Abstract
The E26 transformation-specific (Ets) transcription factor PU.1 is required to generate lymphoid progenitor cells from hematopoietic stem cells, but it is not required to generate B cells from committed B-cell lineage progenitors. We hypothesized that PU.1 function in B-cell differentiation is complemented by the related Ets transcription factor Spi-B. To test this hypothesis, mice were generated lacking both PU.1 and Spi-B in the B-cell lineage. Unlike mice lacking PU.1 or Spi-B, mice deficient in both PU.1 and Spi-B in the B-cell lineage had reduced frequencies of B cells as well as impaired B-cell differentiation. Strikingly, all PU.1 and Spi-B–deficient mice developed pre-B cell acute lymphoblastic leukemia before 30 weeks of age. Pre-B cells accumulated in the thymus resulting in massive thymic enlargement and dyspnea. These findings demonstrate that PU.1 and Spi-B are essential transcriptional regulators of B-cell differentiation as well as novel tumor suppressors in the B-cell lineage.
Introduction
The E26 transformation-specific (Ets) transcription factor PU.1 (encoded by the gene Sfpi1 in mice and SPI1 in humans) is required for generating lymphoid progenitor cells and is a key regulator of B-cell fate specification.1,2 However, conditional deletion of the Sfpi1 gene under control of the B cell–specific Cd19 locus results in minimal perturbation of B-cell development and function.3,4 Spi-B (encoded by Spib) is expressed in developing B cells5 and interacts with DNA binding sites identical to PU.1.6 Several studies suggest that PU.1 and Spi-B functions are complementary. Spi-B can partially replace PU.1 in myeloid and B-cell differentiation.7,8 In addition, Sfpi1+/−Spib−/− mice have severely reduced frequencies of splenic follicular (FO) B cells compared with either Sfpi1+/− or Spib−/− mice.9 We hypothesized that PU.1 is partially redundant after B-cell commitment because of complementation by Spi-B. To test this hypothesis, we generated mice that delete a conditional Sfpi1lox allele under control of the B cell–specific Cd19 locus on a Spib−/− background (CD19+/CreSfpi1lox/loxSpib−/− mice). We report here that CD19+/CreSfpi1lox/loxSpib−/− mice have reduced frequencies of B cells and impaired B-cell differentiation. Strikingly, all CD19+/CreSfpi1lox/loxSpib−/− mice develop pre-B cell acute lymphoblastic leukemia (ALL) with thymic involvement before 30 weeks of age. These findings demonstrate that PU.1 and Spi-B have complementary function as essential transcriptional regulators and novel tumor suppressors in the B-cell lineage.
Methods
Breeding and care of mice
CD19Cre/Cre mice10 were purchased from The Jackson Laboratory (stock no. 006785) and mated to Sfpi1lox/lox mice2 to generate CD19+/CreSfpi1+/lox mice. These were mated to Spib−/− mice11 to generate CD19+/CreSfpi1+/loxSpib+/− mice that were intercrossed to generate CD19+/CreSfpi1lox/loxSpib−/− mice. Mice used in this study were on the C57Bl/6 background and were generated by mating CD19+/CreSfpi1lox/lox males to CD19+/+Sfpi1lox/lox females or by mating CD19+/CreSfpi1lox/loxSpib−/− males to CD19+/+Sfpi1lox/loxSpib−/− females. Mouse care was monitored under an approved animal use subcommittee protocol in accord with the University of Western Ontario Council on Animal Care.
PCR and genotyping
Genotyping of mice was performed using standard PCR. Quantitative PCR (qPCR) was performed using a Rotor-Gene 6000 instrument (Corbett Life Sciences). The relative frequency of intact Sfpi1lox or deleted Sfpi1Δ alleles was normalized to the frequency of β-actin promoter genomic DNA, using the comparative threshold cycle method.12 The relative frequencies of mRNA transcripts were measured by reverse transcription (RT)-qPCR and were normalized to the frequency of glyceraldehyde-3-phosphate dehydrogenase (Gapdh) transcripts. Primers are described in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Histologic and microscopic analysis
Mice were euthanized by lethal intraperitoneal injection of 540 mg/mL sodium pentobarbital (Euthanyl Forte; Bimeda-MTC, Animal Health). Tissues were fixed in 10% neutral buffered formalin. Femurs were decalcified for 96 hours in 26% formic acid (TBD-2; Thermo Fisher Scientific). Tissues were paraffin embedded, sectioned, and stained with hematoxylin and eosin. High-resolution micrographs were captured using a Q-Color3 digital camera (Olympus).
Flow cytometry
Analysis of antibody-stained cells was performed using an FACSCalibur system (BD Biosciences). Sorting of antibody-stained cells was performed using an FACSAria II system. The sorted cells were determined to be of at least 98% purity on reanalysis using the FACSAria II system. Data analysis was performed using FlowJo 9.1 software (TreeStar). Antibodies were purchased from eBioscience and included phycoerythrin (PE)–conjugated anti-CD19 (1D3), allophycocyanin-conjugated anti-B220 (RA3-6B2), PE-Cy5–conjugated anti-IgM (II/41), PE-conjugated anti-CD93 (AA4.1), fluorescein isothiocyanate (FITC)–conjugated anti-CD23 (B3B4), FITC-conjugated anti-CD21 (eBio8D9), FITC-conjugated anti–BP-1 (6C3), biotin-conjugated anti-CD4 (H129.19), allophycocyanin-conjugated anti-CD8a (53-6.7), FITC anti-CD44 (1M7), and PE-Cy5–conjugated anti-CD45.2. (104).
Statistical analysis
All data are reported as mean ± SD of the mean. Statistical significance was determined using a Student t test unless otherwise indicated. P values < .05 were considered significant. For all figures, *P < .05, **P < .01, and ***P < .001. Statistical analysis was performed using Prism 5.0 (GraphPad Software).
Results
CD19+/CreSfpi1lox/loxSpib−/− mice have reduced frequencies and impaired differentiation of B cells
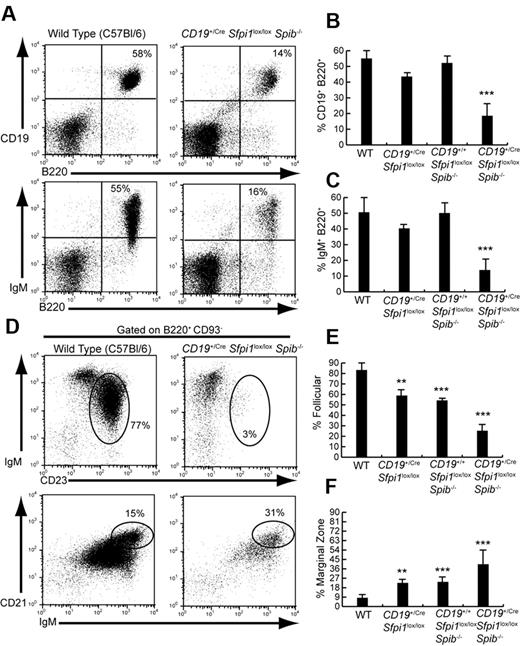
Mice were analyzed lacking PU.1 in the B-cell lineage (CD19+/CreSfpi1lox/lox), Spi-B in the germ line (CD19+/+Sfpi1lox/loxSpib−/−), or Spi-B in the germ line and PU.1 in the B-cell lineage (CD19+/CreSfpi1lox/loxSpib−/−). As reported previously,3,4,11 6- to 10-week-old mice lacking either PU.1 or Spi-B had frequencies of B cells similar to that of C57Bl/6 mice (Figure 1A-C). However, mice lacking both PU.1 and Spi-B in the B-cell lineage had 3-fold reduced frequencies of splenic B cells compared with that of C57Bl/6 mice (Figure 1A-C). Our laboratory recently showed that PU.1 and Spi-B are important for generating splenic FO B cells.9 Therefore, we determined frequencies of IgM+ CD23+ FO and IgMhi CD21hi marginal zone (MZ) B cells in the spleen of CD19+/CreSfpi1lox/loxSpib−/− and control mice. The frequency of splenic IgM+CD23+ FO B cells was reduced more than 3-fold in CD19+/CreSfpi1lox/loxSpib−/− mice compared with C57Bl/6 mice (Figure 1D top panels, E). FO B-cell frequencies were slightly but significantly reduced in mice lacking either PU.1 or Spi-B (Figure 1E). In contrast, the frequencies of splenic IgMhi CD21hi MZ B cells were more than doubled in frequency in CD19+/CreSfpi1lox/loxSpib−/− mice compared with C57Bl/6 mice as well as significantly increased in frequency in mice lacking either PU.1 or Spi-B (Figure 1D bottom panels, F). Therefore, fewer B cells and in particular fewer FO B cells were generated in the absence of PU.1 and Spi-B.
Six- to 10-week-old CD19+/CreSfpi1lox/loxSpib−/− mice have reduced follicular B cell frequencies. (A) Reduced B-cell frequencies in the spleen of CD19+/CreSfpi1lox/loxSpib−/− mice compared with C57Bl/6 mice. Flow cytometric analysis was performed to determine the frequency of B cells expressing CD19 and B220 (top panels) or surface IgM and B220 (bottom panels) in the spleen of mice of the indicated genotypes. (B) Quantitation of the frequency of splenic B cells expressing CD19 and B220 for groups of 5 mice of the indicated genotypes. (C) Quantitation of the frequency of splenic B cells expressing surface IgM and B220 for groups of 5 mice of the indicated genotypes. (D) Reduction of FO B-cell frequency and increase of marginal zone (MZ) B-cell frequency in the spleen of CD19+/CreSfpi1lox/loxSpib−/− mice. Flow cytometric analysis was performed to determine the relative frequency of FO B cells (IgM+CD23+, indicated by ovals in top panels) and MZ B cells (IgMhiCD21hi, indicated by ovals in bottom panels) in the spleen of mice of the indicated genotypes. Results are shown gated on mature B220+CD93− B cells. (E) Quantitation of the frequency of splenic FO B cells (B220+CD93−IgM+CD23+) for groups of 5 mice of the indicated genotypes. (F) Quantitation of the frequency of splenic MZ B cells (B220+CD93−IgMhiCD21hi) for groups of 5 mice of the indicated genotypes.
Six- to 10-week-old CD19+/CreSfpi1lox/loxSpib−/− mice have reduced follicular B cell frequencies. (A) Reduced B-cell frequencies in the spleen of CD19+/CreSfpi1lox/loxSpib−/− mice compared with C57Bl/6 mice. Flow cytometric analysis was performed to determine the frequency of B cells expressing CD19 and B220 (top panels) or surface IgM and B220 (bottom panels) in the spleen of mice of the indicated genotypes. (B) Quantitation of the frequency of splenic B cells expressing CD19 and B220 for groups of 5 mice of the indicated genotypes. (C) Quantitation of the frequency of splenic B cells expressing surface IgM and B220 for groups of 5 mice of the indicated genotypes. (D) Reduction of FO B-cell frequency and increase of marginal zone (MZ) B-cell frequency in the spleen of CD19+/CreSfpi1lox/loxSpib−/− mice. Flow cytometric analysis was performed to determine the relative frequency of FO B cells (IgM+CD23+, indicated by ovals in top panels) and MZ B cells (IgMhiCD21hi, indicated by ovals in bottom panels) in the spleen of mice of the indicated genotypes. Results are shown gated on mature B220+CD93− B cells. (E) Quantitation of the frequency of splenic FO B cells (B220+CD93−IgM+CD23+) for groups of 5 mice of the indicated genotypes. (F) Quantitation of the frequency of splenic MZ B cells (B220+CD93−IgMhiCD21hi) for groups of 5 mice of the indicated genotypes.
It has been reported previously that Tcf3+/− mice have reduced frequencies of FO B cells and increased frequencies of MZ B cells.13 Therefore, the proteins E12 and E47, encoded by Tcf3, might play a role in reduction of FO B-cell frequencies by PU.1 and Spi-B. To test this idea, FO B cells were sorted from the spleen of wild-type, CD19+/+Sfpi1lox/loxSpib−/−, and CD19+/CreSfpi1lox/loxSpib−/− mice. RT-qPCR analysis was performed to measure the relative frequencies of mRNA transcripts encoding E12, E47, Id2, and Id3.14,15 As a control, Fcer2a transcripts encoding CD23 were examined because this gene is activated by PU.1 and Spi-B in FO B cells.9 E12, E47, Id2, Id3, and Fcer2a transcripts were significantly reduced in sorted FO B cells lacking Spi-B, or both PU.1 and Spi-B (supplemental Figure 1A). E12 and E47 transcripts also were reduced in sorted FO B cells from Sfpi1+/−Spib−/− (PUB) mice9 (supplemental Figure 1B). Therefore, FO B-cell frequencies might be reduced in CD19+/CreSfpi1lox/loxSpib−/− mice at least in part as a consequence of reduced levels of E12 and E47.
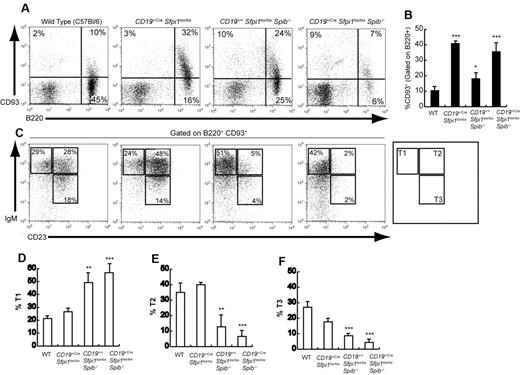
Next, the frequencies of immature transitional B cells expressing B220 and CD93 (AA4.1), a marker of immaturity,16 were examined in CD19+/CreSfpi1lox/loxSpib−/− and control mice. Splenic CD93+ transitional B-cell frequencies were increased ∼ 4-fold by deficiency of PU.1 in B cells, ∼ 2-fold by deficiency of Spi-B, and ∼ 4-fold by deficiency of both PU.1 and Spi-B (Figure 2A-B). Despite the overall increase in transitional B-cell frequencies, the frequencies of IgM+CD23+ transitional-2 or IgMlowCD23+ transitional-3 cells were reduced > 5-fold in the absence of both PU.1 and Spi-B (Figure 2C-F). Therefore, splenic transitional B-cell differentiation, as measured by down-regulation of CD93 and up-regulation of CD23, is impaired in the absence of PU.1 and Spi-B.
Impaired transitional B-cell differentiation in mice deficient in PU.1 and/or Spi-B. (A) Increased splenic transitional B-cell frequencies in mice deficient in PU.1 or Spi-B. Flow cytometric analysis was performed to determine the frequency of transitional B cells expressing both B220 (x-axis) and CD93 (y-axis) in the spleen of 6- to 10-week-old mice of the indicated genotypes (see 4 panels). (B) Quantitation of results shown in Panel A for groups of 5 mice of each indicated genotype. The frequency of CD93+ cells is shown gated on B220+ cells. (C) Altered differentiation of splenic transitional B cells in mice deficient in PU.1 or Spi-B. Flow cytometric analysis was performed to determine the frequency of transitional-1 (IgM+CD23−), transitional-2 (IgM+CD23+), and transitional-3 (IgMlowCD23+) B cells (indicated by boxes and legend in right panel). Results shown were gated on B cells expressing both B220 and CD93. (D-F) Quantitation of results shown in panel C for transitional-1, transitional-2, or transitional-3 B cells for groups of 5 mice of the indicated genotypes.
Impaired transitional B-cell differentiation in mice deficient in PU.1 and/or Spi-B. (A) Increased splenic transitional B-cell frequencies in mice deficient in PU.1 or Spi-B. Flow cytometric analysis was performed to determine the frequency of transitional B cells expressing both B220 (x-axis) and CD93 (y-axis) in the spleen of 6- to 10-week-old mice of the indicated genotypes (see 4 panels). (B) Quantitation of results shown in Panel A for groups of 5 mice of each indicated genotype. The frequency of CD93+ cells is shown gated on B220+ cells. (C) Altered differentiation of splenic transitional B cells in mice deficient in PU.1 or Spi-B. Flow cytometric analysis was performed to determine the frequency of transitional-1 (IgM+CD23−), transitional-2 (IgM+CD23+), and transitional-3 (IgMlowCD23+) B cells (indicated by boxes and legend in right panel). Results shown were gated on B cells expressing both B220 and CD93. (D-F) Quantitation of results shown in panel C for transitional-1, transitional-2, or transitional-3 B cells for groups of 5 mice of the indicated genotypes.
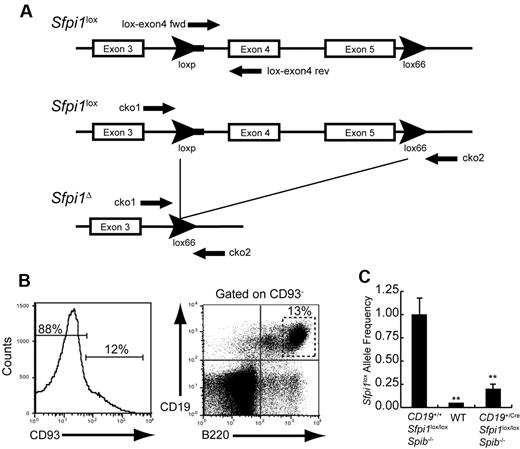
A qPCR assay was designed to determine frequencies of intact (Sfpi1lox) or deleted (Sfpi1Δ) alleles in B cells from CD19+/CreSfpi1lox/loxSpib−/− and control mice (Figure 3A). CD93−B220+ CD19+ B cells were sorted from the spleen of wild-type, CD19+/+Sfpi1lox/loxSpib−/−, or CD19+/CreSfpi1lox/loxSpib−/− mice and used to prepare genomic DNA for qPCR analysis (Figure 3B). The frequency of intact Sfpi1lox alleles was ∼ 20% in B cells from 8-week-old CD19+/CreSfpi1lox/loxSpib−/− mice compared with CD19+/+Sfpi1lox/loxSpib−/− mice, indicating that the Sfpi1lox allele deletion frequency was ∼ 80% (Figure 3C). Thus, Sfpi1lox deletion under the control of CD19Cre was not complete by 6-10 weeks of age.
Measurement of Sfpi1lox allele deletion frequency. (A) Schematic of the Sfpi1lox allele encoding PU.1. Arrowheads indicate loxP or lox66 sequences. The thick line extending from the 5′ loxP sequence indicates unique DNA sequence derived from the targeting vector. Arrows indicate locations recognized by PCR primers. Names of primers are indicated. DNA sequence of the Sfpi1lox allele was determined by cloning and sequencing using primers listed in supplemental Table 1. (B) Enrichment of mature B cells by cell sorting. CD93−CD19+B220+ B cells were enriched from the spleen of 8-week-old wild-type (WT), CD19+/+Sfpi1lox/loxSpib−/−, or CD19+/CreSfpi1lox/loxSpib−/− mice by flow cytometric cell sorting. Genomic DNA was prepared from sorted cells and used as template for qPCR analysis. (C) Measurement of Sfpi1lox allele frequencies using qPCR. Primers lox-exon4 fwd and lox-exon4 rev (supplemental Table 1) were used to determine relative frequencies of intact Sfpi1lox alleles in B cells enriched from mice of the indicated genotypes. The Sfpi1lox allele frequency in CD19+/+Sfpi1lox/loxSpib−/− B cells was set to 1.00 (left bar), and the relative Sfpi1lox allele frequency in CD19+/CreSfpi1lox/loxSpib−/− B cells was expressed as a decimal (right bar). qPCR also was performed on C57Bl/6 B cell genomic DNA (center bar) as an additional negative control to demonstrate specificity of the qPCR reaction.
Measurement of Sfpi1lox allele deletion frequency. (A) Schematic of the Sfpi1lox allele encoding PU.1. Arrowheads indicate loxP or lox66 sequences. The thick line extending from the 5′ loxP sequence indicates unique DNA sequence derived from the targeting vector. Arrows indicate locations recognized by PCR primers. Names of primers are indicated. DNA sequence of the Sfpi1lox allele was determined by cloning and sequencing using primers listed in supplemental Table 1. (B) Enrichment of mature B cells by cell sorting. CD93−CD19+B220+ B cells were enriched from the spleen of 8-week-old wild-type (WT), CD19+/+Sfpi1lox/loxSpib−/−, or CD19+/CreSfpi1lox/loxSpib−/− mice by flow cytometric cell sorting. Genomic DNA was prepared from sorted cells and used as template for qPCR analysis. (C) Measurement of Sfpi1lox allele frequencies using qPCR. Primers lox-exon4 fwd and lox-exon4 rev (supplemental Table 1) were used to determine relative frequencies of intact Sfpi1lox alleles in B cells enriched from mice of the indicated genotypes. The Sfpi1lox allele frequency in CD19+/+Sfpi1lox/loxSpib−/− B cells was set to 1.00 (left bar), and the relative Sfpi1lox allele frequency in CD19+/CreSfpi1lox/loxSpib−/− B cells was expressed as a decimal (right bar). qPCR also was performed on C57Bl/6 B cell genomic DNA (center bar) as an additional negative control to demonstrate specificity of the qPCR reaction.
Expansion of an abnormal pre-B cell population in 11- to 18-week-old CD19+/CreSfpi1lox/loxSpib−/− mice
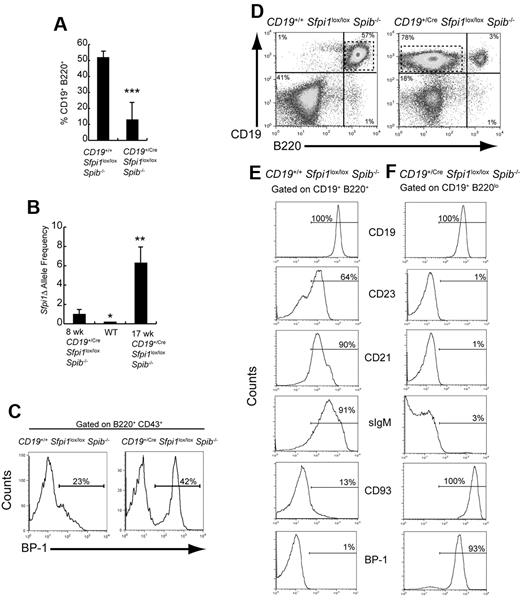
At 11 to 18 weeks of age, the frequency of B220+CD19+ B cells in CD19+/CreSfpi1lox/loxSpib−/− mice was reduced to 13% compared with 18% of splenocytes at 6-10 weeks of age (Figure 4A; compare with Figure 1B). The frequency of deleted (Sfpi1Δ) alleles in sorted splenic B220+CD19+ B cells was increased 6.4-fold in a 17-week-old CD19+/CreSfpi1lox/loxSpib−/− mouse compared with an 8-week-old mouse (Figure 4B), suggesting that the frequency of deletion of Sfpi1lox alleles progressed with age under the control of CD19Cre, as reported previously.3 Next, bone marrow B-cell development was examined. In CD19+/CreSfpi1lox/loxSpib−/− mice, the frequency of bone marrow BP-1+ pre-B cells17 was increased compared with CD19+/+Sfpi1lox/loxSpib−/− mice (Figure 4C). Interestingly, the spleens of 7/9 11- to 18-week-old CD19+/CreSfpi1lox/loxSpib−/− mice contained high frequencies of an abnormal B-cell population that expressed CD19 but little or no B220 (Figure 4D). The lack of B220 expression on abnormal B cells may be the consequence of loss of PU.1-dependent Cd45 transcription.18 Further analysis of these abnormal B cells showed that they did not express CD23, CD21, or surface IgM but that they uniformly expressed high levels of CD93 and BP-1 (Figure 4E-F). A CD19+sIgM−CD93+BP-1+ phenotype is most consistent with that of pre-B cells.16,17 In summary, bone marrow and peripheral B-cell differentiation in CD19+/CreSfpi1lox/loxSpib−/− mice were characterized by an increase in frequency of an abnormal pre-B cell population.
Identification of an abnormal pre-B cell population in 11- to 18-week-old CD19+/CreSfpi1lox/loxSpib−/− mice. (A) B-cell frequencies are severely reduced in 11- to 18-week-old CD19+/CreSfpi1lox/loxSpib−/− mice. Flow cytometric analysis was performed to determine the frequency of splenic B cells expressing CD19 and B220 in the spleen of mice of 11- to 18-week-old mice of the indicated genotypes. Seven mice per group were analyzed. (B) Measurement of deleted (Sfpi1Δ) allele frequencies in B cells from 6- to 10-week-old and 11- to 18-week-old CD19+/CreSfpi1lox/loxSpib−/− mice. Using qPCR, primers cko1 and cko2 (supplemental Table 1) were used to determine relative frequencies of Sfpi1Δ alleles in B cells enriched from mice of the indicated genotype and age. The Sfpi1Δ allele frequency in an 8-week-old CD19+/CreSfpi1lox/loxSpib−/− B cells was set to 1.00 (left bar), and the Sfpi1Δ allele frequency in a 17-week-old CD19+/CreSfpi1lox/loxSpib−/− B cells is expressed as fold increase relative to this value (right bar). qPCR also was performed on C57Bl/6 B cell genomic DNA (center bar) as an additional negative control to demonstrate specificity of the qPCR reaction. (C) Abnormal pre-B cell frequency in bone marrow of a 13-week-old CD19+/CreSfpi1lox/lox Spib−/− mouse. Flow cytometric analysis was used to determine the bone marrow pre-B cell frequencies expressing BP-1 on their cell surface from 13-week-old mice of the indicated genotypes. The histograms shown are gated on B220+CD43+ bone marrow cells. (D) Appearance of an abnormal B cell population expressing low levels of B220. Flow cytometric analysis was performed on splenocytes from 18-week-old mice of the indicated genotypes using antibodies recognizing CD19 or B220. CD19+/CreSfpi1lox/loxSpib−/− mice had a CD19+ B220low population in addition to the CD19+B220+ B-cell population (right panel, dashed box). (E-F) Identification of abnormal CD19+B220low cells as pre-B cells. Flow cytometric analysis using antibodies recognizing the indicated cell surface markers was used to determine the identity of gated CD19+B220+ B cells in CD19+/+Sfpi1lox/loxSpib−/− mice (E) or of gated CD19+B220low B cells in CD19+/CreSfpi1lox/loxSpib−/− mice (F). Gates are shown as dashed boxes in panel A. Results shown are representative of 9 mice analyzed.
Identification of an abnormal pre-B cell population in 11- to 18-week-old CD19+/CreSfpi1lox/loxSpib−/− mice. (A) B-cell frequencies are severely reduced in 11- to 18-week-old CD19+/CreSfpi1lox/loxSpib−/− mice. Flow cytometric analysis was performed to determine the frequency of splenic B cells expressing CD19 and B220 in the spleen of mice of 11- to 18-week-old mice of the indicated genotypes. Seven mice per group were analyzed. (B) Measurement of deleted (Sfpi1Δ) allele frequencies in B cells from 6- to 10-week-old and 11- to 18-week-old CD19+/CreSfpi1lox/loxSpib−/− mice. Using qPCR, primers cko1 and cko2 (supplemental Table 1) were used to determine relative frequencies of Sfpi1Δ alleles in B cells enriched from mice of the indicated genotype and age. The Sfpi1Δ allele frequency in an 8-week-old CD19+/CreSfpi1lox/loxSpib−/− B cells was set to 1.00 (left bar), and the Sfpi1Δ allele frequency in a 17-week-old CD19+/CreSfpi1lox/loxSpib−/− B cells is expressed as fold increase relative to this value (right bar). qPCR also was performed on C57Bl/6 B cell genomic DNA (center bar) as an additional negative control to demonstrate specificity of the qPCR reaction. (C) Abnormal pre-B cell frequency in bone marrow of a 13-week-old CD19+/CreSfpi1lox/lox Spib−/− mouse. Flow cytometric analysis was used to determine the bone marrow pre-B cell frequencies expressing BP-1 on their cell surface from 13-week-old mice of the indicated genotypes. The histograms shown are gated on B220+CD43+ bone marrow cells. (D) Appearance of an abnormal B cell population expressing low levels of B220. Flow cytometric analysis was performed on splenocytes from 18-week-old mice of the indicated genotypes using antibodies recognizing CD19 or B220. CD19+/CreSfpi1lox/loxSpib−/− mice had a CD19+ B220low population in addition to the CD19+B220+ B-cell population (right panel, dashed box). (E-F) Identification of abnormal CD19+B220low cells as pre-B cells. Flow cytometric analysis using antibodies recognizing the indicated cell surface markers was used to determine the identity of gated CD19+B220+ B cells in CD19+/+Sfpi1lox/loxSpib−/− mice (E) or of gated CD19+B220low B cells in CD19+/CreSfpi1lox/loxSpib−/− mice (F). Gates are shown as dashed boxes in panel A. Results shown are representative of 9 mice analyzed.
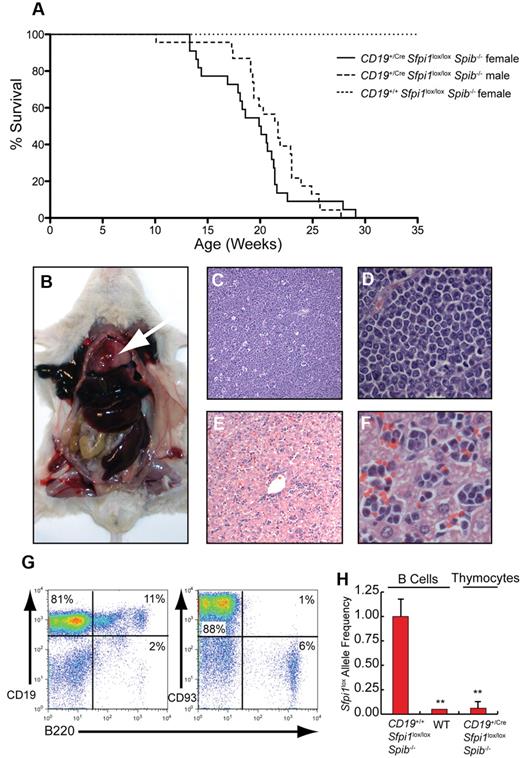
Pre-B ALL with thymic involvement in CD19+/CreSfpi1lox/loxSpib−/− mice
No indications of illness were observed in CD19+/+Sfpi1lox/loxSpib−/− control littermate mice at any age. In contrast, CD19+/CreSfpi1lox/loxSpib−/− mice required euthanasia at a median age of 20 weeks for female mice and 21.7 weeks for male mice because of labored breathing (dyspnea; Figure 5A). The difference between males and females was not statistically significant (P = .0647, using the Mantel-Cox test). Postmortem analysis indicated that dyspnea was caused by an enlarged thymus that filled the chest cavity (Figure 5B). Spleen and liver were consistently enlarged (Figure 5B), whereas lymph nodes were enlarged in one third of mice. To determine the cell type responsible for infiltration and dysfunction of tissues; thymus, spleen, bone marrow, lymph nodes, and blood from moribund CD19+/CreSfpi1lox/loxSpib−/− mice and age-matched CD19+/+Sfpi1lox/loxSpib−/− littermates were examined. Histologic analysis of the thymus revealed that the normal architecture of the organ was no longer recognizable and was replaced with sheets of generally uniform tumor cells (Figure 5C-D). The cells were ∼ 12-15 μm and were loosely arranged. The cells had none to scant lightly basophilic cytoplasm. Nuclei were intermediate in size, with moderate heterochromatin and inconspicuous or occasionally 1 centrally located nucleolus. The rate of mitosis was less than one per high-power field. There was little variation in either cellular or nuclear size. In the thymus, there was a high degree of apoptosis suggested by a “starry sky” appearance caused by invading macrophages (Figure 5C). In the liver there were large numbers of tumor cells expanding sinusoids diffusely with some periportal accumulation (Figure 5E-F). Tumor cells also extensively infiltrated the spleen (supplemental Figure 1A), lymph nodes (supplemental Figure 1B), and bone marrow (supplemental Figure 1C). Lymphocyte counts were increased in the blood of CD19+/CreSfpi1lox/loxSpib−/− mice compared with CD19+/+Sfpi1lox/loxSpib−/− littermates (supplemental Figure 1D).
Pre-B ALL with thymic involvement in CD19+/CreSfpi1lox/loxSpib−/− mice. (A) Kaplan-Meier survival plot for aging mice of the indicated genotype and sex. (B) Enlarged thymus (arrow), enlarged spleen, and enlarged liver are shown in a moribund CD19+/CreSfpi1lox/loxSpib−/− mouse. (C) Histology of the thymus in a moribund CD19+/CreSfpi1lox/loxSpib−/− mouse (original magnification ×4). (D) Enlargement of thymic section shown in panel C, original magnification ×60. (E) Histology of the liver in moribund CD19+/CreSfpi1lox/loxSpib−/− mice. (F) Enlargement of liver section shown in panel E to demonstrate the presence of lymphocytes in sinusoids, original magnification ×60. (G) High frequency of immature B cells in the thymus of a moribund CD19+/CreSfpi1lox/loxSpib−/− mouse. Flow cytometric analysis was performed to determine the frequency of cells expressing the indicated cell surface markers in the thymus of a moribund CD19+/CreSfpi1lox/loxSpib−/− mouse. (H) Measurement of Sfpi1lox allele frequencies in thymic pre-B cells. qPCR was performed to determine the relative frequency of intact Sfpi1lox alleles in pre-B cells from the thymus of a moribund CD19+/CreSfpi1lox/loxSpib−/− mouse (right bar). As a control, the Sfpi1lox allele frequency in splenic B cells enriched from an 8-week-old CD19+/+Sfpi1lox/loxSpib−/− mice was determined and set to 1.00 (left bar). qPCR also was performed on C57Bl/6 B cell genomic DNA (center bar) as an additional negative control to demonstrate specificity of the qPCR reaction.
Pre-B ALL with thymic involvement in CD19+/CreSfpi1lox/loxSpib−/− mice. (A) Kaplan-Meier survival plot for aging mice of the indicated genotype and sex. (B) Enlarged thymus (arrow), enlarged spleen, and enlarged liver are shown in a moribund CD19+/CreSfpi1lox/loxSpib−/− mouse. (C) Histology of the thymus in a moribund CD19+/CreSfpi1lox/loxSpib−/− mouse (original magnification ×4). (D) Enlargement of thymic section shown in panel C, original magnification ×60. (E) Histology of the liver in moribund CD19+/CreSfpi1lox/loxSpib−/− mice. (F) Enlargement of liver section shown in panel E to demonstrate the presence of lymphocytes in sinusoids, original magnification ×60. (G) High frequency of immature B cells in the thymus of a moribund CD19+/CreSfpi1lox/loxSpib−/− mouse. Flow cytometric analysis was performed to determine the frequency of cells expressing the indicated cell surface markers in the thymus of a moribund CD19+/CreSfpi1lox/loxSpib−/− mouse. (H) Measurement of Sfpi1lox allele frequencies in thymic pre-B cells. qPCR was performed to determine the relative frequency of intact Sfpi1lox alleles in pre-B cells from the thymus of a moribund CD19+/CreSfpi1lox/loxSpib−/− mouse (right bar). As a control, the Sfpi1lox allele frequency in splenic B cells enriched from an 8-week-old CD19+/+Sfpi1lox/loxSpib−/− mice was determined and set to 1.00 (left bar). qPCR also was performed on C57Bl/6 B cell genomic DNA (center bar) as an additional negative control to demonstrate specificity of the qPCR reaction.
Flow cytometric analysis showed that 80% to 90% of cells in the thymus, spleen, and lymph nodes of moribund CD19+/CreSfpi1lox/loxSpib−/− mice were CD19+ and expressed the pre-B cell markers BP-1 and CD93 (Figure 5G; supplemental Figure 2). The thymus contained only 5% to 8% T cells expressing CD4 or CD8 and few double-positive cells (supplemental Figure 2A). Pre-B cells in the thymus expressed high levels of the hyaluronic acid receptor CD44, a molecule with an established role in tumor metastasis.19 Thymic pre-B cells expressed reduced levels of total CD45 on their surface (supplemental Figure 2B), consistent with the lack of B220 expression (Figure 5G) and a requirement for PU.1 in activating Cd45 (encoding B220) in B cells.18 Finally, the frequency of intact Sfpi1lox alleles was measured in pre-B cells from the thymus of a 24-week-old CD19+/CreSfpi1lox/loxSpib−/− mouse and determined to be < 1%, indicating near complete deletion in tumor cells (Figure 5H). Together, these results suggest that the disease in moribund CD19+/CreSfpi1lox/loxSpib−/− mice is pre-B ALL.
Transcripts encoding BLNK/SLP-65 are reduced in CD19+/CreSfpi1lox/loxSpib−/− pre-B ALL cells
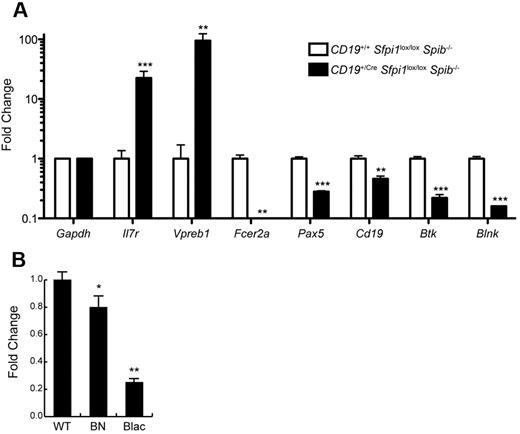
B-cell linker protein ([BLNK] also named SLP-65) is an intracellular adaptor protein that is expressed in B cells and is essential for transduction of signals from the B-cell receptor.20 BLNK is considered a tumor suppressor gene for pre-B ALL.21 Our laboratory previously noted that Blnk transcript levels were reduced in cultured pro-B cell lines lacking PU.1 and Spi-B.22 To determine whether Blnk transcript levels were reduced in pre-B ALL cells, splenic B220lowCD19+ cells were enriched by cell sorting from CD19+/CreSfpi1lox/loxSpib−/− mice. As a control, B220+CD19+ B cells were enriched from the spleen of CD19+/+Sfpi1lox/loxSpib−/− littermates (sorting scheme as shown in Figure 4D). RNA was prepared from sorted cells and used as template for RT-qPCR analysis. As shown in Figure 6A, genes encoding the α subunit of the IL-7 receptor (IL-7Rα, encoded by Il7r) and VpreB (Vpreb1), markers of pre-B cell differentiation,23 were highly expressed in CD19+/CreSfpi1lox/loxSpib−/− leukemia cells compared with controls. In contrast, Fcer2a transcripts (encoding CD23) were nearly undetectable, consistent with flow cytometry results in Figure 4F and a requirement for PU.1 and/or Spi-B in Fcer2a transcription.9 Cd19 transcripts were reduced in CD19+/CreSfpi1lox/loxSpib−/− pre-B ALL cells compared with controls, as expected, because these cells lack one functional Cd19 allele. Finally, transcripts encoding the transcription factor Pax-5 (Pax5), Bruton's tyrosine kinase (Btk), and BLNK (Blnk) were examined. Pax-5 and BLNK are known tumor suppressor genes in human pre-B ALL,21,24 whereas Btk can cooperate with BLNK as a tumor suppressor gene in mouse models.25 Pax-5, Btk, and BLNK were reduced 2-fold, 4-fold, and 6-fold, respectively, in CD19+/CreSfpi1lox/loxSpib−/− pre-B ALL cells compared with controls. To obtain additional evidence that Blnk might be activated by PU.1, we determined relative Blnk transcript levels in cultured pro-B cells that express distinct PU.1 concentrations as a consequence of engineered hypomorphic mutant alleles.26 Compared with wild-type pro-B cells, Sfpi1BN/BN pro-B cells express ∼ 20% of normal PU.1 levels and expressed significantly reduced Blnk transcript levels, whereas Sfpi1Blac/Blac pro-B cells express ∼ 2% of normal PU.1 levels and expressed 4-fold reduced Blnk transcript levels (Figure 6B). Thus, Blnk transcript levels respond to PU.1 in a dose-dependent manner. Together, these results suggest that Blnk transcription is activated by PU.1.
Reduced PU.1 levels induce reduced expression of transcripts encoding BLNK. (A) Changes in frequencies of mRNA transcripts in CD19+/CreSfpi1lox/loxSpib−/− pre-B ALL cells. Cell sorting was used to enrich B220low CD19+ pre-B ALL cells from the spleen of 17-week-old CD19+/CreSfpi1lox/loxSpib−/− mice, and as a control, B220+ CD19+ B cells from the spleen of 17-week-old CD19+/+Sfpi1lox/loxSpib−/− mice (see Figure 4D for illustration). RNA was prepared from each sample, and RT-qPCR was used to measure relative frequencies of steady-state mRNA transcript levels for genes indicated in the x-axis. Transcript frequencies were normalized to Gapdh transcript levels. The y-axis represents fold change in transcripts in CD19+/CreSfpi1lox/loxSpib−/− pre-B ALL cells compared with CD19+/CreSfpi1lox/loxSpib−/− B cells. (B) Dose-dependent reduction of Blnk transcript levels in response to reduced PU.1 levels. RNA was prepared from cultured pro-B cells generated from the fetal wild-type (WT), Sfpi1BN/BN (BN), or Sfpi1Blac/Blac (Blac) mice. RT-qPCR was performed as described above to determine relative frequencies of Blnk transcripts.
Reduced PU.1 levels induce reduced expression of transcripts encoding BLNK. (A) Changes in frequencies of mRNA transcripts in CD19+/CreSfpi1lox/loxSpib−/− pre-B ALL cells. Cell sorting was used to enrich B220low CD19+ pre-B ALL cells from the spleen of 17-week-old CD19+/CreSfpi1lox/loxSpib−/− mice, and as a control, B220+ CD19+ B cells from the spleen of 17-week-old CD19+/+Sfpi1lox/loxSpib−/− mice (see Figure 4D for illustration). RNA was prepared from each sample, and RT-qPCR was used to measure relative frequencies of steady-state mRNA transcript levels for genes indicated in the x-axis. Transcript frequencies were normalized to Gapdh transcript levels. The y-axis represents fold change in transcripts in CD19+/CreSfpi1lox/loxSpib−/− pre-B ALL cells compared with CD19+/CreSfpi1lox/loxSpib−/− B cells. (B) Dose-dependent reduction of Blnk transcript levels in response to reduced PU.1 levels. RNA was prepared from cultured pro-B cells generated from the fetal wild-type (WT), Sfpi1BN/BN (BN), or Sfpi1Blac/Blac (Blac) mice. RT-qPCR was performed as described above to determine relative frequencies of Blnk transcripts.
Discussion
ALL is the most common form of cancer in young children, and despite an 80% cure rate, represents a leading cause of leukemia-related deaths in children and adults.27 Pre-B ALL is frequently associated with mutations affecting genes that encode transcriptional regulators.28 Reduced PU.1 expression has been associated with acute myelogenous leukemia in human patients,29,30 and is sufficient to induce acute myelogenous leukemia in mouse models.31,32 There are several reports of reduced PU.1 levels in lymphoid leukemia or lymphoma.33,34 Our studies establish that reduced expression of PU.1 and Spi-B are sufficient to induce pre-B ALL in mice, suggesting that PU.1 and Spi-B function as tumor suppressor genes in the B-cell lineage. Our studies also establish that PU.1 and Spi-B have complementary function as transcription factors essential for B-cell differentiation.
PU.1 is required for generating lymphoid progenitor cells and is a key regulator of B-cell fate specification.1,2 PU.1 has been reported as regulating many important target genes in the B-cell lineage. Therefore, it was surprising when two groups reported that conditional deletion of the Sfpi1 gene under control of the B-cell–specific Cd19 locus resulted in minimal perturbation of B-cell development and function.3,4 Our studies establish that PU.1 is partially redundant after commitment to the B-cell lineage because of complementation by the related Ets transcription factor Spi-B. Spi-B is expressed throughout B-cell development5 and has DNA binding specificity indistinguishable from that of PU.1.6 Therefore, we would expect that many important PU.1 target genes in the B-cell lineage also would be recognized by Spi-B. It will be important to determine the identities of genes that can be activated by both PU.1 and Spi-B. In addition, there may be further redundancies with the less characterized but highly related Ets transcription factor Spi-C.9
The results presented in this study establish that PU.1 and Spi-B have critical and complementary functions in B-cell differentiation after acquisition of CD19 expression. Frequencies of B cells and especially FO B-cell frequencies declined with age in CD19+/CreSfpi1lox/loxSpib−/− mice. However, low frequencies of B cells expressing both CD19 and B220 persisted in CD19+/CreSfpi1lox/loxSpib−/− mice up to 18 weeks of age (Figure 4D right panel). Because deletion of the Sfpi1lox allele under control of CD19Cre is progressive and incomplete (Figures 3C and 4B), it is possible that these cells persist because they still express significant amounts of PU.1 protein. It is not yet known what target genes are most responsible for promotion of B cell differentiation by PU.1 and Spi-B. Interestingly, E12 and E47 mRNA transcript levels were significantly reduced in sorted FO B cells from either CD19+/+Sfpi1lox/loxSpib−/− or CD19+/CreSfpi1lox/loxSpib−/− mice (supplemental Figure 1). E12 and E47 proteins are critically important for B-cell differentiation,35 and mice with reduced levels of these proteins (Tcf3+/− mice) have impaired FO B-cell differentiation.13 Therefore, PU.1 and Spi-B may directly or indirectly reinforce E12 and E47 expression during B-cell differentiation.
The B-cell linker protein BLNK/SLP-65 is considered a tumor suppressor protein because homozygous null mutation of Blnk is sufficient to induce pre-B cell ALL in mice36 and mutation or aberrant mRNA splicing of Blnk is associated with human pre-B ALL.21,24,36 Interestingly, the incidence of disease is only ∼ 5% in Blnk−/− mice. However, the incidence of pre-B ALL is increased to nearly 80% in mice homozygous for germ line null alleles encoding both BLNK and Btk (Blnk−/−Btk−/− mice).25 In the absence of BLNK and Btk, signals from the pre-B cell receptor are uncoupled from IL-7R–induced proliferation, resulting in a failure of pre-B cells to exit the cell cycle.25,37 We noted previously that Sfpi1−/−Spib−/− pro-B cells cultured on stromal layers with IL-7 proliferate more rapidly than wild-type pro-B cells, differentiate into pre-B–like cells, and express reduced levels of Blnk transcripts.22 Therefore, pre-B ALL in CD19+/CreSfpi1lox/loxSpib−/− mice might be caused in part by reduced Blnk expression in response to reduced PU.1 and Spi-B.
There are several differences between pre-B ALL that develops in Blnk−/− or Blnk−/−Btk−/− mice and what we report in CD19+/CreSfpi1lox/loxSpib−/− mice. First, disease occurs in 100% of CD19+/CreSfpi1lox/loxSpib−/− mice, whereas it occurs in < 80% of Blnk−/−Btk−/− mice by 5 months of age.25 Second, disease in CD19+/CreSfpi1lox/loxSpib−/− mice is characterized by massive thymic enlargement, which was not reported in Blnk−/−Btk−/− mice. The molecular basis of migration of pre-B ALL cells into the thymus in CD19+/CreSfpi1lox/loxSpib−/− mice is not known. However, we note that cultured pro-B cells expressing reduced PU.1 levels ectopically express a number of T cell–specific genes.38 Therefore, it is possible that pre-B cells lacking PU.1 and Spi-B express genes that promote homing to the thymus. More work will be necessary to identify the exact genes and proteins involved.
In summary, our work establishes PU.1 and Spi-B as complementary tumor suppressors in developing B cells. The mouse model that we describe should be useful for identifying pathways of leukemogenesis that can be targeted at the molecular level by novel therapies.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank H. Leighton Grimes (Cincinnati Children's Hospital) for providing Sfpi1+/lox mice, Li X. Xu and Ali Abbas for genotyping of mice, and Kristin Chadwick of the London Regional Flow Cytometry facility for cell sorting. They also thank Ewa Cairns (University of Western Ontario) and Anargyros Xenocostas (London Regional Cancer Center) for critically reading the manuscript.
This work was supported by Canadian Institutes of Health Research grants 103028 and 106581 (R.P.D.), Natural Sciences and Engineering Research Council of Canada grant 386046 (R.P.D.), and a Curtis Cadman memorial studentship award (M.R.G.).
National Institutes of Health
Authorship
Contribution: K.M.S., S.K.H.L, and R.P.D. designed, performed, and analyzed experiments; M.R.G. performed experiments; I.W. and H.-A.T.C.-P. performed analysis and interpretation; and R.P.D. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rodney P. DeKoter, Department of Microbiology and Immunology, Schulich School of Medicine & Dentistry, The University of Western Ontario, London, ON, Canada N6A 5C1; e-mail: rdekoter@schulich.uwo.ca.