Abstract
Heterozygous deletions spanning chromosome 5q31.2 occur frequently in the myelodysplastic syndromes (MDS) and are highly associated with progression to acute myeloid leukemia (AML) when p53 is mutated. Mutagenesis screens in zebrafish and mice identified Hspa9 as a del(5q31.2) candidate gene that may contribute to MDS and AML pathogenesis, respectively. To test whether HSPA9 haploinsufficiency recapitulates the features of ineffective hematopoiesis observed in MDS, we knocked down the expression of HSPA9 in primary human hematopoietic cells and in a murine bone marrow–transplantation model using lentivirally mediated gene silencing. Knockdown of HSPA9 in human cells significantly delayed the maturation of erythroid precursors, but not myeloid or megakaryocytic precursors, and suppressed cell growth by 6-fold secondary to an increase in apoptosis and a decrease in the cycling of cells compared with control cells. Erythroid precursors, B lymphocytes, and the bone marrow progenitors c-kit+/lineage−/Sca-1+ (KLS) and megakaryocyte/erythrocyte progenitor (MEP) were significantly reduced in a murine Hspa9-knockdown model. These abnormalities suggest that cooperating gene mutations are necessary for del(5q31.2) MDS cells to gain clonal dominance in the bone marrow. Our results demonstrate that Hspa9 haploinsufficiency alters the hematopoietic progenitor pool in mice and contributes to abnormal hematopoiesis.
Introduction
The myelodysplastic syndromes (MDS) are a group of clonal hematopoietic disorders exhibiting ineffective hematopoiesis characterized by peripheral blood cytopenias. Whole chromosome 5 loss or interstitial deletions spanning 1 copy of 5q31.2 are among the most common abnormalities detected by conventional cytogenetic analysis in de novo (6%-20% of patients) and therapy-related MDS (up to 40% of patients).1-3 The commonly deleted segment in 5q31.2 has been mapped,4,5 and mutational analysis of the residual nondeleted allele has not identified a classic tumor-suppressor gene.4-6 Therefore, we and others hypothesize that haploinsufficiency of a gene(s) in 5q31.2, not the loss of both alleles, may be a critical initiating event in MDS pathogenesis and may contribute to acute myeloid leukemia (AML) transformation when combined with additional cooperating mutations.
The minimally deleted region on chromosome 5q31.2 is distinct from the minimally deleted region on 5q33.1,7 which is associated with the 5q minus syndrome and carries a lower risk of progressing to AML compared with 5q31.2 deletions. Haploinsufficiency of the RPS14 gene, located in the 5q33.1 region, causes abnormal erythroid differentiation and accelerated apoptosis both in vitro and in a murine knockout model, supporting its role in the pathogenesis of the 5q minus syndrome.8,9 Deletions on the long arm of chromosome 5 typically encompass both 5q31.2 and 5q33.1 minimally deleted regions, suggesting that haploinsufficiency of multiple genes may be necessary to recapitulate the full spectrum of abnormalities observed in patients with MDS. Haploinsufficiency of Egr1 (located on 5q31.2), Apc (located proximal to the 5q31.2 minimally deleted region), or Npm (located distal to the 5q31.2 minimally deleted region) are associated with abnormal hematopoiesis in mice, further supporting the possibility that the deletion of multiple 5q genes may cooperate in MDS initiation or progression.10-14
We hypothesized that haploinsufficiency of HSPA9, a gene located on the 5q31.2 interval, may also influence hematopoiesis in MDS patients based on several observations. HSPA9 mRNA levels are reduced by 50% in CD34+-purified hematopoietic progenitors isolated from patients with del(5q) MDS compared with MDS patients without del(5q) and normal control CD34+ cells, consistent with haploinsufficient expression levels in del(5q) patients.6 An N-ethyl-N-nitrosourea mutagenesis screen in zebrafish identified that a bi-allelic mutation in Hspa9 resulted in anemia, dysplastic immature erythroblasts, accelerated apoptosis, and leukopenia.15 Heterozygous Hspa9 mutant fish also displayed accelerated apoptosis in erythroid cells consistent with ineffective hematopoiesis.15 In mice, Hspa9 is a gene in the 5q31.2 interval that is a common retroviral insertional mutagenesis site associated with development of AML (see the Mouse Retrovirus Tagged Cancer Gene Database at: http://rtcgd.ncifcrf.gov/).16 Finally, HSPA9 was identified as a mediator of erythropoietin (EPO) signaling in primary human progenitor cells.17 These data suggest that haploinsufficiency of HSPA9 may contribute to the ineffective hematopoiesis observed in early-stage MDS and possibly to AML progression.
To directly test the effects of HSPA9 haploinsufficiency on hematopoiesis, we measured cell differentiation, apoptosis, and cell-cycle status in primary human CD34+ progenitor cells after knocking down HSPA9 expression using short hairpin RNAs (shRNAs). Next, we modeled haploinsufficiency in mice using shRNA knockdown of Hspa9 in a bone marrow transduction/transplantation model. Both approaches resulted in abnormal erythropoiesis and a reduction in hematopoietic progenitors following knockdown. Our results indicate that the loss of 1 copy of HSPA9 may contribute to abnormal hematopoiesis, and suggest that haploinsufficiency of multiple genes may be the critical genetic event that is necessary to fully recapitulate MDS pathogenesis in a model organism.
Methods
Isolation of human hematopoietic progenitor cells
Isolation of umbilical cord blood mononuclear cells was performed by Ficoll gradient centrifugation followed by CD34+ cell selection using the autoMACS positive-selection system to achieve > 90% purity of CD34+ cells (Miltenyi Biotec). Tissue-acquisition protocols were approved by the Washington University School of Medicine Institutional Review Board.
Mice
C57BL/6 mice (Ly5.2 allele), a wild-type congenic strain of C57BL/6 that have the Ly5.1 allele (B6.SJL-PtPrc*Pep3bBoyJ), and Tp53 knockout mice in the C57BL/6 background (Ly5.2 allele; B6.129S2-Trp53tm1Tyj/J) were obtained from The Jackson Laboratory. All animal work was approved by the Washington University School of Medicine Animal Studies Committee.
Lentiviral shRNA vectors and production
pLKO.1 vectors were used for lentiviral-mediated gene knockdown in human cell cultures.18 The Fcy-si vector was used for in vivo shRNA lentiviral mouse studies, as described previously.19,20 The human and murine shRNA sequences used are listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). For lentiviral production, human embryonic kidney 293T (HEK 293T) cells were transfected with the pLKO.1 or Fcy-si vector, packaging vector (p.delta8.9 or p.delta8.2A), and an envelope vector (pMD.G encoding VSV-G) using TransIT-LT1 (Mirus Bio) transfection reagents; by calcium phosphate transfection according to the protocols of the manufacturer and the RNAi Consortium (available from: http://www.broadinstitute.org/rnai/public/resources/protocols); or as previously reported.20 Conditioned medium was collected 48, 72, and 96 hours after transfection. Relative titers of pLKO.1 virus were determined using puromycin selection (5 μg/mL) in NIH 3T3 cells followed by alamarBlue cell-viability determination. For Fcy-si virus, titers were determined by flow cytometric measurement of the percentage of yellow fluorescent protein-positive (YFP+) HEK 293T cells. An average Fcy-si viral titer of 5 × 108/mL was achieved after concentration.
Human CD34+ progenitor cell lentiviral shRNA transduction and culture
CD34+ progenitor cells were primed in X-VIVO medium (Lonza Walkersville) containing human cytokines (50 ng/mL of stem-cell factor [SCF], 50 ng/mL of Fms-related tyrosine kinase 3 [FLT-3] ligand, 50 ng/mL of thrombopoietin [TPO], 50 ng/mL of interleukin 3 [IL3], l-glutamine, and antibiotics) for 16 hours before lentiviral transduction. Cells were spinoculated in the presence of polybrene (2-4 μg/mL) and incubated overnight at 37°C in 5% CO2. Cells were washed and incubated in erythroid, granulocytic, or megakaryocytic unilineage differentiation media. For erythroid differentiation, cells were cultured in serum-free expansion medium (StemCell Technologies) with 25 ng/mL of SCF, 10 ng/mL of IL3, 10 ng/mL of IL6, 0.5 units of EPO, 100 U/mL of penicillin/streptomycin (P/S), and l-glutamine for 7 days. For granulocytic cultures, cells were plated in Iscove medium supplemented with 20% fetal calf serum, 10 ng/mL of SCF, 100 ng/mL of granulocyte-colony stimulating factor (Amgen), 200 ng/mL of transferrin (Sigma-Aldrich), 100 ng/mL of insulin (Sigma-Aldrich), 50uM β-mercaptoethanol, 100 U/mL of P/S, and l-glutamine for 7 days. To promote megakaryocyte differentiation, cells were grown in serum-free expansion medium with the addition of 100 ng/mL of SCF, 10 ng/mL of IL3, 100 ng/mL of TPO, 100 U/mL of P/S, and l-glutamine for 12 days. All cytokines were obtained from Peprotech.
Murine bone marrow lentiviral transduction/transplantation and homing assay
Six to 10 C57BL/6 Ly5.1 donor mice were killed to obtain bone marrow cells for each cohort. Bone marrow cells were lineage depleted (phycoerythrin [PE]-conjugated Ter119+, Gr-1+, CD3e+, B220+) using an autoMACS (Miltenyi Biotec) and lentivirally transduced with a multiplicity of infection (MOI) of 20 in α-minimum essential medium supplemented with 15% fetal calf serum, 10 ng/mL of TPO, 10 ng/mL of IL3, 50 ng/mL of FLT3 ligand, and 100 ng/mL of SCF for 24 hours. All cytokines were obtained from Peprotech. TP53−/− donor mice (C57BL/6 Ly5.2) have been described previously.21 Congenic C57Bl/6 Ly5.2 or Ly5.1 recipient mice were irradiated with a single dose of 1000 cGy, and 250 000-500 000 cells were injected retro-orbitally into each mouse. A small portion of transduced donor cells (20 000-50 000 cells) was plated in culture for 2-4 days to assess the percentage of transduced YFP+ cells in combination with allophycocyanin (APC)–conjugated CD117 antibody (BD Pharmingen) and 7-amino-actinomycin D (7-AAD) labeling for dead-cell exclusion. The bone marrow–homing efficiency of transduced donor cells was measured by counting the percentage of YFP+ methylcellulose colonies from pre-injected donor cells compared with the percentage of YFP+ methylcellulose colonies from bone marrow cells harvested from recipient mice 24 hours after injection, as described previously (M3434; StemCell Technologies).20
Western blotting
Cell lysates were prepared in radioimmunoprecipitation assay buffer as described previously.22 The following antibodies were used: Mcl-1 (sc-819; Santa Cruz Biotechnology), Bcl-x (610211; BD Transduction Laboratories), Bcl-2 (sc7382; Santa Cruz Biotechnology), Bak (06-536; Upstate Biotechnology), Bim (202000; Calbiochem), Bax (554104; BD Pharmagen), HSPA9 (MA3-028; Affinity BioReagents), β-actin (A5441; Sigma-Aldrich), and tubulin (sc-5286; Santa Cruz Biotechnology).
Hematopoietic progenitor assays
Transduced human progenitor cells or YFP+ bone marrow cells isolated from individual mice were counted using a hemocytometer and plated, in duplicate, in 1.3 mL of complete methylcellulose medium (H4434 or M3434, respectively; StemCell Technologies) or in methylcellulose medium supplemented with 3 units/mL of EPO (M3234; StemCell Technologies). Colonies with > 30 cells were counted on days 10-14.
Human cell flow cytometry
Flow cytometry antibodies included anti–human CD71 (fluorescein isothiocyanate [FITC]), glycophorin A (PE), CD34 (APC), CD66 (PE), CD61 (FITC), CD41a (PE), CD42b (PE; all from BD Pharmingen), and CD15 (FITC; eBioscience). Cells were incubated for 1 hour with bromodeoxyuridine (BrdU), stained with cell-surface antibodies, fixed/permeabilized, and stained with Alexa Fluor 647-conjugated anti–BrdU antibody (Invitrogen) according to the manufacturer's protocol (BD Biosciences). Cells were then labeled with 7-AAD (Calbiochem) and analyzed for cell cycle. Cells were labeled with PE-conjugated annexin V and 7-AAD (BD Biosciences) and PE-conjugated active caspase-3 (BD Biosciences), and the JC-1 flow kit (Invitrogen) was used to measure mitochondrial depolarization according to the manufacturer's protocols. All data were acquired using a 5-color FACScan flow cytometer (BD Biosciences) and analyzed using FlowJo 8.8.7 software (TreeStar).
Murine cell flow cytometry
Cells were pre-incubated with CD16/32 (eBioscience), followed by staining with antibodies directed against murine CD45.1 (APC), CD45.2 (PerCP-Cy5.5), Gr-1 (PE), CD11b (APC–Alexa Fluor 750), B220 (PE), CD3e (PE), Ter119 (APC–Alexa Fluor 750; all from BD Pharmingen), CD71 (Alexa Fluor 647; AbD Serotec), and 7-AAD for dead-cell exclusion. These data were acquired using the 5-color FACScan flow cytometer and analyzed using FlowJo 8.8.7 software. To profile the progenitor pools of lentivirally transduced YFP+ cells, cells were stained with the antibody cocktail of biotin-conjugated anti–CD34, followed by secondary staining of PE-anti–streptavidin, PE-Cy7 lineage cocktail (Gr-1, Ter119, B220, CD19, CD3, CD4, CD8, IL7R), Sca-1 (APC), CD117 (APC–Alexa Fluor 750), and Fc-gammaR (PE-Cy5.5; eBioscience). Analysis of c-kit+/lineage−/Sca-1+ (KLS), common myeloid progenitors (CMPs), granulocyte/macrophage progenitors (GMPs), and megakaryocyte/erythroid progenitors (MEPs) was performed as described previously.23
Cell sorting
Bone marrow cells were harvested, washed, and stained with 7-AAD for dead-cell exclusion, and YFP+ cells were sorted into sterile medium and processed as outlined in “Hematopoietic progenitor assays,” “Western blotting,” and “Human cell flow cytometry,” respectively for methylcellulose colony assays, BrdU assays, and for protein lysates (iCyt). To assess the cell-cycle status of bone marrow cells, mice were injected intraperitoneally with 2 mg of BrdU (10 mg/mL) at 24 and 12 hours prior to harvesting the bone marrow. YFP+-sorted cells were stained with an Alexa Fluor 647–conjugated anti–BrdU antibody (Invitrogen) and analyzed by flow cytometry.
Statistics
All data are presented as the means ± SD, and P values were determined using a 2-tailed Student t test in Prism 5 software (GraphPad).
Results
HSPA9 knockdown delays the erythroid maturation of primary human CD34+ cells
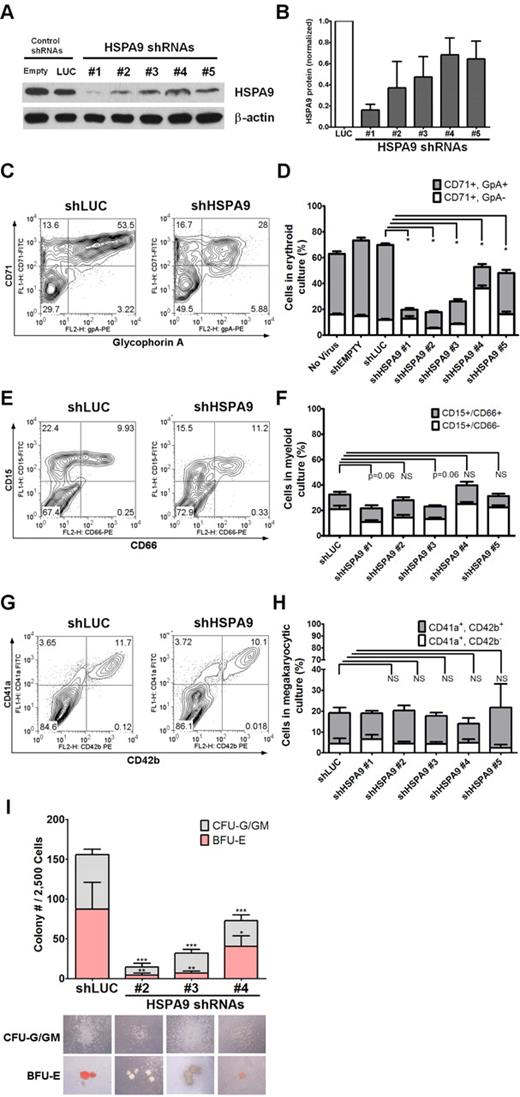
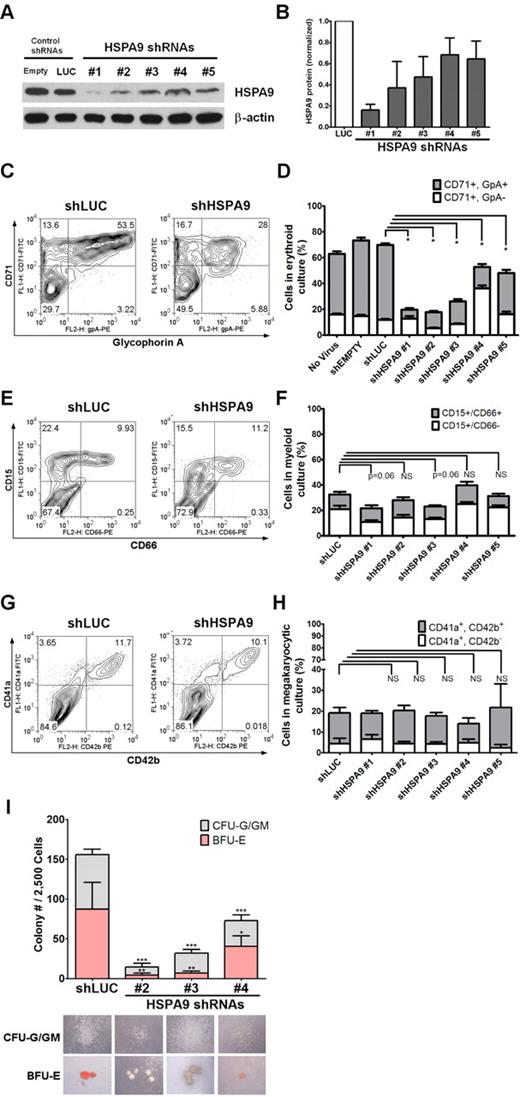
We purified human CD34+ hematopoietic progenitors from cord blood samples (> 90% purity) and infected them individually with 5 unique lentiviral shRNAs targeting HSPA9 (achieving 30%-90% knockdown of protein levels) and 2 control shRNAs that also carry the puromycin resistance gene (Figure 1A-B). The relative viral titers for all constructs were similar. Transduced cells were maintained in puromycin selection medium; grown in unilineage culture conditions that supported erythroid, myeloid, or megakaryocytic cell growth; and analyzed on days 7-12 of culture. Maturing erythroid precursors (CD71+/glycophorin A+) were significantly reduced in all HSPA9-knockdown cultures compared with control cells when grown in erythroid unilineage differentiation conditions (7.1%-32% vs 58%, respectively; 1.8-8.2-fold reduction; n = 4 for each shRNA; P < .0001; Figure 1C-D). Using myeloid unilineage culture conditions, the percentage of cells expressing the myeloid antigen CD15+ was not significantly reduced (21.8%-39.7% vs 32.4%; 0.81-1.5-fold reduction; n = 6 for each shRNA; P = .06-.85; Figure 1E-F). Similarly, HSPA9 knockdown did not result in a reduction in the percentage of CD41a+ cells after 12 days of culture in megakaryocyte medium compared with control cells (13.9%-21.7% vs 19.1%, respectively; 0.88-1.37-fold reduction; n = 2 for each shRNA; P = .39-.96; Figure 1G-H). Next, we assessed the affect of HSPA9 knockdown on progenitor cells by seeding an equal number of transduced CD34+ cells in complete methylcellulose medium containing puromycin. There was a greater reduction in burst-forming unit-erythroid (BFU-E) colonies (average 5.1-fold reduction; P ≤ .04) compared with the reduction in colony-forming unit–granulocyte (CFU-G)/granulocyte-macrophage (GM) colonies (3.1-fold reduction; P < .001) compared with control cells (Figure 1I). BFU-E colonies generated from HSPA9-knockdown cells also contained reduced levels of hemoglobin on visual inspection (Figure 1I). These results indicate that erythropoiesis was affected to a greater degree than other lineages after HSPA9 knockdown.
Knockdown of HSPA9 in human hematopoietic progenitor cells alters their erythroid differentiation. (A) Representative immunoblot of HSPA9 protein in CD34+ cells transduced with shRNAs targeting control genes (empty or luciferase) or HSPA9 after 7 days in erythroid culture conditions. (B) Quantification of HSPA9 protein levels in cells normalized to the luciferase shRNA (n = 2-5). (C) Representative flow cytometric plots for CD71 (transferrin receptor) and glycophorin A (GpA) expression in control and HSPA9-knockdown cells. Numbers represent the percentage of cells in each quadrant. CD71−/GpA− cells are the most immature, CD71+/GpA− are intermediate, and CD71+/GpA+ are the most mature cells.8 (D) Quantification of CD71+ cells (n = 4; *P < .0001). (E) Representative flow cytometric plots for CD15 and CD66 expression in control and HSPA9-knockdown cells. Numbers represent the percentage of cells in each quadrant. CD15−/CD66− cells are the most immature, CD15+/CD66− are intermediate, and CD15+/CD66+ are the most mature cells. (F) Quantification of CD15+ cells (n = 6). (G) Representative flow cytometric plots for CD41a and CD42b expression in control and HSPA9-knockdown cells. Numbers represent the percentage of cells in each quadrant. (H) Quantification of CD41a+ cells (n = 2). (I) Transduced cells were plated in methylcellulose medium containing puromycin, and the number of CFU-G plus CFU-GM colonies and BFU-E colonies were counted on day 12 (n = 2; *P = .04; **P = .003; ***P < .001). Pictures of representative CFU and BFU colonies are shown below the appropriate shRNA construct. Hemoglobin appears red in the representative BFU-E colonies; empty mean no shRNA sequence was inserted into the pLKO.1 vector. LUC indicates luciferase; and NS, not significant. All data represent the means ± SD.
Knockdown of HSPA9 in human hematopoietic progenitor cells alters their erythroid differentiation. (A) Representative immunoblot of HSPA9 protein in CD34+ cells transduced with shRNAs targeting control genes (empty or luciferase) or HSPA9 after 7 days in erythroid culture conditions. (B) Quantification of HSPA9 protein levels in cells normalized to the luciferase shRNA (n = 2-5). (C) Representative flow cytometric plots for CD71 (transferrin receptor) and glycophorin A (GpA) expression in control and HSPA9-knockdown cells. Numbers represent the percentage of cells in each quadrant. CD71−/GpA− cells are the most immature, CD71+/GpA− are intermediate, and CD71+/GpA+ are the most mature cells.8 (D) Quantification of CD71+ cells (n = 4; *P < .0001). (E) Representative flow cytometric plots for CD15 and CD66 expression in control and HSPA9-knockdown cells. Numbers represent the percentage of cells in each quadrant. CD15−/CD66− cells are the most immature, CD15+/CD66− are intermediate, and CD15+/CD66+ are the most mature cells. (F) Quantification of CD15+ cells (n = 6). (G) Representative flow cytometric plots for CD41a and CD42b expression in control and HSPA9-knockdown cells. Numbers represent the percentage of cells in each quadrant. (H) Quantification of CD41a+ cells (n = 2). (I) Transduced cells were plated in methylcellulose medium containing puromycin, and the number of CFU-G plus CFU-GM colonies and BFU-E colonies were counted on day 12 (n = 2; *P = .04; **P = .003; ***P < .001). Pictures of representative CFU and BFU colonies are shown below the appropriate shRNA construct. Hemoglobin appears red in the representative BFU-E colonies; empty mean no shRNA sequence was inserted into the pLKO.1 vector. LUC indicates luciferase; and NS, not significant. All data represent the means ± SD.
Accelerated apoptosis and altered cell cycle in HSPA9-knockdown erythroid cultures
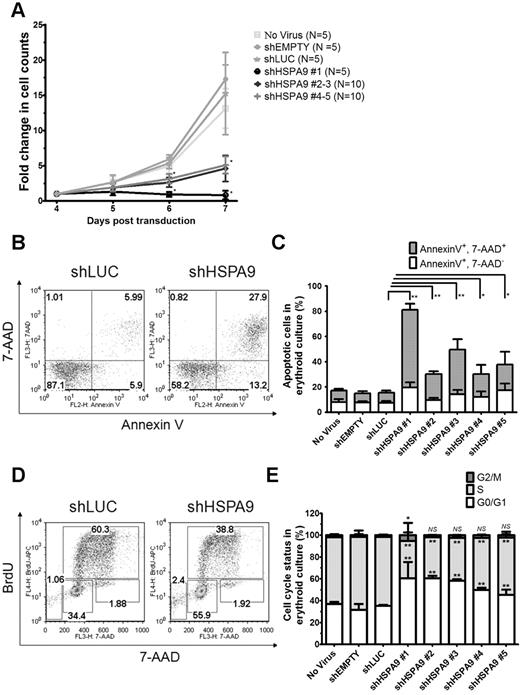
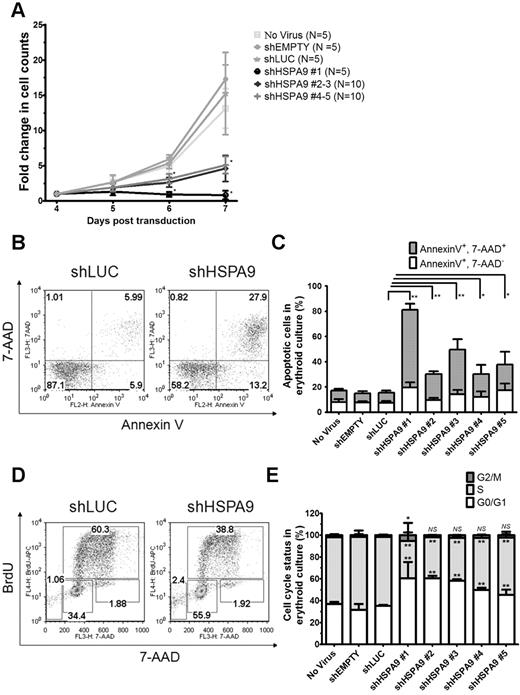
The cell density of erythroid cultures was normalized after 4 days in puromycin selection, and apoptosis (annexin V/7-AAD) and cell-cycle status (BrdU/7-AAD) were measured on day 7 using flow cytometry. Cells transduced with control shRNAs expand 15.2-fold from days 4-7 compared with only a 0.8–5.2-fold expansion in HSPA9-knockdown cells (n = 5 for each shRNA; P < .001; Figure 2A). The reduced number of cells in HSPA9-knockdown cultures was associated with an increase in apoptosis and a decrease in the number of cells entering the cell cycle compared with control shRNA-expressing cells: 8.2% of cells in control cultures were annexin V+/7-AAD+ compared with 18%-62% of cells in HSPA90knockdown cultures (n = 5 for each shRNA; P ≤ .03; Figure 2B-C). Increased mitochondrial membrane depolarization and active/cleaved caspase-3 expression were also observed with HSPA9 knockdown (supplemental Figure 1A-C). The proportion of cells containing active caspase-3 was higher in the CD34− fraction compared with the CD34+ fraction in shHSPA9 cultures, suggesting that differentiating cells may be more susceptible to apoptosis following HSPA9 knockdown (supplemental Figure 1C). Interrogation of anti- and pro-apoptotic BCL2 family members revealed that cleavage of BAX to the apoptosis-inducing p18 form occurred in HSPA9 knockdown cells, further implicating activation of the intrinsic apoptosis pathway following HSPA9 knockdown (supplemental Figure 2). Following 1 hour of BrdU exposure, 63% of cells in shLUC control cultures were in the S phase of the cell cycle compared with only 34%-52% of HSPA9-knockdown cultures (n = 5 for each shRNA; P < .006). Knockdown of HSPA9 was associated with an increase of cells in the G0/G1 phase of the cycle (P < .006; Figure 2D-E). These results suggest that knockdown of HSPA9 in primary hematopoietic cells results in an accelerated apoptosis commonly observed with ineffective erythropoiesis and cell-cycle changes.
Decreased proliferation of primary human hematopoietic progenitor cells, increased apoptosis, and reduction of cells in the S phase following knockdown of HSPA9. (A) Transduced cells were seeded at equal concentrations after 4 days in erythroid culture conditions, and the total number of cells was counted for 3 consecutive days in the presence of puromycin selection medium. The fold change in cell counts was calculated relative to the number of cells on day 4 (n = 5 for each shRNA; *P < .001). (B) Representative flow cytometric plots for annexin V and 7-AAD in control and HSPA9-knockdown cells. Numbers represent the percentage of cells in each quadrant. (C) Quantification of annexin V+/7-AAD+ cells (a surrogate for apoptosis; n = 5 for each shRNA; *P ≤ .03; **P < .0001). (D) Representative flow cytometric plots for BrdU and 7-AAD in control and HSPA9-knockdown cells. Numbers represent the percentage of cells in each gate. Sub-G0 cells are in the low left gate. Cells in G0/G1 of the cell cycle are BrdU−/7-AAD−, S-phase cells are BrdU+, and G2/M cells are BrdU−/7-AAD+. (E) Quantification of the cell-cycle phases after exclusion of cells in the sub-G0 fraction (n = 5 for each shRNA; *P < .02; **P < .006). Empty mean no shRNA sequence was inserted into the pLKO.1 vector. LUC indicates luciferase; and NS, not significant. All data represent the means ± SD.
Decreased proliferation of primary human hematopoietic progenitor cells, increased apoptosis, and reduction of cells in the S phase following knockdown of HSPA9. (A) Transduced cells were seeded at equal concentrations after 4 days in erythroid culture conditions, and the total number of cells was counted for 3 consecutive days in the presence of puromycin selection medium. The fold change in cell counts was calculated relative to the number of cells on day 4 (n = 5 for each shRNA; *P < .001). (B) Representative flow cytometric plots for annexin V and 7-AAD in control and HSPA9-knockdown cells. Numbers represent the percentage of cells in each quadrant. (C) Quantification of annexin V+/7-AAD+ cells (a surrogate for apoptosis; n = 5 for each shRNA; *P ≤ .03; **P < .0001). (D) Representative flow cytometric plots for BrdU and 7-AAD in control and HSPA9-knockdown cells. Numbers represent the percentage of cells in each gate. Sub-G0 cells are in the low left gate. Cells in G0/G1 of the cell cycle are BrdU−/7-AAD−, S-phase cells are BrdU+, and G2/M cells are BrdU−/7-AAD+. (E) Quantification of the cell-cycle phases after exclusion of cells in the sub-G0 fraction (n = 5 for each shRNA; *P < .02; **P < .006). Empty mean no shRNA sequence was inserted into the pLKO.1 vector. LUC indicates luciferase; and NS, not significant. All data represent the means ± SD.
Transplantation and homing of shRNA-transduced murine hematopoietic progenitors
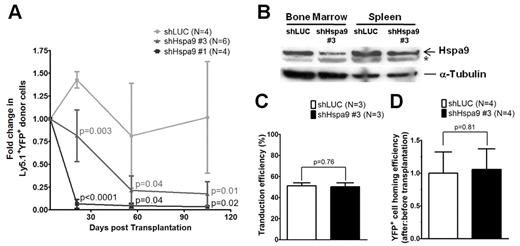
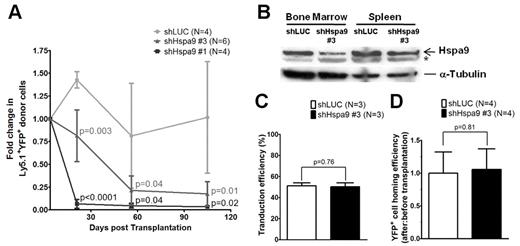
Murine bone marrow–progenitor cells were infected with lentiviral shRNAs co-expressing YFP. Four independent lentiviral shRNAs targeting murine Hspa9 were produced, and knockdown of Hspa9 was first assessed in the murine BaF3 cell line. Two shRNAs were identified that produced 40%-90% knockdown of Hspa9 in BaF3 cells compared with a control shRNA targeting the luciferase gene (murine shRNAs no. 3 and no. 1, respectively). To achieve adequate transduction of primary murine bone marrow cells in vivo, we used an MOI of 20. shRNAs no. 1 and no. 3 were used in vivo to knock down Hspa9 in murine bone marrow cells, followed by transplantation of infected cells into wild-type congenic recipient mice. Following knockdown of Hspa9 in murine hematopoietic progenitor cells, there was a time-dependent loss of YFP+ peripheral blood cells over several months, with the greatest reduction in YFP+ cells occurring with the more severe Hspa9 knockdown (shRNA no. 1; Figure 3A). To model Hspa9 haploinsufficiency, we subsequently characterized the hematopoiesis in mice transduced with murine Hspa9 shRNA no. 3, which produced ∼ 50% knockdown of Hspa9 in murine bone marrow and spleen cells harvested from recipient mice 8 weeks after transplantation (Figure 3B).
Transplantation and homing of transduced murine hematopoietic progenitor cells. (A) Short-term engraftment of shRNA-transduced murine hematopoietic cells (YFP+) in the peripheral blood of recipient mice (n = 4-6 mice each). (B) Immunoblot of Hspa9 protein in transduced YFP+ cells isolated from the bone marrow and spleens of recipient mice 8 weeks after transplantation. (C) Transduction efficiency was measured as the percentage of YFP+ murine bone marrow progenitor cells after 2-4 days in culture (n = 3). (D) Homing efficiency of transduced cells was measured as the ratio of after-transplantation to before-transplantation YFP+ CFU-Cs (n = 4). *Nonspecific band in murine bone marrow and spleen cells. LUC indicates luciferase. All data represent the means ± SD.
Transplantation and homing of transduced murine hematopoietic progenitor cells. (A) Short-term engraftment of shRNA-transduced murine hematopoietic cells (YFP+) in the peripheral blood of recipient mice (n = 4-6 mice each). (B) Immunoblot of Hspa9 protein in transduced YFP+ cells isolated from the bone marrow and spleens of recipient mice 8 weeks after transplantation. (C) Transduction efficiency was measured as the percentage of YFP+ murine bone marrow progenitor cells after 2-4 days in culture (n = 3). (D) Homing efficiency of transduced cells was measured as the ratio of after-transplantation to before-transplantation YFP+ CFU-Cs (n = 4). *Nonspecific band in murine bone marrow and spleen cells. LUC indicates luciferase. All data represent the means ± SD.
Given the reduction in YFP+ cells following transplantation of Hspa9 knockdown cells, we measured the ability of transplanted cells to home to the bone marrow using a methylcellulose colony assay. First, we determined that the transduction efficiency of the shLUC and shHspa9 no. 3 viruses were equivalent in primary murine bone marrow cells grown in culture for several days (51% vs 50%, respectively; P = .76; n = 3 for each shRNA; Figure 3C). To assess the homing of transduced/transplanted murine cells to the bone marrow, we next measured the percentage of YFP+ methylcellulose colonies that were derived from an aliquot of pre-injected cells compared with the percentage of YFP+ colonies that were harvested from the bone marrow of recipient mice 24 hours after injection, as described previously.20 There was no difference in the ratio of YFP+ colonies from post-injected/pre-injected Hspa9 cells compared with the ratio in control knockdown mice, indicating that the time-dependent reduction in YFP+ cells was not due to a defect in homing of transplanted cells to the bone marrow (1.0 vs 1.06, respectively; P = .81; n = 4 for each shRNA; Figure 3D). In summary, the control (luciferase) and Hspa9 (no. 3) shRNAs were used at the same MOI, had equal transduction efficiencies, and had similar homing abilities to the bone marrow following transplantation, suggesting that the reduction in YFP+ cells was due to a cell-intrinsic defect. Assessment of the long-term engraftment of Hspa9 shRNA–transduced cells at 24 weeks was not possible due to the severe reduction in YFP+ cells over time. Therefore, all subsequent analysis addressed the short-term engraftment of cells at 3-8 weeks after transplantation using 2 independent transduction/transplantation cohorts for each shRNA (control luciferase shRNA, n = 8 mice total; Hspa9 no. 3 shRNA, n = 9 mice total).
Altered hematopoiesis in mice following knockdown of Hspa9 in hematopoietic progenitor cells
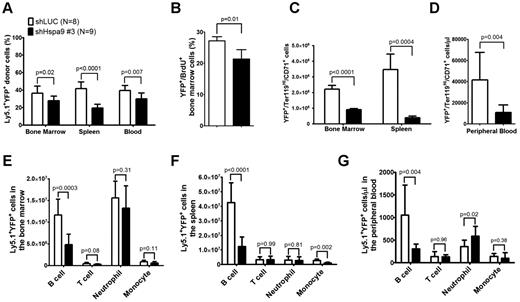
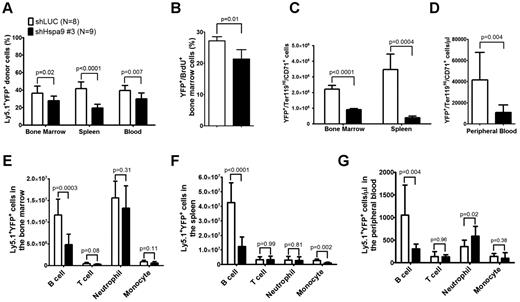
The total number of bone marrow, spleen, or peripheral blood cells was similar in mice receiving the Hspa9 knockdown or control cells (supplemental Figure 3). The percentage of donor Ly5.1 cells in the peripheral blood, bone marrow, and spleens was also similar between the control and Hspa9-knockdown mice (mean range = 85%-92%, -fold change = 0.96-1.0; supplemental Figure 4). However, the percentage of Ly5.1+/YFP+ donor cells was significantly reduced in the bone marrow (27.9% vs 36.3%; P = .02), spleen (19.5% vs 41.6%; P < .0001), and peripheral blood (29.5% vs 39.5%; P = .007) of mice receiving Hspa9-knockdown cells compared with mice receiving control knockdown cells, which is consistent with in vitro results and suggests that Hspa9-knockdown cells are outcompeted by the nontransduced normal cells (Figure 4A). In addition to the reduction in Hspa9 shRNA YFP+ cells compared with the control shRNA, the percentage of YFP+ cells in the S phase was significantly reduced in Hspa9-knockdown cells compared with control knockdown cells in the bone marrow (21.3% vs 27.2%, respectively; n = 4 each; P = .01; Figure 4B).
Reduced erythroid and B lymphocytes in the bone marrow, spleens, and peripheral blood of mice receiving Hspa9-shRNA–transduced progenitor cells. (A) The percentage of donor (Ly5.1+)–transduced (YFP+) cells was measured in the bone marrow, spleens, and peripheral blood of recipient mice up to 8 weeks after transplantation (n = 8-9). (B) Mice were given 2 intraperitoneal injections of BrdU 12 and 24 hours prior to isolating YFP+ bone marrow cells and staining them for BrdU incorporation as a measure of cells in the S phase of the cell cycle. (n = 4). (C-D) The contribution of YFP+ erythroid precursors was measured in the bone marrow, spleens, and peripheral blood of recipient mice up to 8 weeks after transplantation (n = 8-9). (E-G) The contribution of YFP+ B cells, T cells, neutrophils, and monocytes was measured in the bone marrow, spleens, and peripheral blood of recipient mice using flow cytometry up to 8 weeks after transplantation (n = 8-9). All data represent the means ± SD.
Reduced erythroid and B lymphocytes in the bone marrow, spleens, and peripheral blood of mice receiving Hspa9-shRNA–transduced progenitor cells. (A) The percentage of donor (Ly5.1+)–transduced (YFP+) cells was measured in the bone marrow, spleens, and peripheral blood of recipient mice up to 8 weeks after transplantation (n = 8-9). (B) Mice were given 2 intraperitoneal injections of BrdU 12 and 24 hours prior to isolating YFP+ bone marrow cells and staining them for BrdU incorporation as a measure of cells in the S phase of the cell cycle. (n = 4). (C-D) The contribution of YFP+ erythroid precursors was measured in the bone marrow, spleens, and peripheral blood of recipient mice up to 8 weeks after transplantation (n = 8-9). (E-G) The contribution of YFP+ B cells, T cells, neutrophils, and monocytes was measured in the bone marrow, spleens, and peripheral blood of recipient mice using flow cytometry up to 8 weeks after transplantation (n = 8-9). All data represent the means ± SD.
We next examined the short-term repopulating ability of transduced donor cells by examining YFP expression in erythroid, myeloid, and lymphoid cells in the bone marrow, spleens, and peripheral blood of recipient mice. The absolute number of transduced Ter119High/CD71+ erythroid cells was reduced by 2.5-fold in the bone marrow (8.97 × 105 vs 2.21 × 106; P < .0001), by 8.9-fold in the spleen (3.87 × 105 vs 3.46 × 106; P = .0004; Figure 4C), and by 3.8-fold in the peripheral blood (10.95 × 103/μL vs 41.60 × 103/μL; P = .004; Figure 4D), whereas the total number of transduced myeloid cells (CD11b+/Gr-1− monocytes and CD11b+/Gr-1+ neutrophils) in the 3 compartments was not reduced in mice receiving Hspa9 compared with control knockdown cells, respectively (except monocytes in the spleen; Figure 4E-G). Flow cytometric analysis of the different stages of erythroid maturation was performed as described previously.24,25 All stages of erythroid maturation were affected in Hspa9-knockdown cells, while the spleen showed the largest reduction in total cells (supplemental Figure 5). The morphology of erythroid precursors was not altered following Hspa9 knockdown (supplemental Figure 6). Compared with controls, the absolute number of transduced B lymphocytes, but not T lymphocytes, was reduced by 2.4-fold in the bone marrow (1.16 × 107 vs 4.81 × 106; P = .0003), by 3.4-fold in the spleen (4.26 × 108 vs 1.24 × 108; P < .0001), and by 3.5-fold in the peripheral blood (1.06 × 103/μL vs 3.05 × 102/μL; P = .004) of Hspa9-knockdown mice, respectively (Figure 4E-G). These results suggest that Hspa9-knockdown cells cycle less frequently and that erythroid cells and B cells are preferentially affected by Hspa9 knockdown compared with myeloid cells and T cells.
Hematopoietic progenitor pools are altered in the bone marrow of mice receiving Hspa9-knockdown cells
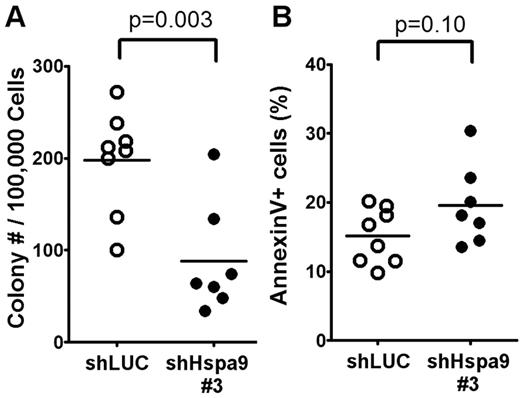
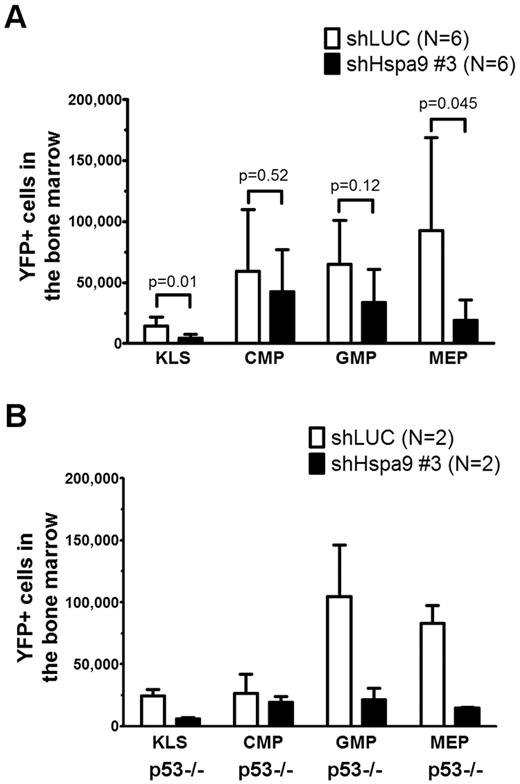
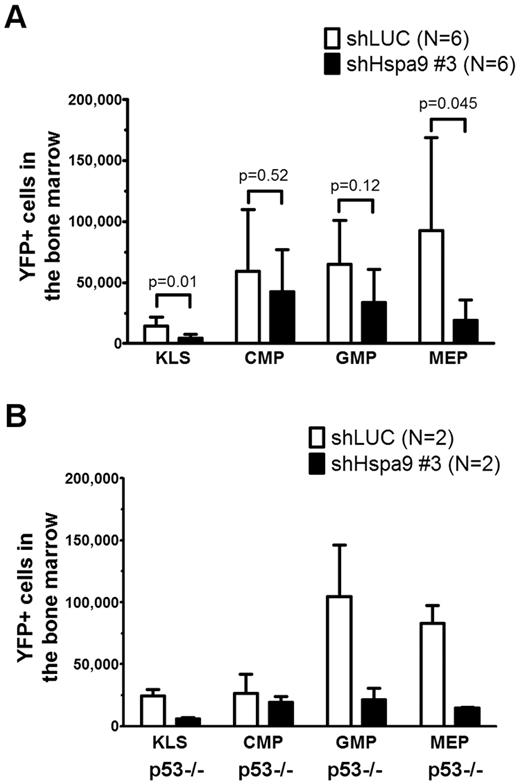
We flow sorted YFP+ bone marrow cells from mice transplanted with Hspa9 and control shRNAs and plated an equal number of cells in complete methylcellulose medium for 10 days. There were fewer CFU-C colonies obtained from Hspa9-knockdown bone marrow cells, suggesting that Hspa9 haploinsufficiency may affect progenitor cells (2.2-fold reduction; P = .003; Figure 5A). In addition, Hspa9-knockdown cells harvested from methylcellulose plates tended to have an increase in the percentage of cells that were annexin V+ compared with control cells, indicating that Hspa9-progenitor cells may also have increased levels of apoptosis (19.6% vs 15.1%, respectively; P = .10; Figure 5B). We also observed a 2-fold reduction in BFU-E methylcellulose colonies obtained from Hspa9-knockdown bone marrow cells (supplemental Figure 7). To assess whether a specific progenitor population was preferentially affected, we measured the total number of YFP+ bone marrow–progenitor cells using flow cytometry.23 The total number of YFP+/IL7Ralpha−/ KLS cells in the bone marrow were significantly reduced in Hspa9-knockdown mice compared with controls (4.60 × 103 vs 14.19 × 103, respectively; P = .014; n = 6 for each shRNA), indicating that Hspa9 knockdown alters an early progenitor pool that contains stem cells (Figure 6A). MEPs (YFP+/IL7R-alpha−/lineage−/Sca-1−/c-kit+/CD34−/Fc-gammaRlo) were affected to a larger degree in the Hspa9-knockdown mice (19.22 × 103 vs 92.40 × 103; 4.8-fold reduction; P = .045) compared with the GMPs (YFP+/IL7R-alpha−/lineage−/Sca-1−/c-kit+/CD34+/Fc-gammaRhi) and CMPs (YFP+/IL7R-alpha−/lineage−/Sca-1−/c-kit+/CD34+/Fc-gammaRlo; 33.64 × 103 vs 64.84 × 103; 1.93-fold reduction; P = 0.12; 42.65 × 103 vs 59.31 × 103; 1.39-fold reduction; P = 0.52, respectively; Figure 6A and supplemental Table 2). These results suggest that MEPs may be preferentially susceptible to altered Hspa9 levels and this may account for the reduction in erythroid precursors that we observed in Hspa9-knockdown mice.
Measurement of bone marrow CFU-Cs following Hspa9 knockdown. (A) YFP+ cells were isolated from the bone marrow of recipient mice and CFU-Cs were measured (n = 7-8). (B) All of the methylcellulose colonies derived from a mouse were harvested, pooled, and the percentage of annexin V+ cells was measured by flow cytometry (n = 7-8).
Measurement of bone marrow CFU-Cs following Hspa9 knockdown. (A) YFP+ cells were isolated from the bone marrow of recipient mice and CFU-Cs were measured (n = 7-8). (B) All of the methylcellulose colonies derived from a mouse were harvested, pooled, and the percentage of annexin V+ cells was measured by flow cytometry (n = 7-8).
Bone marrow hematopoietic progenitors are reduced in mice transplanted with Hspa9-knockdown cells. (A) KLS cells, CMPs, GMPs, and MEPs were measured in the bone marrow of recipient mice using flow cytometry (n = 6). (B) TP53-homozygous–null donor bone marrow cells (p53−/−) were transduced with control or Hspa9 shRNA, and YFP+ KLS, CMPs, GMPs, and MEPs were measured in the bone marrow of recipient mice using flow cytometry (n = 2). All data represent the means ± SD.
Bone marrow hematopoietic progenitors are reduced in mice transplanted with Hspa9-knockdown cells. (A) KLS cells, CMPs, GMPs, and MEPs were measured in the bone marrow of recipient mice using flow cytometry (n = 6). (B) TP53-homozygous–null donor bone marrow cells (p53−/−) were transduced with control or Hspa9 shRNA, and YFP+ KLS, CMPs, GMPs, and MEPs were measured in the bone marrow of recipient mice using flow cytometry (n = 2). All data represent the means ± SD.
Knockdown of Hspa9 in p53−/− donor bone marrow cells
Several lines of evidence have suggested that the progenitor phenotype observed with Hspa9 knockdown may be dependent on wild-type p53 function. HSPA9 binds and sequesters p53 in the cytoplasm, and knockdown of HSPA9 results in nuclear localization of p53.26,27 Redistribution of p53 following HSPA9 knockdown inhibits the colony formation of MCF7 and U2OS cell lines, similar to the reduced methylcellulose colonies observed in our HSPA9-knockdown cells.28 Therefore, if wild-type p53 function mediates this effect, then the loss or mutation of p53 could allow Hspa9-haploinsufficient progenitors to survive. To test directly whether the Hspa9-progenitor phenotype was p53 dependent, we repeated the transduction/transplantation experiments using p53-homozygous–null donor bone marrow cells. Surprisingly, the reduction in KLS and MEP progenitor cells observed with Hspa9 knockdown was not rescued in p53-homozygous–null donor cells (4.1- and 5.6-fold reduction, respectively), suggesting that the altered hematopoietic progenitor pool observed in Hspa9-knockdown mice may be p53 independent (Figure 6B).
Discussion
In this study, we used primary human hematopoietic progenitor cells and mice to study the effects of HSPA9 haploinsufficiency on hematopoiesis. There was a strong dose-dependent inhibition of erythroid growth and differentiation observed following knockdown of HSPA9 in primary human hematopoietic cells using multiple shRNAs. shRNA knockdown of Hspa9 in murine bone marrow cells, followed by transplantation into wild-type recipient mice, recapitulated the erythroid phenotype observed in vitro. Both in vitro and in vivo models implicate HSPA9 as a gene that contributes to normal erythropoiesis, and suggest that HSPA9 haploinsufficiency may alter the hematopoietic progenitor pool by activating apoptosis and altering the cell cycle. Unlike Rps14 (a 5q33.1 gene), the progenitor phenotype in Hspa9-knockdown mice is not corrected by the loss of p53, implicating a nonoverlapping mechanism for the altered hematopoiesis observed with Hspa9 haploinsufficiency.9 These results suggest that HSPA9 haploinsufficiency may contribute to the features of ineffective erythropoiesis observed in patients with MDS, and support the hypothesis that haploinsufficiency of multiple genes on the 5q interval are likely to be important and cooperate during MDS initiation and AML progression.
HSPA9 (mortalin, GRP75, PBP74, MTHSP75) is a highly conserved HSP70 family member that binds proteins located in the mitochondria, cytoplasm, and centrosome as a chaperone.29 Originally characterized as a mitochondrial heat-shock protein that is part of the translocase of the inner mitochondrial membrane complex, HSPA9 supplies the ATP power to import proteins through the inner mitochondrial membrane.30 Not surprisingly, loss of HSPA9 contributes to mitochondrial dysfunction and abnormal mitochondrial biogenesis in Caenorhabditis elegans.31 In humans, inherited variants in ALAS2, a mitochondrial-localized heme-synthesis enzyme, cause a congenital hematopoietic disorder affecting erythropoiesis.32 Therefore, it is possible that altered mitochondrial import of a heme-synthesis enzyme could contribute to an erythroid phenotype in HSPA9-knockdown cells. Mitochondrial dysfunction can also result from mitochondrial DNA deletions that occur in the Pearson marrow-pancreas syndrome, a bone marrow failure syndrome in children with typical features of MDS, including bone marrow dysplasia and sideroblastic anemia.33 These examples suggest that mitochondrial dysfunction resulting from HSPA9 knockdown may contribute to the erythroid phenotype we observed.
HSPA9 is also located in the cytoplasm, where it can bind p53.26-28 Knockdown of HSPA9 in cell lines causes p53 to redistribute from the cytoplasm to the nucleus, resulting in cell-cycle arrest and apoptosis, both features observed in hematopoietic cells following HSPA9 knockdown. HSPA9 also binds p53 and MPS1 at the centrosome during cell-cycle progression.34,35 MPS1 is a kinase that regulates centrosome duplication, and knockdown of HSPA9 results in a reduction in centrosome reduplication, which is necessary for normal cell division and chromosomal stability.34 These functional alterations induced by HSPA9 knockdown may preferentially affect erythroid cells, because EPO stimulation of primary human progenitor cells normally induces up-regulation of HSPA9.17 If HSPA9 up-regulation is blunted in haploinsufficient cells, then the ability of HSPA9 to chaperone key erythroid regulatory proteins may not occur during differentiation and may lead to ineffective erythropoiesis. This is plausible because HSP70 (the defining HSPA9 family member) can bind and protect GATA-1 from caspase-3 cleavage during erythroid differentiation.36,37 Although MDS patients have a reduction in B-cell progenitors,38-41 it is not known whether del(5q) directly contributes to this phenotype. Analysis of recent gene-expression profiling data from CD34+ cells harvested from del(5q) MDS patients identified dysregulation of B-cell–signaling genes, which supports the possibility that haploinsufficiency of del(5q) genes may alter B-cell progenitors.42
A major question in the field remains how do early-stage del(5q) MDS cells gain clonal dominance in the bone marrow and ultimately transform into AML? It is possible that the accelerated apoptosis commonly seen in early MDS may preferentially affect maturing cells and spare del(5q) stem cells, allowing these cells to become clonally dominant after they acquire additional non-del(q5) gene mutations. Our in vitro culture data are consistent with this possibility, because early progenitor cells may be less susceptible to apoptosis than maturing cells following HSPA9 knockdown. The time-dependent reduction in YFP+ peripheral blood cells following Hspa9 knockdown may also be due to the effects of Hspa9 knockdown on differentiating cells in vivo. The association of del(5q) and TP53 mutations in advanced MDS/AML, and the recent finding that the progenitor defect observed in mice with a heterozygous deletion in 8 del(5q33.1) genes was p53 dependent,9 suggests that mutations in p53 may be necessary to overcome the accelerated apoptosis and cell-cycle changes observed in HSPA9-knockdown cells.43-45 Surprisingly, the progenitor defects that we observed following Hspa9 knockdown were p53 independent in mice. However, our findings are consistent with a recent report indicating that up-regulation of the p53 pathway was only observed in CD34+ cells harvested from 5q minus syndrome patients and not in CD34+ cells from non-5q–minus syndrome patients that harbor a deletion of chromosomes 5q.46 Although the loss of p53 may not be the key cooperating mutation with HSPA9 haploinsufficiency that is necessary for progenitors to survive and potentially confer clonal dominance in the murine shRNA model, other unknown non-del(5q) mutations could cooperate with HSPA9 loss.
An alternative hypothesis is that haploinsufficiency of additional 5q31.2 genes (ie, CTNNA1 or EGR1) may cooperate to allow growth of HSPA9-haploinsufficient cells and contribute to the full phenotype of early-stage MDS, including clonal dominance. It is critical to remember that del(5q) always deletes multiple 5q genes, and that haploinsufficiency of HSPA9 alone (due to a mutation or small deletion) has not been found to occur in MDS.6,47 Support for the hypothesis that multiple 5q genes may cooperate comes from modeling haploinsufficiency of 5q33.1 genes that are commonly deleted in the 5q minus syndrome. Recent studies in mice have shown that haploinsufficiency of miR-145 and miR-146a contributes to the thrombocytosis observed in the 5q minus syndrome, while the deletion of 8 genes, including Rps14, contributes to the development of anemia.9,48 Therefore, it is possible that a combination of haploinsufficient 5q31.2 genes, each conferring a unique phenotype, could cooperate to produce the abnormal clonal hematopoiesis observed in MDS and could potentially contribute to AML transformation. These genes (including Hspa9) will require validation using single-gene–knockout models to precisely study haploinsufficiency.
HSPA9, EGR1, and CTNNA1 are del(5q31.2) genes that have been implicated in the development of AML. Hspa9 is a 5q31.2 gene in the Retrovirus Tagged Cancer Gene Database that is a common integration site associated with murine myeloid leukemia. The retroviral integrations in AML cells occur in the inverted orientation relative to the Hspa9 transcriptional direction and Hspa9 mRNA is not overexpressed in these AML samples, suggesting that Hspa9 insertions are inactivating events leading to haploinsufficiency (data not shown). Our group and others have not identified a bi-allelic mutation in HSPA9 in human MDS or AML samples, which is also consistent with haploinsufficiency.4,6,47 Haploinsufficiency of Egr1 cooperates with ethylnitrosourea mutagenesis to induce murine AML, confirming that EGR1 also plays a role during transformation.10 CTNNA1 is an appealing cooperating gene during progression to AML, because methylation of CTNNA1 is increased in high-risk del(5q) MDS/AML compared with low-risk MDS, and methylation is associated with reduced mRNA levels.11,49 Our data and the literature indicate that accurate modeling of MDS may require the cooperation of several del(5q31.2) genes, including HSPA9 haploinsufficiency, as well as additional mutations in non-del(5q) genes.
Our long-term goal is to identify the full complement of mutations located on or off of the del(5q) interval that initiate MDS and lead to the progression from MDS to AML. Discovery of all of the genetic mutations is now possible using next-generation sequencing technologies to decipher cancer genomes.50 As the list of mutant genes in MDS and AML continues to grow, identifying mutations that are functionally important will likely require modeling mutations in organisms, including 5q31.2 gene haploinsufficiency. The challenge moving forward will be to combine haploinsufficiency/loss-of-function mutations and gain-of-function mutations into an accurate preclinical model of MDS that could be used to study the biology of MDS and to test novel therapeutics. As a first step toward this goal, a better understanding of the normal functions of individual del(5q31.2) genes may allow us to specifically target MDS clones early in the disease and prevent AML transformation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Sheila Stewart (and the Children's Discovery Institute at Washington University) and Jeff Millbrandt for providing pLKO.1 and Fcy-si shRNA vectors, respectively; the Siteman Cancer Center High-Speed Cell Sorting Core for outstanding technical assistance (P30 CA091842); the St Louis Cord Blood bank for supplying samples; and James Thompson, Tim Graubert, Dan Link, Mark Sands, and Tim Ley for helpful scientific discussions.
This work was supported by National Institutes of Health Grant HL082973 and the Howard Hughes Medical Institute Physician-Scientist Early Career Award (to M.J.W.).
National Institutes of Health
Authorship
Contribution: T.H.-P.C., A.K., and M.J.W. designed the research; T.H.-P.C., A.K., K.K., M.A.W., G.R., J.F.T., and M.J.W. performed the research; T.H.-P.C., A.K., K.K., M.A.W., G.R., J.F.T., and M.J.W. collected the data; T.H.-P.C., A.K., K.K., M.A.W., G.R., J.F.T., and M.J.W. analyzed the data; and T.H.-P.C. and M.J.W. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Matthew J. Walter, MD, Washington University School of Medicine, Division of Oncology, Section of Stem Cell Biology, Campus Box 8007, 660 South Euclid Ave, St Louis, MO 63110; e-mail: mjwalter@dom.wustl.edu.