Although platelets are anucleate, they have long been known to contain genomic material, specifically RNA, but the question remains; why should anyone care about an exquisitely detailed profile of RNA from a cell without a nucleus? For decades, the general observation of platelet RNA and the characterization of reticulated platelets have been used to estimate the rate of thrombopoiesis; however, the actual transcripts represented by this RNA were of little interest with the assumption that the RNA was merely a remnant from the precursor megakaryocyte.
Over the past decade, there have been finite but steady observations adding support to the concept that platelet RNA is biologically and patho-physiologically meaningful. Although human platelets are anucleate cells, they retain mRNA, functional splicing, and translational machinery1 and there is a strong correlation between transcript abundance and protein expression.2,3 In the clinical literature, the majority of peripheral blood transcript studies have used whole blood or leukocytes; however, select studies examining RNA from isolated platelets have demonstrated that there is a specific platelet phenotype that is directly associated with athero-thrombotic disease.4-6 Compared with other cells, platelet RNA is particularly intriguing because it is unique in representing a nearly fixed transcriptome. While it is believed that there may be some flux or degradation, compared with a nucleated cell, the change may be minimal and significant turnover in transcript levels is reflected in days compared with hours for nucleated cells.
In the current issue of Blood, Rowley et al extend our understanding of the platelet molecular signature using next generation RNA sequencing to more completely characterize the transcriptome of human and mouse platelets.7 Similar to what has been observed in functional studies; there are many similarities and some differences between mice and man. Reassuringly, the differences between the transcripts parallel known functional differences such as with the protease-activated receptor. Specifically, 58% of the mRNAs expressed by human platelets were found in mouse platelets while 83% of transcripts expressed by mouse platelets are found in human platelets.
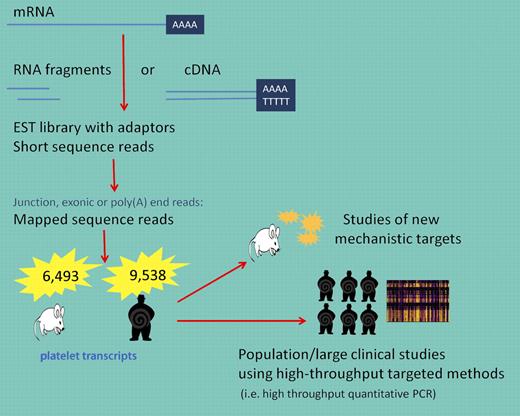
Why is this new dataset of interest? First, and consistent with current principles, Rowley and colleagues have made these data publically available and accessible. In addition, although limited by very small numbers and the study of a select species, these data can serve as a reference point for understanding differences when mechanistic models are pursued. The transcriptome of any given cell is complex and deeper characterization is essential for interpreting functional elements of the genome and identifying relevant mechanistic applications. There are various techniques that have been developed to explore and quantify the transcriptome including hybridization-based approaches such as microarrays. High-density arrays have been developed but have inherent limitations including reliance on existing knowledge of genome sequences and technical issues such as cross-hybridization and narrower range of detection. Sequence-based approaches, such as the one used in the study by Rowley et al, are not limited to detecting transcripts that correspond to existing genomic sequences (see figure). However, there are limitations including the need to construct cDNA libraries and significant bioinformatics challenges such as the need to have efficient methods to store, retrieve, and process vast amounts of data essential to reduce errors in analysis and remove low-quality or false reads.8 Because large genes must be fragmented before reads have been obtained, alignment must be done to ensure that a sequence read matches multiple locations of the genome.
Using RNA-seq, long RNAs are converted into a library of cDNA fragments and sequences are obtained from each cDNA using high-throughput sequencing technology. The resulting sequence reads are aligned with the reference transcriptome. These data can be used to understand new mechanistic pathways in animal, cell culture, or in vitro experiments or applied to larger populations to answer clinically relevant and statistically meaningful questions about pathophysiologic states.
Using RNA-seq, long RNAs are converted into a library of cDNA fragments and sequences are obtained from each cDNA using high-throughput sequencing technology. The resulting sequence reads are aligned with the reference transcriptome. These data can be used to understand new mechanistic pathways in animal, cell culture, or in vitro experiments or applied to larger populations to answer clinically relevant and statistically meaningful questions about pathophysiologic states.
As made clear in the study by Rowley and colleagues, these data will be invaluable for identifying new mechanistic targets that can be furthered studied in animal or in vitro models (see figure). For clinical studies, the use of genome-wide RNA-seq data will be more challenging. The number of data points generated per sample is enormous and the true importance of these findings in small clinical studies will likely draw comparisons in relevance to early SNP studies with copious false-positive data being generated. However, if used judiciously, these data may be linked to individual genomic data as well as used to identify novel transcripts associated with disease. Subsequent validation and testing using a targeted approach (ie, high-throughput quantitative RT-PCR) in a much larger sample set will reveal the true clinical relevance of these detailed transcript findings. Lastly, as with genome-wide association studies, RNA sequencing in a much larger population will need to be performed before we can establish what is a normal platelet transcript.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■