Abstract
Natural killer (NK) cell–mediated antibody-dependent cellular cytotoxicity involving FcγRIIIa (CD16) likely contributes to the clinical efficacy of rituximab. To assess the in vivo effects of CD16 polymorphisms on rituximab-induced NK activation, blood was evaluated before and 4 hours after initiation of the initial dose of rituximab in 21 lymphoma subjects. Rituximab induced NK activation and a drop in circulating NK-cell percentage in subjects with the high-affinity [158(VF/VV)] but not the low-affinity [158(FF)] CD16 polymorphism. There was no correlation between NK-cell activation or NK-cell percentage and polymorphisms in CD32A, C1q, or CH50. We conclude that NK activation occurs within 4 hours of rituximab infusion in subjects with the high-affinity CD16 polymorphism but not those with the low-affinity CD16 polymorphism. This finding may help explain the superior clinical outcome seen in the subset of high-affinity CD16 polymorphism lymphoma patients treated with single-agent rituximab.
Introduction
Despite the remarkable success of rituximab in treating CD20+ malignancies,1,2 there is still much we do not know about why patients respond, or do not respond, to therapy. Evidence that antibody-dependent cellular cytotoxicity plays a major role in the clinical activity of rituximab comes from several sources, including data exploring the impact of genetic polymorphisms in FcγR on rituximab effects. CD16 with valine at codon 158 (V) binds with higher affinity to human IgG1 than does CD16 with phenylalanine at codon 158 (F).3,4 In vitro, rituximab-coated target cells activate natural killer (NK) cells from subjects with the V polymorphism (VV/VF) at lower rituximab concentrations than (FF) subjects.5 The higher-affinity polymorphism also correlates with a better clinical response rate to single-agent rituximab.6-9 However, it is not known whether rituximab-induced NK-cell activation varies as a function of CD16 polymorphisms in vivo. In the present study, we evaluated NK cells from lymphoma subjects before and 4 hours after initiation of their first dose of rituximab therapy and assessed how CD16 polymorphisms affect NK-cell number and NK activation phenotype.
Methods
Subject eligibility
Subjects who met the following criteria were eligible for enrollment: (1) B-cell proliferative disorder with < 5000 B cells per cubic millimeter in blood; (2) no rituximab therapy in the past 6 months; (3) scheduled to receive rituximab at the standard dose (375 mg/m2), either as a single agent or as part of combination therapy; (4) if the patient was to receive combination therapy, the regimen allowed rituximab to be given before other antilymphoma drugs during the first course of therapy; and (5) provided informed consent as approved by the University of Iowa Institutional Review Board in accordance with the Declaration of Helsinki. Subject characteristics are summarized in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Sample collection and analysis
Blood was obtained immediately before and 4 hours after initiation of rituximab infusion, administered by the standard procedure followed at the University of Iowa. Analysis included the following: (1) complete blood cell count (CBC); (2) NK-cell percentage and NK activation based on surface expression of CD56, CD16, and CD54, as described previously5,10,11 ; (3) genetic polymorphisms in CD16 (position 158),5,7 C1q (position 276),12,13 and CD32A (position 131)7,14 by PCR with genomic DNA (pretherapy sample only); and (4) CH50 (Diamedix).
Statistical analysis
Means and SE were computed for changes in NK-cell activation and are reported separately for high- and low-affinity CD16 polymorphisms. Significance of mean changes and associations between markers were evaluated by paired t tests and Pearson correlation coefficients, respectively. All statistical tests were 2-sided and assessed for significance at .05 levels with the SAS 9.2 software package.
Results and discussion
Rituximab-induced NK-cell activation was evaluated in 21 subjects with various B-cell disorders. Only 1 subject was CD16 homozygous for V (VV) and was grouped with VF subjects for analysis. Clinical signs of infusion reaction15 were noticed in 8 subjects (supplemental Table 1) but did not correlate with the measured parameters. The majority of subjects had both the pretherapy and 4-hour postrituximab samples obtained before any other treatment. Four subjects had chemotherapy before rituximab, and 3 subjects had dexamethasone premedication before rituximab. There were no significant differences in any of the parameters measured between subjects who received chemotherapy or dexamethasone before rituximab and those who did not.
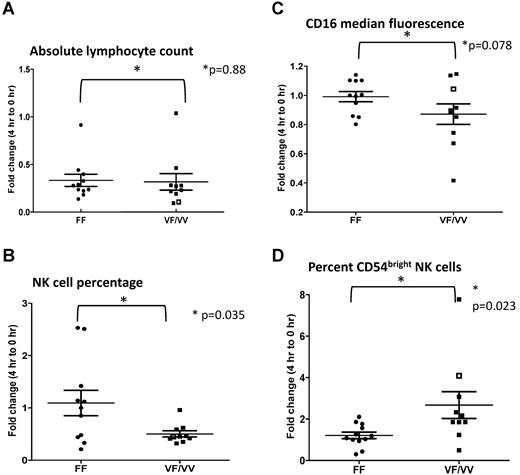
Rituximab treatment decreased total lymphocyte count within 4 hours compared with baseline in the majority of subjects (P < .0001), with a similar effect in both VF/VV and FF subjects (VF/VV versus FF P = .8837; Figure 1A; supplemental Figure 1A). In contrast, the percentage of NK cells decreased in VF/VV subjects (P < .0001) but not in FF subjects (P = .70). The difference between VF/VV and FF subjects in the drop in NK-cell percentage was statistically significant (P = .035; Figure 1B; supplemental Figure 1B).
Fold change in the observed parameters at 4 hours after the initiation of rituximab infusion compared with the baseline (0 hours). (A) Absolute lymphocyte count. (B) Percentage of NK cells in circulation. (C) NK-cell CD16 median fluorescence intensity. (D) Percentage of CD54bright NK cells. The single subject with a VV polymorphism is represented as an open square (□).
Fold change in the observed parameters at 4 hours after the initiation of rituximab infusion compared with the baseline (0 hours). (A) Absolute lymphocyte count. (B) Percentage of NK cells in circulation. (C) NK-cell CD16 median fluorescence intensity. (D) Percentage of CD54bright NK cells. The single subject with a VV polymorphism is represented as an open square (□).
We previously demonstrated a strong in vitro correlation between rituximab-induced CD16 down-modulation, CD54 (intercellular adhesion molecule-1) up-regulation, and NK-mediated antibody-dependent cellular cytotoxicity.5 We therefore evaluated NK-cell CD16 and CD54 expression after rituximab therapy. A trend toward CD16 down-modulation was seen in VF/VV subjects (P = .08) but not in FF subjects (P = .83). The drop in CD16 expression for VF/VV versus FF subjects was of borderline significance (P = .078; Figure 1C; supplemental Figure 1C). Rituximab induced a 3-fold increase in the percentage of CD54bright NK cells (P = .029) in VF/VV subjects but no significant increase in CD54bright NK cells in FF subjects (P = .51). The difference between VF/VV subjects and FF subjects in fold change in CD54bright NK cell percentage was statistically significant (P = .023; Figure 1D; supplemental Figure 1D).
When all subjects were considered, and when FF subjects were considered separately, there was a significant correlation between CD16 down-modulation and CD54 up-regulation (FF subjects R = −0.72, P = .018; Table 1), which indicates mAb-induced CD16 down-modulation can result in NK-cell activation even in subjects with the low-affinity CD16 polymorphism. There was also a trend toward a correlation between CD54 up-regulation and a drop in lymphocyte count and between CD16 down-modulation and a decrease in NK-cell percentage. No significant association was seen between NK activation and different lymphoma subtypes (data not shown). In addition, NK activation did not correlate with other biomarkers, including CD32A and C1q polymorphisms, or serum complement levels (data not shown).
On the basis of these results, we hypothesize that CD54 up-regulation alters NK-cell trafficking to the tumor, and this, along with enhanced NK-cell activation, contributes to the enhanced therapeutic response reported in subjects with the VF/VV polymorphism. Indeed, we previously reported in preliminary studies that rituximab treatment may lead to trafficking of mononuclear cells into involved lymph nodes.16
There are limitations to the present study. The lack of enough VV subjects prevented us from assessing whether this group of subjects responds differently to rituximab. We evaluated NK-cell response just 4 hours after initiation of rituximab but did not evaluate later time points because of possible late effects of chemotherapy. The ability to correlate the effects of CD16 polymorphisms on longer-term changes in NK-cell number, NK-cell activation, or most importantly, clinical response and outcome, will be an important next step, even if it is complicated by concomitant chemotherapy. In addition, in the absence of more rigorous imaging studies or actual biopsy samples to test NK activation in the tumor per se, it is not possible to know whether the NK cells that left the circulation actually trafficked to involved lymph nodes, became activated at the site of tumor, or contributed to the antitumor effects. Finally, these findings may not be relevant in circumstances in which polymorphisms in Fc receptors have not been shown to have an impact on prognosis, such as chronic lymphocytic leukemia.
Several next-generation anti-CD20 antibodies are in various stages of development.17-20 Several of these have been modified so they have enhanced affinity to CD16 and are more effective than rituximab in vitro at activating NK cells and inducing antibody-dependent cellular cytotoxicity irrespective of CD16 polymorphisms.17,20,21 It will be interesting to determine whether these antibodies are able to induce rapid NK-cell activation in vivo in individuals with the FF polymorphism and, most importantly, whether this translates into an improved clinical outcome.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This research was supported in part by National Institutes of Health grants P50 CA97274 and R01 CA137198 and a translational research grant from the Leukemia & Lymphoma Society.
National Institutes of Health
Authorship
Contribution: S.V. performed research, analyzed data, and wrote the manuscript; S.-Y.W., C.D., and S.B. performed research; L.J. and T.K. collected research specimens; A.B. performed statistical analysis; B.K.L. designed research; and G.J.W. provided oversight of the research, designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: G.J.W. and B.L. serve as consultants for Genentech. The remaining authors declare no competing financial interests.
Correspondence: George J. Weiner, MD, Holden Comprehensive Cancer Center, University of Iowa, Iowa City, IA 52242; e-mail: george-weiner@uiowa.edu.