Abstract
Natural killer (NK) cells are classically viewed as effector cells that kill virus-infected and neoplastic cells, but recent studies have identified a rare mucosal NK- cell subpopulation secreting the TH17 cytokine IL-22. Here, we report identification of 2 distinct lineages of mucosal NK cells characterized as NKG2A+NFIL3+RORC– and NKp44+NFIL3+RORC+. NKG2A+ NK cells were systemically distributed, cytotoxic, and secreted IFN-γ, whereas NKp44+ NK cells were mucosae-restricted, noncytotoxic, and produced IL-22 and IL-17. During SIV infection, NKp44+ NK cells became apoptotic, were depleted, and had an altered functional profile characterized by decreased IL-17 secretion; increased IFN-γ secretion; and, surprisingly, increased potential for cytotoxicity. NKp44+ NK cells showed no evidence of direct SIV infection; rather, depletion and altered function were associated with SIV-induced up-regulation of inflammatory mediators in the gut, including indoleamine 2,3-dioxygenase 1. Furthermore, treatment of NKp44+ NK cells with indoleamine 2,3-dioxygenase 1 catabolites in vitro ablated IL-17 production in a dose-dependent manner, whereas other NK-cell functions were unaffected. Thus lentiviral infection both depletes and modifies the functional repertoire of mucosal NK cells involved in the maintenance of gut integrity, a finding that highlights the plasticity of this rare mucosal NK-cell population.
Introduction
Natural killer (NK) cells are typically characterized as the primary effector cells of the innate immune system because they are able to lyse virus-infected or tumor cells without prior antigen sensitization. NK cells have evolved multiple mechanisms for the lysis of virus-infected or tumorigenic cells, involving interactions among signals mediated by activating NK receptors, inhibitory NK receptors, and cytokines.1 Activating NK receptors include the natural cytotoxicity receptors NKp30, NKp44, NKp46, and NKp80 as well as a subset of killer immunoglobulin-like receptors (KIRs) and NKG2D. Inhibitory NK receptors include the CD94/NKG2A heterodimers and multiple KIR family members. Through the integration of signals delivered by the activating and inhibitory receptors, NK cells are able to maintain a balance between tolerance to self and cytotoxicity against diseased or allogeneic cells.
Although NK cells are best characterized for their cytotoxicity, mounting evidence suggests they are far more heterogeneous than previously appreciated and that they also can regulate antiviral and inflammatory responses. Several different subsets of NK cells with diverse functions have been identified in humans.2 The dominant subset in peripheral blood is CD16+CD56dim, and it primarily mediates cytotoxic activity but secretes relatively little IFN-γ. In contrast, a distinct subset of CD16−/lowCD56hi NK cells displays little cytotoxic activity but secretes relatively large amounts of IFN-γ, TNF-α, and MIP-1β. Analysis of NK cells in humans has been limited by the fact that most experiments have focused on cells from blood, whereas most effector and regulatory functions are likely to occur in tissues where NK cells would encounter neoplastic or virus-infected cells. To this end, we and others have recently reported a systemic but heterogeneous distribution of NK cells throughout the gastrointestinal tract as well as vagina and cervical tissues of humans and nonhuman primates.3-5 Tissue NK- cell subsets, like their peripheral blood counterparts, are functional, lyse the classic NK target cell line K562, express granzymes and perforin, and secrete of IFN-γ and TNF-α. However, recently a lymphocyte population expressing NK markers but also secreting IL-22 and IL-17, cytokines generally associated with TH17 cells,6 was identified in human tonsillar tissue and murine lamina propria.3,7,8 These cells, termed NK-22 cells, also share similar transcriptional profiles with TH17 cells and express high levels of the transcription factors RAR-related orphan γ receptor (RORγt), aryl hydrocarbon receptor (AHR), and interferon regulatory factor 4 (IRF4).3,7,8 Furthermore, NK-22 cells are ontogenically distinct from classic NK cells but share a common lineage with lymphoid tissue-inducing cells (LTi cells).9
NK cells are critical for defense against many viral infections and also have been linked to control of HIV infection.10 NK cells have been shown to inhibit HIV replication ex vivo through secretion of β-chemokines, cytolysis of infected cells via NKG2D ligand-mediated killing, and antibody dependent cell-mediated cytotoxicity via virus-specific antibodies.11-16 Acute HIV infection is associated with expansion of CD56dimCD16+ NK cells in humans, suggesting a virus-driven increase in cytolytic activity,17 and long-term nonprogressors have increased NK- cell cytotoxicity compared with viremic individuals.18 A genetic linkage study has demonstrated that KIR3DS1 and its putative ligand HLA-B Bw4-80I are associated with slower disease progression,19 and the interaction of the 2 molecules leads to inhibition of HIV-1 replication in vitro.20 Several studies suggest that NK cells also help mediate control of simian immunodeficiency virus (SIV) infection. Acute infection with SIVmac251 induced activation of macaque NK cells, defined in this study as CD3−CD16+, which was reflected by up-regulation of CD69 expression and increased lysis of K562 cells.21 NK cells have been shown to lyse SIV-infected cells or cells pulsed with SIV.16 In pig-tailed macaques infected with SIV, an inverse association between the magnitude of the NK response and the probability of development of neuroAIDS was observed.16 Longitudinal studies of NK responses after SIV infection of sooty mangabeys and rhesus macaques (Macaca mulatta) demonstrated early onset NK responses in sooty mangabeys (Cercocebus atys) as well as in a subset of SIV-infected rhesus macaques that subsequently controlled SIV infection, suggesting that early NK responses were associated with a lack of disease progression in both species.22 A reduced frequency of NK cells and decreased NK activity in peripheral blood have been associated with poor control of SIV infection, and a subset of macaque KIR3DL alleles was associated with a higher SIV viral load.23
Efforts to better understand the specific roles that NK cells play during lentivirus infections have been hindered by a lack of data on NK cell–virus interactions in the mucosae, where HIV transmission and replication primarily take place.24 In light of the localization of NK-22 cells in the mucosae and their proposed role in maintaining gut integrity, elucidation of the interplay between HIV/SIV and NK-22 cells is of considerable interest but has not yet been explored. Here, we characterize anatomically, phenotypically, and functionally the macaque counterparts to human NK-22 cells and address the effects of chronic SIV infection on each of these parameters.
Methods
Animals and SIV infections
In total, 50 Indian rhesus macaques were sampled for this study: 29 SIV-naive and 21 infected chronically with SIV; 13 were infected intravenously with SIVmac239 and 8 were infected with SIVmac251, either intravaginally (n = 2) or intrarectally (n = 6). All animals were free of simian retrovirus type D and simian T-lymphotrophic virus type 1 and were housed at the New England Primate Research Center and maintained in accordance with the guidelines of the Committee on Animals of the Harvard Medical School and the Guide for the Care and Use of Laboratory Animals.
Cell collection and processing
Processing of blood and tissue samples was carried out using assays optimized in our laboratory as described previously.5,25 In brief, total peripheral blood mononuclear cells were isolated from EDTA-treated venous blood by density gradient centrifugation over lymphocyte separation media (MP Biomedicals), and a hypotonic ammonium chloride solution was used to lyse contaminating red blood cells. For biopsy collection, macaques were first anesthetized with ketamine HCl (10-20 mg/kg i.m.) or tiletamine/zolazepam (Telazol, 5 mg/kg i.m.), and 10-15 rectal biopsies were collected using 1.9-mm fenestrated endoscopic biopsy forceps (Olympus). Mononuclear cell isolation from biopsy pieces was performed using mechanical and enzymatic disruption as described previously.25
Polychromatic flow cytometry
Flow cytometry staining of mononuclear cells was carried out for cell surface and intracellular molecules (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) using standard protocols.5 LIVE/DEAD Aqua dye (Invitrogen) and isotype-matched controls and/or fluorescence-minus-one controls26 were included in all assays. Acquisitions were made on an LSR II flow cytometer (BD Biosciences) and analyzed using FlowJo Version 8.86 software (TreeStar). Multiparametric analyses were performed using Pestle (Version 1.6.2) and SPICE (Version 5.1).27
In vitro cultures with 3-HAA
Mononuclear cells were isolated from mesenteric lymph node of 3 normal rhesus macaques. Cells (2 × 106) were cultured for 24 hours at 37°C in 5% CO2 in RPMI 1640 containing 10% FBS (media) or with exogenous 3-hydroxyanthranilic acid (3-HAA; Sigma-Aldrich) at final concentrations of 0.01, 0.1, and 1.0μM. Cultures with DMSO alone (vehicle in which 3-HAA was dissolved) were used as additional controls. After 24 hours, cultures were stimulated with phorbol 12-myristate 13-acetate and ionomycin, and intracellular cytokine staining (ICS) analysis was performed.
Plasma viral load quantification
Total RNA copy number equivalents were determined in EDTA-treated plasma using a standardized quantitative real-time RT-PCR assay based on amplification of conserved gag sequences as described previously.28
Cell sorting
Live CD45+CD3+CD4+, CD45+CD3–NKG2A+NKp44–, and CD45+CD3–NKG2A–NKp44+ cell subsets were sorted from bulk mucosal mononuclear cells using an FACSAria cell sorter (BD Biosciences). Sorts were routinely > 99% pure for all populations, and cell yields generally ranged between 103 and 105 cells.
Statistical analyses
All statistical and graphical analyses were done using Prism Version 5.0 software (GraphPad Software). Nonparametric Wilcoxon matched pairs, Mann-Whitney, and Spearman correlation tests were used where indicated, and P < .05 were assumed to be significant.
Results
Mucosal tissues contain 2 distinct lineages of NK cells
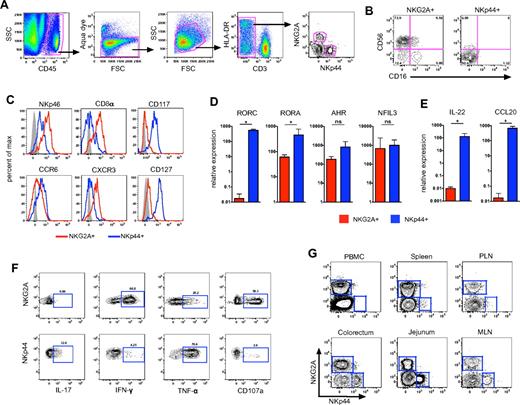
To identify mucosal NK-cell subpopulations, we first analyzed lymphocytes isolated from colorectal biopsies of normal rhesus macaques using polychromatic flow cytometry. We first gated on CD45+ leukocytes to exclude contaminating epithelial cells and then excluded dead cells using a vital stain. Among live CD45+CD3– colorectal mononuclear cells, we found 2 mutually exclusive populations of cells expressing the NK cell-related markers NKG2A and NKp44, respectively (Figure 1A). As we have reported previously,5 most NKG2A+ cells in the gut-associated lymphoid tissue expressed the characteristic NK molecule CD56, with a subset of cells expressing the FcγRIII receptor CD16 (Figure 1B). In contrast, NKp44+ NK cells expressed very little CD56 and were negative for CD16. Furthermore, NKp46 and CD8α, 2 molecules often used to delineate NK cells in macaques,5,29 were expressed at high levels on NKG2A+ NK cells but were dimly expressed on NKp44+ NK cells (Figure 1C). We also found greater levels of the chemokine receptor CCR6 on NKp44+ NK cells compared with NKG2A+ NK cells, whereas CXCR3 was expressed at higher levels on NKG2A+ NK cells, similar to published reports for human NK cells.3
Gut-associated lymphoid tissues contain 2 distinct lineages of NK cells. (A) Representative gating strategy to identify NKG2A+ and NKp44+ NK cells among live mononuclear cells in tissues in rectal mucosa specimens. (B) Flow cytometry plots demonstrating disparate expression of CD56 and CD16 on NKG2A+ and NKp44+ NK cells. (C) Representative histogram overlays depicting expression of NKp46, CD8α, CD117, CD127, CCR6, and CXCR3 on the 2 NK-cell subpopulations. Isotype-matched controls are shown in gray. RT-PCR analysis was used to quantify mRNA transcripts of transcription factors (D) and regulatory cytokines IL-22 and CCL20 (E) in NKG2A+ and NKp44+ NK cells sorted from rectal mucosa of normal macaques. Mann-Whitney U test (*P < .05). (F) Flow cytometry plots depicting CD107a and intracellular cytokine responses in the 2 NK-cell subpopulations after mitogen stimulation. Polychromatic flow cytometry and ICS experiments are representative of 8-16 animals per group; molecular analyses show geometric means ± SEM of 4 animals per group. (G) Flow cytometry plots comparing distribution of NKG2A+ and NKp44+ NK cells in blood and various tissues of rhesus macaques using the gating strategy shown in panel A; data are representative of > 12 animals. MLN indicates mesenteric lymph node; and PLN, peripheral lymph node.
Gut-associated lymphoid tissues contain 2 distinct lineages of NK cells. (A) Representative gating strategy to identify NKG2A+ and NKp44+ NK cells among live mononuclear cells in tissues in rectal mucosa specimens. (B) Flow cytometry plots demonstrating disparate expression of CD56 and CD16 on NKG2A+ and NKp44+ NK cells. (C) Representative histogram overlays depicting expression of NKp46, CD8α, CD117, CD127, CCR6, and CXCR3 on the 2 NK-cell subpopulations. Isotype-matched controls are shown in gray. RT-PCR analysis was used to quantify mRNA transcripts of transcription factors (D) and regulatory cytokines IL-22 and CCL20 (E) in NKG2A+ and NKp44+ NK cells sorted from rectal mucosa of normal macaques. Mann-Whitney U test (*P < .05). (F) Flow cytometry plots depicting CD107a and intracellular cytokine responses in the 2 NK-cell subpopulations after mitogen stimulation. Polychromatic flow cytometry and ICS experiments are representative of 8-16 animals per group; molecular analyses show geometric means ± SEM of 4 animals per group. (G) Flow cytometry plots comparing distribution of NKG2A+ and NKp44+ NK cells in blood and various tissues of rhesus macaques using the gating strategy shown in panel A; data are representative of > 12 animals. MLN indicates mesenteric lymph node; and PLN, peripheral lymph node.
In humans and mice, NKp44+ NK cells express high levels of CD117 (c-kit) as well as the IL-7 receptor CD127,3,7,9 and we observed a similar expression pattern on macaque NKp44+ NK cells (Figure 1C). In contrast, little to no expression of CD117 and CD127 was found on NKG2A+ NK cells. Because both molecules are generally associated with less differentiated lymphocyte populations,3,4,7,9 this disparate expression pattern suggested NKG2A+ cells are more differentiated than NKp44+ NK cells.
To further confirm the identities of macaque mucosal NK-cell subpopulations, we quantified mRNA transcripts of selected transcription factors in sorted NKp44+ and NKG2A+ NK cells. As reported for humans, NKp44+ NK cells expressed high levels of the transcription factors RORγt (RORC), RORα (RORA), and AHR (Figure 1D), which are characteristically expressed in TH17 cells.6 In contrast, NKG2A+ NK cells had low expression of each of these factors, most notably a virtual absence of RORγt. However, NKp44+ and NKG2A+ NK cells expressed similar levels of the transcription factor NFIL3 (also known as E4BP4), which is required for NK-cell development,30 but previously not described in mucosal NKp44+ NK cells. Thus, based on transcriptional profiling, NKp44+ NK cells share features of both TH17 and classic NK cells.
In both humans and mice, NKp44+ NK cells have been termed NK-22 cells based on their ability to secrete IL-22, a feature not shared with classic NK cells.3 Unfortunately, no human IL-22–specific antibodies tested were found to be cross-reactive in rhesus macaques (data not shown). However, in sorted NKp44+ NK cells, we found high levels of IL-22 mRNA expressed constitutively with virtually no expression in NKG2A+ NK cells (Figure 1E). Consistent with human and mouse data, we also observed a similar pattern of constitutive expression of the CCR6 ligand CCL20 in NKp44+ NK cells but not NKG2A+ NK cells. These data suggested NKp44+ NK cells are poised to secrete both IL-22 and CCL20 and may home to mucosal sites via the CCL20-CCR6 axis. Next, we further evaluated the functionality of NKG2A+ and NKp44+ NK cells in a 4-function ICS assay adapted from previous assays optimized in our laboratory.5 After mitogen stimulation, NKp44+ NK cells secreted the regulatory cytokines IL-17 and TNF-α but little IFN-γ, and they expressed only low levels of the degranulation marker CD107a (Figure 1F). In strong contrast, on stimulation NKG2A+ NK cells secreted no IL-17, significant amounts of IFN-γ, and up-regulated CD107a. However, unlike classic NKG2A+ NK cells, NKp44+ NK cells from both naive and SIV-infected macaques were nonresponsive to the MHC-devoid cell line 721.221 (supplemental Figure 1). In addition, unlike NKG2A+ NK cells, NKp44+ did not express any KIR2D molecules as evidenced by a lack of binding by pan-KIR2D antibodies (Miltenyi Biotec; data not shown), which could suggest they do not express KIRs in general. The functional profiles of mucosal NKp44+ and NKG2A+ NK cells are therefore inherently distinct, with the former being regulatory and proinflammatory and the latter probably being directed toward virus inhibition and cell killing.
Previous studies of mucosal NKp44+ NK cells in humans have been restricted to tonsillar tissues (where they were first described), ileal Peyer patches, and lamina propria lymphocytes.3,31 Because of the restricted access to human mucosal tissues, this issue can be addressed far more comprehensively using macaque models. As we have reported previously,5 NKG2A+ NK cells were found in virtually all tissues examined (Figure 1G). In contrast, NKp44+ NK cells were absent from peripheral blood and spleen but were abundant in colorectal tissue, jejunum, as well as mesenteric, but not peripheral lymph nodes (Figure 1G). We also identified NKp44+ NK cells in cervical and vaginal tissue (data not shown). Overall, these data suggest that although classic NKG2A+ NK cells are systemically distributed, NKp44+ NK cells are compartmentalized to mucosal tissues.
Numeric loss and altered phenotype of mucosal NKp44+ NK cells during chronic SIV infection
Because little is known about the effects of chronic lentivirus infection on mucosal NKG2A+ NK cells, and nothing is currently known about the effects on NKp44+ NK cells, we quantified NK cells in colorectal biopsies and found that frequencies of both subpopulations were reduced in SIV-infected compared with naive macaques (Figure 2A). However, because standard flow cytometric analyses address relative abundance and can be confounded by influx or efflux of other cell types, we next determined absolute numbers of cells using a bead-based flow cytometric assay. A modest reduction of NKG2A+ NK cells was observed in SIV-infected macaques compared with naive controls (medians, 91 and 150 cells/mg of tissue, respectively), but NKp44+ NK cells were reduced ∼ 9-fold (medians, 18 cells/mg and 161 cells/mg, respectively; Figure 2B). Reductions in both NK-cell populations tended to be negatively associated with plasma viral loads (supplemental Figure 2), and the loss of NKp44+ NK cells also strongly correlated (Rs = 0.877; P < .0001) with the loss of total gut CD4+ T cells (data not shown). However, the loss of NKp44+ NK cells in the gut was greater than the ∼ 5-fold loss of total CD4+ T cells in SIV-infected macaques, which was characterized by specific reductions in both CCR5+CD4+ memory T cells and CD4+ TH17 cells (supplemental Figure 3A-B). By comparison, no significant change in numbers of CD8+ T cells was observed in SIV-infected macaques.
Chronic SIV infection induces numeric and phenotypic alterations in gut-associated NK cells. (A) Frequencies of NKp44+ and NKG2A+ NK cells among live CD45+CD3− mononuclear cells in rectal biopsies of naive and SIV-infected macaques. Horizontal lines indicate medians of 16-18 animals per group. (B) Absolute numbers of NKp44+, NKG2A+, and CD4+ and CD8+ T cells per milligram of rectal biopsy tissue in naive and chronically SIV-infected macaques were enumerated using a bead-normalized flow cytometry assay. (C) Expression of intracellular Ki67 and caspase-3 or cell surface annexin V binding was determined on NKp44+ and NKG2A+ NK cells. (D) Median fluorescence intensities (MFI) of cell surface molecules on NKG2A+ and NKp44+ NK cells in naive and SIV-infected macaques. Bars represent means ± SEM of 8-14 animals per group. Asterisks above columns reflect naïve versus SIV statistical comparisons, whereas those below the axes reflect NKG2A+ versus NKp44+ NK cells statistical comparisons in normal animals. Mann-Whitney U tests were used for naïve versus SIV comparisons and Wilcoxon matched pairs tests were used to compare NKG2A+ and NKp44+ NK cells (*P < .05, **P < .01, ***P < .001).
Chronic SIV infection induces numeric and phenotypic alterations in gut-associated NK cells. (A) Frequencies of NKp44+ and NKG2A+ NK cells among live CD45+CD3− mononuclear cells in rectal biopsies of naive and SIV-infected macaques. Horizontal lines indicate medians of 16-18 animals per group. (B) Absolute numbers of NKp44+, NKG2A+, and CD4+ and CD8+ T cells per milligram of rectal biopsy tissue in naive and chronically SIV-infected macaques were enumerated using a bead-normalized flow cytometry assay. (C) Expression of intracellular Ki67 and caspase-3 or cell surface annexin V binding was determined on NKp44+ and NKG2A+ NK cells. (D) Median fluorescence intensities (MFI) of cell surface molecules on NKG2A+ and NKp44+ NK cells in naive and SIV-infected macaques. Bars represent means ± SEM of 8-14 animals per group. Asterisks above columns reflect naïve versus SIV statistical comparisons, whereas those below the axes reflect NKG2A+ versus NKp44+ NK cells statistical comparisons in normal animals. Mann-Whitney U tests were used for naïve versus SIV comparisons and Wilcoxon matched pairs tests were used to compare NKG2A+ and NKp44+ NK cells (*P < .05, **P < .01, ***P < .001).
To address the mechanism of loss of NK cells in the gut, we analyzed expression of the proliferation marker Ki67 and the apoptotic molecules caspase-3 and annexin-V. In naive macaques, NKG2A+ NK cells expressed significantly greater levels of Ki67, caspase-3, and annexin-V than NKp44+ NK cells, suggesting that even under normal conditions, classic NKG2A+ NK cells have higher turnover rates, whereas NKp44+ NK cells are relatively quiescent (Figure 2C; supplemental Figure 4). In SIV-infected macaques, Ki67 expression increased in NKG2A+ NK cells, whereas rates of apoptosis were relatively unaffected. However, the opposite was true for NKp44+ NK cells: apoptosis increased without any change in Ki67 expression.
Although both NK-cell populations were significantly reduced during SIV infection, there was no change in phenotypic markers such as NKp46, CD8α, and CCR6 (Figure 2D). There was also no significant change among the 4 NKG2A+ NK-cell subsets based on CD16 and CD56 expression (data not shown). However, CD117 and CD127 were significantly down-regulated on NKp44+ NK cells (Figure 2D), suggesting that the remaining cells might be more differentiated. A similar loss of CD117 and CD127 expression has been associated with ex vivo activation and differentiation of these cells in human studies.9
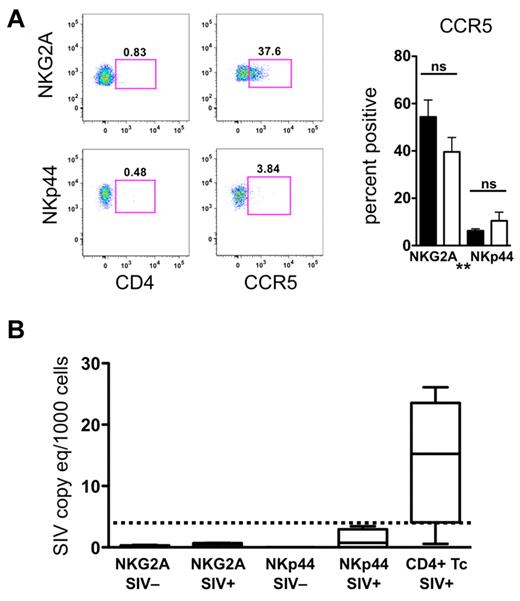
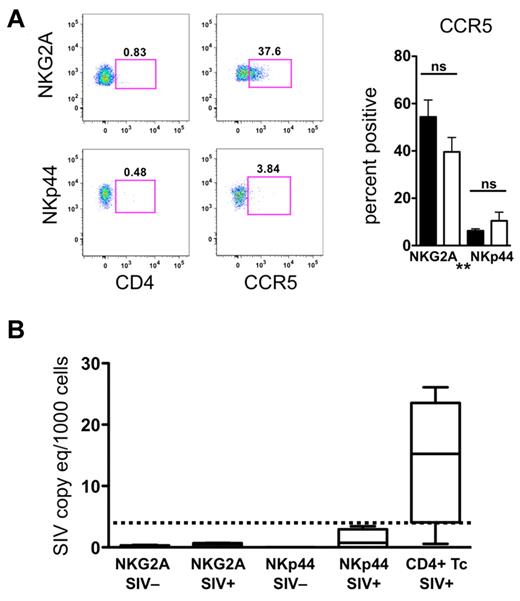
Because of the numeric loss and increased apoptosis of NKp44+ NK cells, we questioned whether NKG2A+ or NKp44+ NK cells might be targets of direct SIV infection and replication. However, the fact that neither of these cell subpopulations expressed CD4, and only NKG2a+ NK cells expressed detectable levels of CCR5, suggested these cells were unlikely to become infected in vivo (Figure 3A). Furthermore, we were unable to detect SIV RNA transcripts in any NKG2A+ or NKp44+ NK-cell samples (Figure 3B). These data indicated that the loss of NKp44+ cells in the gut of chronically SIV-infected animals is unlikely to be because of direct infection but may involve altered differentiation or increased apoptosis.
Mucosal NK cells are not infected by SIV in vivo. (A) Cell surface expression of CD4 and CCR5 on mucosal NKG2A+ and NKp44+ NK cells. Bars represent means ± SEM of 8 animals per group. Mann-Whitney U tests were used for naïve versus SIV comparisons and Wilcoxon matched pairs tests were used to compare NKG2A+ and NKp44+ NK cells (*P < .05, **P < .01, ***P < .001). (B) RT-PCR analysis was used to quantify SIV RNA in NKG2A+ and NKp44+ NK cells and CD4+ T cells sorted from rectal mucosa of normal and SIV-infected macaques. The lower limit of detection of the assay was 3 SIV RNA copies/1000 cells and is indicated by the dashed bar.
Mucosal NK cells are not infected by SIV in vivo. (A) Cell surface expression of CD4 and CCR5 on mucosal NKG2A+ and NKp44+ NK cells. Bars represent means ± SEM of 8 animals per group. Mann-Whitney U tests were used for naïve versus SIV comparisons and Wilcoxon matched pairs tests were used to compare NKG2A+ and NKp44+ NK cells (*P < .05, **P < .01, ***P < .001). (B) RT-PCR analysis was used to quantify SIV RNA in NKG2A+ and NKp44+ NK cells and CD4+ T cells sorted from rectal mucosa of normal and SIV-infected macaques. The lower limit of detection of the assay was 3 SIV RNA copies/1000 cells and is indicated by the dashed bar.
Altered function of mucosal NK cells during SIV infection
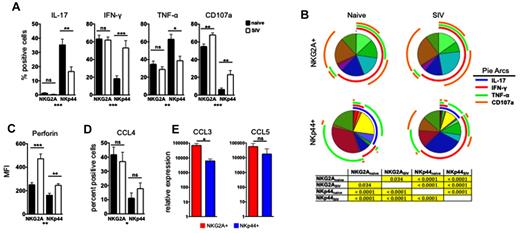
Because chronic SIV infection induced both numeric and phenotypic changes in mucosal NK cells, we next addressed whether these cells exhibited any functional alterations. On stimulation, NKp44+ NK cells from SIV-infected animals had significantly reduced production of both IL-17 and TNF-α, coupled with increased production of IFN-γ and increased expression of CD107a, suggesting that although these cells had impaired regulatory cytokine production, antiviral and cytolytic functions increased (Figure 4A). Furthermore, multiparametric analysis revealed that although all NKp44+ IL-17+ multifunctional cells were reduced in frequency, IL-17+ monofunctional cells were ablated (Figure 4B). Multiparametric analysis also showed that the NKp44+ CD107a+ degranulating cells were primarily monofunctional or were bifunctional IFN-γ–producing cells. These data suggest that there is significant functional heterogeneity within the NKp44+ NK-cell population and that these functionally distinct groups may be affected differently during SIV infection. In contrast, the functional profiles of NKG2A+ NK cells were only modestly modulated during SIV infection, with only CD107a expression increasing, as we reported previously for peripheral blood NK cells during chronic SIV infection.5 As expected in naive animals, NKG2A+ NK cells expressed higher intracellular levels of the cytolytic molecule perforin than NKp44+ NK cells, but consistent with increased CD107a expression, perforin was significantly up-regulated in both NK subpopulations in SIV-infected macaques (Figure 4C). Although performing classic cytotoxicity assays with the limited cell numbers was not possible, increased perforin expression has been shown to correlate with increased killing activity in several experimental models,32,33 suggesting that the NKp44+ NK-cell subset, which primarily secretes regulatory cytokines in naive animals, may acquire cytotoxic functions during chronic SIV infection.
Chronic SIV infection alters functional profiles of gut NK cells. (A) Colorectal mononuclear cells were stimulated with phorbol 12-myristate 13-acetate/ionomycin for 12 hours and then IL-17, IFN-γ, and TNF-α production and CD107a expression were measured on NKG2A+ and NKp44+ NK cells in naive and SIV-infected macaques. The monofunctional profiles of each subpopulation were determined by expressing each response as a proportion of the total cell population. The means ± SEM for 12 animals per group are shown. (B) Multiparametric analyses on the data shown in Figure 6A were performed with SPICE 5.0 software. Pies indicate means of 12 animals per group. Tables show results of 1-sided permutation tests comparing each of the pies as calculated by SPICE; P < .05 are considered significant and are highlighted in yellow. (C) Intracellular perforin expression was determined in NK cells ex vivo; bars represent means ± SEM for 13-16 animals per group. (D) Expression of CCL4 (MIP-1β) was determined by ICS. (E) Quantitative RT-PCR analysis of constitutively expressed CCL3 (MIP-1α) and CCL5 (RANTES) in NKG2A+ and NKp44+ NK cells sorted from rectal biopsies of normal macaques. Mann-Whitney U tests were used for naïve versus SIV comparisons, and Wilcoxon matched pairs tests were used to compare NKG2A+ and NKp44+ NK cells (*P < .05, **P < .01, ***P < .001. MFI indicates median fluorescence intensity.
Chronic SIV infection alters functional profiles of gut NK cells. (A) Colorectal mononuclear cells were stimulated with phorbol 12-myristate 13-acetate/ionomycin for 12 hours and then IL-17, IFN-γ, and TNF-α production and CD107a expression were measured on NKG2A+ and NKp44+ NK cells in naive and SIV-infected macaques. The monofunctional profiles of each subpopulation were determined by expressing each response as a proportion of the total cell population. The means ± SEM for 12 animals per group are shown. (B) Multiparametric analyses on the data shown in Figure 6A were performed with SPICE 5.0 software. Pies indicate means of 12 animals per group. Tables show results of 1-sided permutation tests comparing each of the pies as calculated by SPICE; P < .05 are considered significant and are highlighted in yellow. (C) Intracellular perforin expression was determined in NK cells ex vivo; bars represent means ± SEM for 13-16 animals per group. (D) Expression of CCL4 (MIP-1β) was determined by ICS. (E) Quantitative RT-PCR analysis of constitutively expressed CCL3 (MIP-1α) and CCL5 (RANTES) in NKG2A+ and NKp44+ NK cells sorted from rectal biopsies of normal macaques. Mann-Whitney U tests were used for naïve versus SIV comparisons, and Wilcoxon matched pairs tests were used to compare NKG2A+ and NKp44+ NK cells (*P < .05, **P < .01, ***P < .001. MFI indicates median fluorescence intensity.
Mucosal NK cells are a potent source of β-chemokines
As we have shown previously,5 classic NKG2A+ NK cells from peripheral blood can produce CCL4 (MIP-1β), a β-chemokine that can potently inhibit HIV/SIV infection of target cells. Because HIV/SIV primarily replicate in the gut, we next investigated whether either of the NK-cell subpopulations in the mucosa could secrete β-chemokines, thereby potentially inhibiting SIV replication directly in situ. Although both NKG2A+ and NKp44 NK cells produced CCL4, production was significantly higher in NKG2A+ NK cells (Figure 4D). We next measured the levels of CCL3 (MIP-1α) and CCL5 (RANTES) mRNA in sorted NKG2A+ and NKp44+ NK cells after mitogen stimulation. NKG2A+ NK cells had ∼ 1 log greater CCL3 mRNA compared with NKp44+ NK cells and also expressed higher levels of CCL5 (Figure 4E). β-chemokine production by these cells was not altered by SIV infection. However, because of the numeric decrease of both NK subpopulations in SIV-infected macaques, SIV infection is likely to induce a net loss of NK-associated β-chemokine production in the gut.
Chronic SIV infection perturbs the cytokine milieu of the gut microenvironment
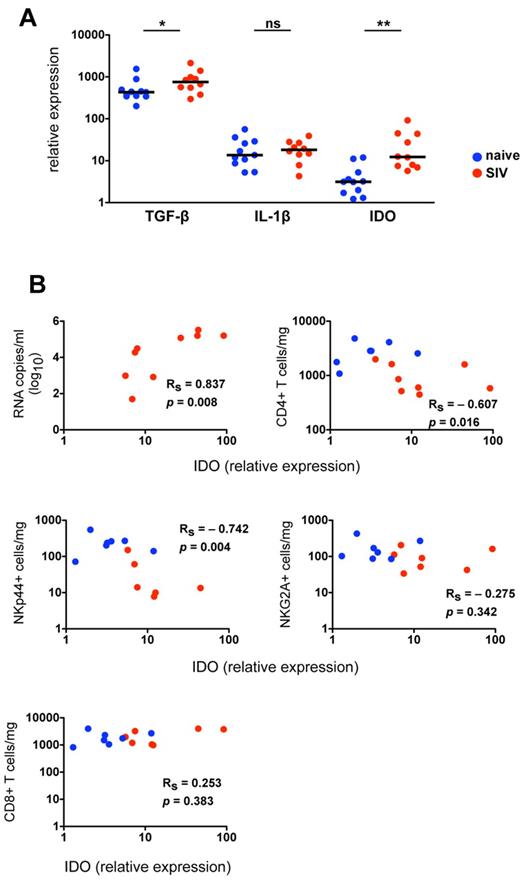
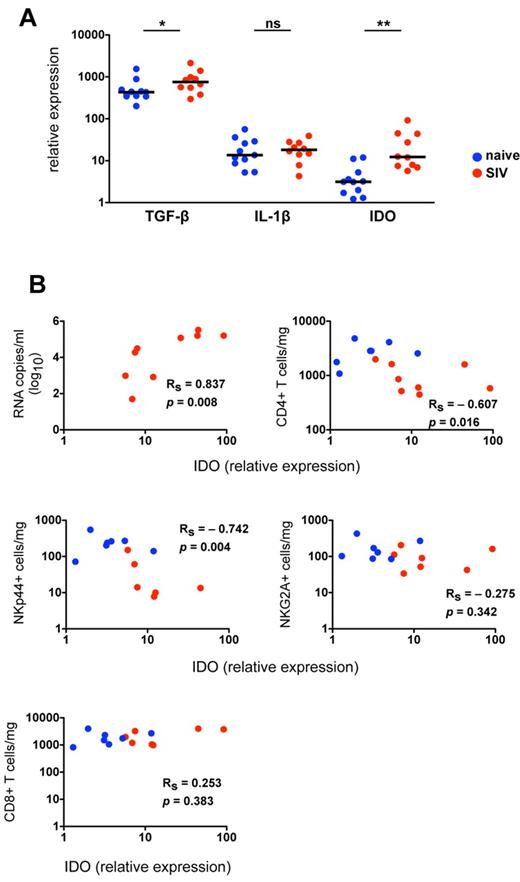
Because chronic lentivirus infections induce substantial inflammatory and immunologic changes in the gut mucosae, characterized by increased inflammation, increased cell death, and a breakdown of the epithelial barrier,34 we hypothesized that cytokine signals necessary for NK-cell development and function might be altered in SIV-infected macaques. To address this hypothesis, we analyzed expression of mediators of NK-cell differentiation in colorectal biopsies from normal and SIV-infected animals. Because several cytokines known to affect differentiation of TH17 cells also have been shown to modulate NKp44+ differentiation or function,31,35,36 we also examined expression of mediators of TH17 development. Based on quantitative RT-PCR analysis of mRNA transcripts in whole biopsies, we found no significant differences between normal and SIV-infected macaques in expression of IL-1β, IL-23, IL-12a, or IL-6, all of which regulate either function or differentiation of NKG2A+ and NKp44+ NK cells in humans (Figure 5A; supplemental Figure 5).3,31,37,38 IFN-α, which activates classic NK cells and can induce apoptosis, was elevated in a subgroup of infected animals but did not reach statistical significance. By comparison, TGF-β, a major mediator of inflammation and TH17 development,6,35,36 was detected at high levels in colorectal biopsies of naive macaques, but it was significantly elevated in SIV-infected macaques (Figure 5A). Most notably, indoleamine 2,3-dioxygenase 1 (IDO1) expression in gut biopsies of SIV-infected macaques was increased 6-fold over normal controls and correlated positively with plasma viral loads (Figure 5B). These data are consistent with a recent study of HIV-infected humans that found increased IDO1 in the rectal mucosa was associated with a loss of CD4+TH17 cells in the gut and subsequent immune activation.39
IDO1 is up-regulated in the gut mucosa of SIV-infected macaques and is associated with a loss of NKp44+ NK cells. (A) Quantitative RT-PCR analysis of cytokine transcripts was performed on rectal biopsies collected from naive and SIV-infected macaques. Mann-Whitney U tests were used for naïve versus SIV comparisons (*P < .05, **P < .01, ***P < .001). (B) Relative expression of IDO1 in rectosigmoid biopsies was correlated with plasma viral loads (top left) and absolute numbers of NKp44+ and NKG2A+ NK and CD4+ and CD8+ T cells quantified in biopsy specimens of the same animals as shown in Figure 4B. P values < .05 are considered significant.
IDO1 is up-regulated in the gut mucosa of SIV-infected macaques and is associated with a loss of NKp44+ NK cells. (A) Quantitative RT-PCR analysis of cytokine transcripts was performed on rectal biopsies collected from naive and SIV-infected macaques. Mann-Whitney U tests were used for naïve versus SIV comparisons (*P < .05, **P < .01, ***P < .001). (B) Relative expression of IDO1 in rectosigmoid biopsies was correlated with plasma viral loads (top left) and absolute numbers of NKp44+ and NKG2A+ NK and CD4+ and CD8+ T cells quantified in biopsy specimens of the same animals as shown in Figure 4B. P values < .05 are considered significant.
Loss and functional perturbation of NKp44+ NK cells is related to changes in the gut microenvironment
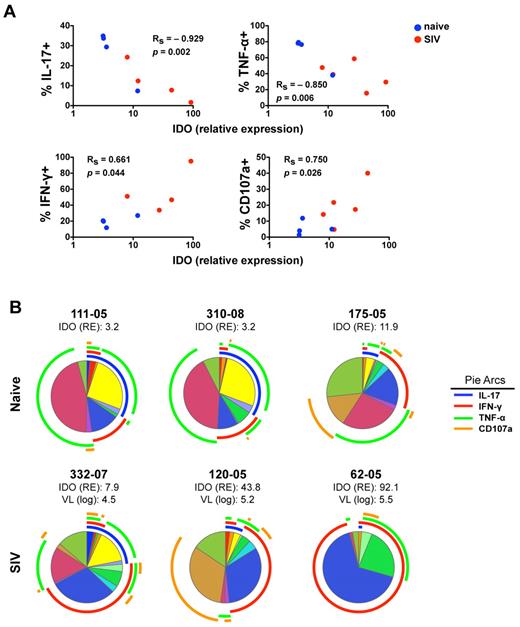
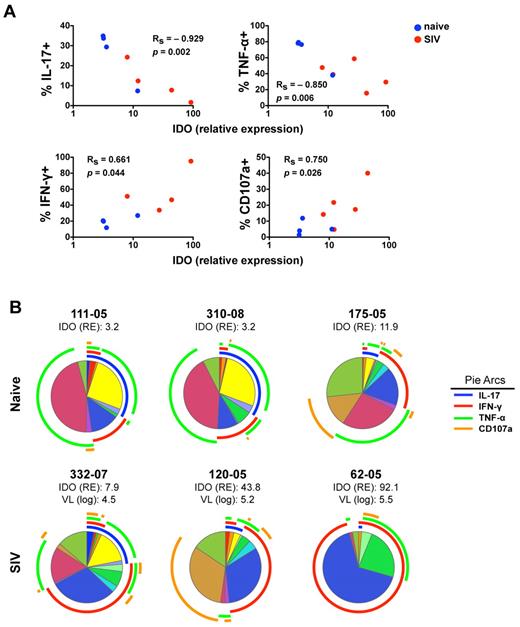
Because IDO1 expression in the gut has been linked to suppression of TH17 cells, which share many features with NKp44+ NK cells, we next investigated the impact of SIV-induced increases in IDO1 on NKp44+ NK cells. Interestingly, increasing expression of IDO1 in the gut showed a strong negative correlation with absolute numbers of CD4+ T cells and NKp44+ NK cells but not with NKG2A+ NK or CD8+ T cells (Figure 5B). This suggested IDO1 expression could be a negative regulator for maintenance, development, or both of NKp44+ but not classic NKG2A+ NK cells. Furthermore, increased IDO1 expression was negatively associated with IL-17 and TNF-α production but positively associated with secretion of IFN-γ and CD107a expression (Figure 6A). These data suggested that IDO1 may be involved in the SIV-induced shift in the NKp44+ cell repertoire away from regulatory function to antiviral and cytolytic activities. Although the increased IDO1 expression was SIV-induced, we reasoned that if IDO1 is a regulator of NKp44+ NK-cell function, the effect would be observed independently of SIV infection status. Indeed, in animals 111-05 and 310-08, which had very low expression of IDO1 in the gut, NKp44+ NK cells secreted copious amounts of IL-17 and TNF-α, but IFN-γ production and CD107a expression on stimulation were virtually absent (Figure 6B). Comparatively, macaque 175-05, which had the highest IDO1 expression in our naive cohort, had low secretion of IL-17 and TNF-α, but IFN-γ secretion and CD107a expression were more similar to that of infected animals. In macaque 332-07, which had the lowest expression of IDO1 in our SIV-infected cohort, NKp44+ NK cells had relatively intact secretion of IL-17 and TNF-α in contrast to another infected macaque, 120-05, which had high IDO1 expression, reduced IL-17 and TNF-α production, and greater secretion of IFN-γ and CD107a expression. Furthermore, in macaque 62-05, which had the highest detectable levels of IDO1 of any animal, IL-17 production was ablated, and the functional response to stimulation was almost completely dominated by IFN-γ production. This suggested that IDO1 not only contributes to regulation of NKp44+ NK-cell function but also does so in a dose-dependent manner not necessarily dependent on SIV infection.
Increased IDO1 expression in the gut mucosa is associated with modulation of IL-17 and IFN-γ secretion by NKp44+ cells. (A) Monofunctional analyses of stimulated NKp44+ NK cells as shown in Figure 6A were correlated with relative expression values of IDO in whole biopsy specimens of the same animals. P values < .05 are considered significant. (B) Multiparametric analyses of NKp44+ NK cells function using SPICE 5.0 for individual naive and SIV-infected animals. Corresponding relative expression (RE) of IDO1 and plasma viral loads (VL) of infected animals are shown.
Increased IDO1 expression in the gut mucosa is associated with modulation of IL-17 and IFN-γ secretion by NKp44+ cells. (A) Monofunctional analyses of stimulated NKp44+ NK cells as shown in Figure 6A were correlated with relative expression values of IDO in whole biopsy specimens of the same animals. P values < .05 are considered significant. (B) Multiparametric analyses of NKp44+ NK cells function using SPICE 5.0 for individual naive and SIV-infected animals. Corresponding relative expression (RE) of IDO1 and plasma viral loads (VL) of infected animals are shown.
Because TGF-β was also increased in the gut during chronic SIV infection, we evaluated whether there was any relationship between levels of TGF-β and NKp44+ NK cells. Although no correlation was found between expression of TGF-β and absolute numbers of NKp44+ NK cells, there were significant negative correlations between levels of TGF-β and both IL-17 and TNF-α secretion by NKp44+ NK cells (supplemental Figure 6). However, TGF-β expression was not significantly related to secretion of IFN-γ or CD107a expression on NKp44+ NK cells. A recent report demonstrated that although TGF-β is a necessary factor for TH17 cell development, overexpression suppresses development and function by antagonizing RORγt.40 Although a similar regulatory mechanism could exist for NKp44+ NK cells in vivo because of their high expression of RORγt, in vitro treatment of NKp44+ NK cells with exogenous TGF-β had no significant effect on function (data not shown).
The IDO1 catabolite 3-HAA specifically suppresses IL-17 production by NKp44+ NK cells
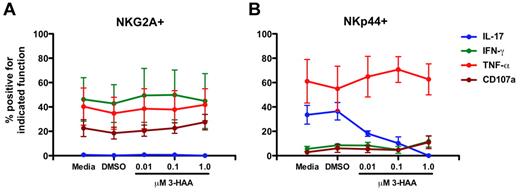
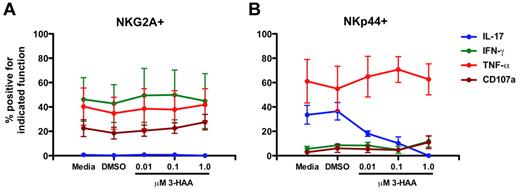
Because increased expression of IDO1 was associated with perturbations of NKp44+ NK-cell function, and IDO1 has been previously associated with suppression of TH17 cells,39 we next evaluated the direct effects of the IDO1 catabolite 3-HAA, which has been shown previously to suppress TH17 development,39 on mucosal NK cells in vitro. Addition of 3-HAA to mucosal samples had no obvious effect on NKG2A+ NK-cell functions (Figure 7A). In contrast, IL-17 production by NKp44+ NK cells was suppressed by addition of 3-HAA in a dose-dependent manner (Figure 7B) similar to the observed effects of 3-HAA on TH17 cells,39 although occurring at lower doses. However, TNF-α production was unaffected and IFN-γ and CD107a expression were only modestly increased at the highest concentration of 3-HAA. These data corroborate the in vivo findings that IDO1 has a significant impact on NKp44+ NK-cell function but also suggest that other factors could be contributing to the perturbed function and phenotype.
3-HAA suppresses IL-17 production by NKp44+ NK cells in vitro in a dose-dependent manner. Mononuclear cells isolated from mesenteric lymph nodes of SIV-naive rhesus macaques were cultured in the presence of increasing concentrations of 3-HAA for 24 hours and then mitogen-stimulated. NKG2A+ (A) and NKp44+ (B) NK cells were analyzed functionally as shown in Figures 1 and 6. Means ± SEM for 3 animals are shown.
3-HAA suppresses IL-17 production by NKp44+ NK cells in vitro in a dose-dependent manner. Mononuclear cells isolated from mesenteric lymph nodes of SIV-naive rhesus macaques were cultured in the presence of increasing concentrations of 3-HAA for 24 hours and then mitogen-stimulated. NKG2A+ (A) and NKp44+ (B) NK cells were analyzed functionally as shown in Figures 1 and 6. Means ± SEM for 3 animals are shown.
Discussion
Here we describe the anatomic compartmentalization and functional characterization of 2 distinct lineages of primate NK cells, distinguishable by their mutually exclusive expression of NKG2A and NKp44. Furthermore, we describe a phenomenon whereby chronic SIV infection induces specific alterations in the cytokine milieu of the gut mucosa, the primary site of virus replication, which subsequently disrupt the functional niche of the novel mucosal NKp44+ NK-cell subpopulation. This perturbation is 2-fold: (1) a massive numeric loss of NKp44+ NK cells and (2) a significant reshaping of the NKp44+ NK-cell functional repertoire characterized by reduced IL-17 secretion coupled with increased IFN-γ secretion and expanded potential for cytotoxicity.
NK-cell diversity and heterogeneity have been significantly underappreciated, and we now delineate phenotypic and transcriptional profiles of 2 highly dichotomous lineages of NKG2A+NFIL3+RORC– and NKp44+NFIL3+RORC+ NK cells in the gut mucosae. The latter probably represents the macaque counterpart to NK-22 cells in humans and mice,3,7 indicating that this unique cell type is evolutionarily conserved among primates and rodents. Human NKp44+ NK cells have been found previously to form an ontogenic lineage with LTi cells independently of classic NK cells,9 suggesting that NKp44+ NK cells are not NK cells in the classic sense but rather are part of a larger group of “NK-like” cells recently termed “innate lymphoid cells.”41 The most convincing evidence in support of this is that the disparate transcriptional profiles of the NKG2A+ and NKp44+ lineages translated into highly bifurcated functional repertoires of IL-17–IL-22–IFNγ++perforin+ and IL-17+IL-22+IFN-γ+/–perforin– cell populations, respectively. This dichotomy suggests that as the ontogenic relationships of these distinct cell types are further clarified, NKp44+ and classic NK cells will be confirmed as 2 independent lineages that fill distinct functional niches in mucosal immunology.
HIV and SIV infection result in massive virus replication in the gut, rapidly inducing the collapse of structural, immune, and cytokine networks. Accordingly, multiple mechanisms probably contribute to the loss of NKp44+ NK cells. Although perturbations of the gut cytokine milieu during lentivirus infections are manifold,42,43 recent research has found the effects of the enzyme IDO1, which metabolizes tryptophan, to have multiple regulatory effects on immune cells of the gut microenvironment.44 In the rectal mucosa of HIV-1–infected subjects, increased IDO1 and accumulation of tryptophan metabolites correlate with loss of TH17 cells,39 and we observed a similar relationship with increased IDO1 in the gut mucosae of SIV-infected macaques and loss of NKp44+ NK cells. Because of the transcriptional similarities between TH17 and NKp44+ NK cells, the mechanism described by Favre et al39 may suppress IL-17 production and deplete NKp44+ NK cells: increased IDO1 causes tryptophan deprivation and an increase in toxic metabolites that suppress RORγt expression.39,40,45,46 Indeed, increases in IDO1 were not only associated with numeric loss of NKp44+ NK cells but also decreased IL-17 production (Figures 5 and 6). Furthermore, in our in vitro experiments we found that addition of the IDO1 catabolite 3-HAA specifically suppressed IL-17 production by NKp44+ NK cells but left other functions intact. The source(s) of increased IDO1 is unclear but is often derived from activated macrophages, myeloid dendritic cells, and plasmacytoid dendritic cells,39,47,48 the latter of which we have found at increased frequencies in the colorectum of SIV-infected animals (data not shown). Gut TGF-β also increased in SIV-infected macaques, and although required for TH17 cell differentiation,36 at high concentrations TGF-β inhibits TH17 development by antagonizing RORγt,40 and could similarly suppress NKp44+ NK-cell development. If these factors resulted in a generalized suppression of RORγt, NKp44+ NK cells would be expected to be more mature, a hypothesis supported by down-regulation of CD127 and CD117 on NKp44+ NK cells in infected animals. Finally, the observation that NKp44+ NK cells acquired potential for cytotoxicity in SIV-infected macaques, as evidenced by increased intracellular perforin and CD107a expression on stimulation, is particularly notable. In classic NK-cell differentiation, perforin expression is a late event,49 and although additional studies will be required to delineate the mechanism, this could suggest NKp44+ NK cells are differentiating toward a phenotype more similar to classic NK cells. One could speculate that if RORγt expression declines, expression of NFIL3 in NKp44+ NK cells allows NKp44+ NK cells to acquire a more classic NK-cell phenotype. However, as shown in supplemental Figure 1, even in SIV-infected animals, NKp44+ NK cells were nonresponsive to a lack of MHC, suggesting that the acquired cytotoxic functions are probably not triggered by MHC-KIR interactions. Regardless, these data highlight the fact that the regulation, development, and differentiation of the NKp44+ NK-cell subpopulation are probably dependent on a complex combination of factors. However, future studies will be needed to address whether the observed plasticity of NKp44+ NK cells is mediated at the single cell or population level.
Loss of TH17 cells during SIV infection has been associated previously with breakdown of gut integrity, which depends on IL-17 and IL-22 for homeostasis, resulting in microbial translocation and chronic immune activation,42 a pathology to which loss of NKp44+ NK cells may also contribute. Although no direct evidence currently exists for a loss of NKp44+ NK contributing to loss of gut integrity in primates, recent studies have shown that the murine counterpart to this cell type is necessary to maintain mucosal integrity, as well as to defend against the intestinal pathogen Citrobacter rodentium.3,50 Although gut TH17 cells outnumber NKp44+ NK cells, because NKp44+ NK cells produce IL-17 and IL-22 constitutively, their contribution to homeostasis may be disproportionate to actual cell number. Regardless, it is clear that chronic lentivirus infection creates an inflammatory gut environment that is hostile toward both the development and normal functionality of the plastic NKp44+ NK-cell population.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Elaine Roberts for dedicated animal care; Drs Dan Barouch, Ron Desrosiers, and David T. Evans for access to macaque samples; Drs Jeff Lifson and Michael Piatak Jr and the Quantitative Molecular Diagnostics Core of the AIDS and Cancer Virus Program, SAIC-Frederick Inc, National Cancer Institute at Frederick, for plasma SIV RNA determinations; Dr Michael Murphey-Corb for the PBS-U5 plasmid and the cell-associated SIV real-time PCR assay; and Dr Arnaud Colantonio for helpful discussions.
This work was supported through National Institutes of Health grants AI071306, AI090735, and RR00168, and Center for HIV/AIDS Vaccine Immunology/HIV Vaccine Trials Network Early Career Investigator award U19 AI 067854-04 to R.K.R.
National Institutes of Health
Authorship
Contribution: R.K.R, T.I.E., J.G., and F.E.W. processed samples and performed ICS and flow cytometry experiments; P.A.R. performed gene expression analyses; M.C. performed flow cytometric analysis and cell sorting; Y.V.K. developed primer and probe sets; A.C. managed animal protocols and performed rectal biopsies; R.K.R, P.A.R., and R.P.J. analyzed the data; and R.K.R. and R.P.J. designed the experiments and wrote the paper.
Conflict-of-interest disclosure: R.P.J. and R.K.R. have filed a patent application on bead-based quantification of mucosal lymphocytes. The remaining authors declare no competing financial interests.
Correspondence: R. Paul Johnson, Division of Immunology, New England Primate Research Center, Harvard Medical School, One Pine Hill Dr, Southborough, MA 01772-9102; e-mail: paul_johnson@hms.harvard.edu.