center (M103KV010013-04K2201-0131) grant (D.P.) from the Ministry of Education, Science & Technology, Republic of Korea.
Authorship
Contribution: C.K., H.S.L., and D.L. designed the research, performed experiments, analyzed data, and wrote the paper; E.-G.C. designed the research and analyzed data; S.D.L., S.J.Y., and S.B.K. performed experiments; and D.P. and M.G.K. analyzed data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for C.K. is Department of Medicine, University of California San Diego, La Jolla, CA. The current affiliation for E.-G.C. is Center for Neuroscience and Aging, Sanford-Burnham Medical Research Institute, La Jolla, CA.
Correspondence: Dongeun Park, School of Biological Sciences, Seoul National University, Seoul 151-747, Republic of Korea; e-mail: depark@snu.ac.kr; and Moon Gyo Kim, Department of Biological Sciences, Inha University, Yonghyun-Dong, Incheon, Republic of Korea; e-mail: mgkim@inha.ac.kr.
References
Author notes
C.K., H.S.L., and D.L. contributed equally to this study.