Abstract
The primitive hematopoietic stem/progenitor cells (HSPCs) during embryonic hematopoiesis are thought to be short-lived (SL) with limited self-renewal potential. The fate and consequence of these short-lived HSPCs, once reprogrammed into “long-lived” in a living animal body, remain unknown. Here we show that targeted expression of a dominant-negative C/ebpα (C/ebpαDN) in the primitive SL-HSPCs during zebrafish embryogenesis extends their life span, allowing them to survive to later developmental stage to colonize the definitive hematopoietic sites, where they undergo a proliferative expansion followed by erythropoietic dysplasia and embryonic lethality because of circulation congestion. Mechanistically, C/ebpαDN binds to a conserved C/EBP-binding motif in the promoter region of bmi1 gene, associated with a specific induction of bmi1 transcription in the transgenic embryos expressing C/ebpαDN. Targeted expression of Bmi1 in the SL-HSPCs recapitulates nearly all aberrant phenotypes induced by C/ebpαDN, whereas knockdown of bmi1 largely rescues these abnormalities. The results indicate that Bmi1 acts immediately downstream of C/ebpαDN to regulate the survival and self-renewal of HSPCs and contribute to the erythropoietic dysplasia.
Introduction
The ontogeny of vertebrate hematopoietic system is a strictly regulated process. The hematopoietic stem cells (HSCs) are able to self-renew and take stepwise differentiation to generate multipotential hematopoietic progenitors and finally produce functional mature cells of all lineages. Another hallmark of HSCs is their mysterious migration and sequential shifts between different organs/locations during early embryonic development.1,2 Like mammals, the zebrafish hematopoiesis can be divided into 2 successive phases: the primitive and definitive waves. The primitive wave is transient and begins in 2 anatomically and functionally distinct regions: the anterior- and posterior-bilateral plate mesoderm at 12 hours postfertilization (hpf).3-5 The anterior-bilateral plate mesoderm gives rise to the primitive macrophages and head vasculature,6-8 and the posterior-bilateral plate mesoderm consists of putative hemangioblasts that migrates medially in an anterior to posterior wave to form the intermediate cell mass (ICM) at 18 hpf. The intraembryonic ICM is thought to be the equivalent of mammalian extraembryonic yolk sac blood island and contains hematopoietic stem/progenitor cells (HSPCs) that are able to generate multipotent blood cell lineages, including a large population of primitive erythrocytes, neutrophilic granulocytes, and platelets. Although expressing hematopoietic transcription factors scl, lmo2, gata1, and c/ebpα, these ICM-derived HSPCs are short-lived and possess only short-term or transient hematopoietic activity.7,9,10 As a result, this primitive hematopoiesis wave gradually wanes after the onset of circulation at 24 hpf. Recently, an anatomic location, termed posterior blood island (PBI) that is the portion of ICM behind the yolk sac extension, has also been shown to initiate bilineage hematopoiesis between 24 and 36 hpf through committed erythromyeloid progenitors (EMPs). The bipotent EMPs also express scl and lmo2 transcripts and possess transient proliferative potential but do not seed the hematopoietic pronephros or thymus.11 The zebrafish definitive wave is initiated at the ventral wall of dorsal aorta, an evolutionarily conserved anlage of mammalian aorta-gonad-mesonephros (AGM).12 Distinct from the ICM/PBI-derived short-term HSPCs, the AGM-derived HSPCs possess long-term multipotent hematopoietic activity and self-renewal potential and are able to migrate to the caudal hematopoietic tissue (CHT, equivalent of mammalian fetal liver) through circulation for proliferative expansion, and finally colonize the kidney marrow (equivalent of mammalian bone marrow) and thymus.13,14 Recently, an elegant study showed a subset of HSPCs expressing c-myb and CD41 migrate anteriorly along the bilateral pronephric ducts (PDs) to colonize the developing pronephros.15 These PD-HSPCs appeared to originate from the dorsal AGM-HSPCs, whether alternative locations such as PBI or ICM also contribute to the PD-HSPCs is unclear. These observations that the HSPCs' frequent migration to anatomically distinct organs/locations may reflect the fact that the short-term HSPCs have to explore novel hematopoietic microenvironment as more habitable “niche” capable of providing extrinsic signals (eg, Wnt, TGF-β, and SHH) to support their survival and self-renewal.
The CCAAT enhancer binding protein-α (C/EBPα) is an essential transcription factor for HSPCs self-renewal and granulopoiesis.16,17 Heterozygous mutations in CEBPA were found in approximately 10% acute myeloid leukemia patients.18,19 Most of these mutations affect the amino terminus and eliminate expression of the wild-type 42-kDa protein but do not affect a 30-kDa isoform initiated farther downstream. This truncated protein cannot only inhibit the DNA binding of the wild-type C/EBPα in a dominant-negative (DN) manner but seems to have its own functions and potential targets.20,21 Our previous work showed that the protein and genomic structures of C/EBPα are highly conserved throughout vertebrate evolution. The expression of zebrafish c/ebpa is readily detected in the developing HSPCs in the P-LPM/ICM and colocalizes with other hematopoietic stem/progenitor markers, such as scl, lmo2, pu.1, and gata-1.7 A single cytosine deletion mutation (zD420) in zebrafish that mimics the C/EBPα deletion mutation (hD395) found in human with M2-subtype myeloid leukemia shows similar DN activities to its human counterpart hD395 mutant.7,18 The results suggest that the C/EBPα may play a crucial role in regulating the development and self-renewal of these short-lived HSPCs during embryogenesis, and targeted interference of the C/EBPα signaling through expressing its DN isoform in the developing short-lived HSPCs may provide invaluable insights into the molecular and developmental mechanisms underlying the HSPC self-renewal and diseases.
Although the importance of extrinsic signals in the maintenance of HSPC stemness was clarified, some of the intrinsic factors responsible for HSPC survival and self-renewal have also been identified. Epigenetic regulators, such as the polycomb family proto-oncogene Bmi1, have been shown to be important in regulating HSPC proliferation and self-renewal activity. Ablation of Bmi1 function results in the disruption of self-renewal capacity of normal and leukemia stem cells.22,23 Overexpression of Bmi1 has been found in human high-risk leukemia and other malignancies and is related to unfavorable prognosis.24,25 Although several targets downstream of Bmi1, including p16Ink4a and p19Arf, have been identified,23 the upstream signaling pathways regulating Bmi1 expression remain largely unknown.
Here we show that targeted expression of C/ebpαDN in the ICM/PBI-derived, short-lived, and scl-positive HSPCs with limited self-renewal potential extends their life span, allowing them to colonize the bilateral PDs and CHT, where they proliferate and undergo aberrant differentiation along the erythroid lineage, resulting in a massive increase of immature erythrocytes in the circulation and embryonic lethality because of circulation congestion before 14 days postfertilization (dpf). Furthermore, the EGFP-tagged C/ebpαDN directly interacts with an evolutionarily conserved C/EBP binding motif within the promoter region of bmi1, concomitant with a specific induction of bmi1 transcripts in the ICM/PBI-located HSPCs expressing C/ebpαDN. Targeted expression of Bmi1 in the same population of HSPCs recapitulates nearly all C/ebpαDN-induced phenotypes, and knockdown of bmi1 largely corrects these abnormalities. These studies uncover a mechanistic link between C/ebpα and Bmi1 synergistically to regulate HSPC homeostasis and potential involvement in human erythropoietic disorders.
Methods
Fish care
Zebrafish maintenance, breeding, and staging were performed as previously described.26 The zebrafish study was approved by the Institute of Health Sciences institutional review board.
Establishment of zebrafish transgenic lines and genomic PCR
Tg (lmo2:Cre) transgenic zebrafish was established in our laboratory as previously described.27 A 2.5-kb zebrafish lmo2 promoter fragment28 was used to direct the expression of LDL-EGFP, LDL-EGFP-C/ebpαDN,7 and LDL-Bmi1-EGFP. Transgenic plasmids flanked by I-SceI29 were prepared with an endotoxin-free kit (Promega). Microinjection was performed at one-cell stage embryos with 1- to 1.5-nL of injection solution containing 80 pg/nL of DNA, 0.5× I-SceI buffer and 0.5 U/μL I-SceI meganuclease (New England Biolabs). Injected embryos were raised to sexual maturity (F0 founders) and crossed to wild-type zebrafish to generate F1 progeny, which were screened for the DsRed expression in the ICM/PBI at 22 hpf. The DsRed+ F1 embryos were raised to adults to establish the stable transgenic lines. The adult F1 transgenic zebrafish Tg(lmo2:LDL-EGFP), Tg(lmo2:LDL-EGFP-C/ebpαDN), and Tg(lmo2:LDL-Bmi1-EGFP) were crossed to the Tg (lmo2:Cre) adults to detect the red-green fluorescence shift using a fluorescent stereomicroscope Zeiss SteREO Discovery V20 (Carl Zeiss) with a 1.5× Plan-Apo S objective and AxioVision, Version Rel4.6 software. All the micrographs were taken using 3% methylcellulose as medium. Genomic PCR followed by sequencing was used to validate the Cre recombinase-mediated genomic recombination as previously described.27 Primer sequences are available in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The movies of circulation (supplemental Videos 1-3) were taken by a microscope Olympus IX81 with a 20× DIC objective and Image-Pro Plus Version 5.1 software.
Genomic nested PCR
Genomic DNA from the trunk muscle, tail tip, CHT, and the CHT-released blood cells was extracted by Genomic DNA Isolating Kit (BioDev). PCR reactions using specific nested PCR primers indicated in Figure 1C (black and red arrows) were performed as follows: 95°C, 10 minutes; 38 cycles of 94°C, 30 seconds; 55°C, 30 seconds; 68°C, 2.5 minutes; and then 68°C for10 minutes. The PCR products of first round were purified as the templates for a second round reaction: 95°C, 10 minutes; 38 cycles of 94°C, 30 seconds; 55°C, 30 seconds; 68°C, 1.5 minutes; and then 68°C for 10 minutes. Nested PCR products were separated on a 1.5% agarose gel. Primer sequences are available in supplemental Table 1.
Generation and characterization of stable C/ebpαDN transgenic lines with Cre-loxp strategy. (A-C) Schematic representations of transgenic lines Tg(lmo2:Cre) (A), Tg(lmo2:LDL-EGFP) (B), and Tg(lmo2:LDL-EGFP-C/ebpαDN) (C). All transgenes were under the control of a 2.5-kb lmo2 promoter. The EGFP or EGFP-tagged C/ebpαDN gene was separated from the lmo2 promoter by loxp-DsRed-loxp (LDL) element. Truncated zebrafish C/ebpαDN was in-frame fused with EGFP. Arrows at the bottom of panel C indicate the position of primers used in genomic PCR for genotyping. pA indicates SV40 polyadenylation site. (D-G) Representative 22 hpf progeny of mating Tg(lmo2:LDL-EGFP) with either wild-type (D) or Tg(lmo2: Cre) heterozygote (E), and mating Tg(lmo2:LDL-EGFP-C/ebpαDN) with either wild-type (F) or Tg(lmo2:Cre) heterozygote (G). Red-green fluorescence shifts can only be detected in the double-transgenic embryos carrying the Cre recombinase gene (E,G). The stars indicate the residual red fluorescence after Cre-mediated genomic recombination. HV indicates head vasculature. (H) Western blot analyses confirmed the expression of EGFP (27 kDa) and EGFP-tagged C/ebpαDN fusion protein (54 kDa) in the transgenic embryos presented in panels D to G corresponding to lines D′ to G′.
Generation and characterization of stable C/ebpαDN transgenic lines with Cre-loxp strategy. (A-C) Schematic representations of transgenic lines Tg(lmo2:Cre) (A), Tg(lmo2:LDL-EGFP) (B), and Tg(lmo2:LDL-EGFP-C/ebpαDN) (C). All transgenes were under the control of a 2.5-kb lmo2 promoter. The EGFP or EGFP-tagged C/ebpαDN gene was separated from the lmo2 promoter by loxp-DsRed-loxp (LDL) element. Truncated zebrafish C/ebpαDN was in-frame fused with EGFP. Arrows at the bottom of panel C indicate the position of primers used in genomic PCR for genotyping. pA indicates SV40 polyadenylation site. (D-G) Representative 22 hpf progeny of mating Tg(lmo2:LDL-EGFP) with either wild-type (D) or Tg(lmo2: Cre) heterozygote (E), and mating Tg(lmo2:LDL-EGFP-C/ebpαDN) with either wild-type (F) or Tg(lmo2:Cre) heterozygote (G). Red-green fluorescence shifts can only be detected in the double-transgenic embryos carrying the Cre recombinase gene (E,G). The stars indicate the residual red fluorescence after Cre-mediated genomic recombination. HV indicates head vasculature. (H) Western blot analyses confirmed the expression of EGFP (27 kDa) and EGFP-tagged C/ebpαDN fusion protein (54 kDa) in the transgenic embryos presented in panels D to G corresponding to lines D′ to G′.
Western blot
For Western blot, embryos at indicated developmental stages were deyolked as previously described.30 Embryos were homogenized in lysis buffer (20mM Tris HCl [pH 7.4], 150mM NaCl, 5mM EDTA, 10% glycerol, and 0.1% Triton X-100). Signals were detected with rabbit anti-EGFP polyclonal antibody (1:1000, Clontech) for overnight at 4°C, followed by incubation with HRP-conjugated secondary antibody (1:10 000).
Cytology assay
Blood cell isolation was performed as described.31 The 0.9× PBS containing 50 U/mL heparin, 5% FBS, and 0.006% tricaine was used as isolation buffer. For cytospin, the cell collections were centrifuged at 100g for 5 minutes onto glass slides. Wright-Giemsa stain was performed according to the manufacturer's instructions (Baso). The micrographs were taken by a microscope Nikon ECLIPSE 80i with 100× oil immersion lens and Nikon ACT-1 software.
Morpholino oligonucleotide knockdown
Zebrafish bmi1 morpholino oligonucleotides were purchased from Gene Tools and diluted with nuclease-free water. Microinjection of morpholino oligonucleotides was performed at one- or two-cell stage. All injections were performed with Harvard Apparatus microinjector. Morpholino sequences are available in supplemental Table 1.
WISH
Whole-mount mRNA in situ hybridization (WISH) was performed as described previously.32 For embryos older than 48 hpf, high resolution of WISH was performed as previously described.13 For 2-color in situ hybridization, the scl antisense RNA probe was labeled with digoxigenin (Roche Diagnostics), whereas antisense probes against EGFP were labeled with fluorescein (Roche Diagnostics). The purple color was developed first with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate (Vector Laboratories) as a substrate, followed by the development of red color with Fast Red tablets (Roche Diagnostics). The results were imaged using a stereomicroscope Nikon SMZ1500 with 1× HR Plan Apo objective and ACT-1 vision software. Embryos were mounted in 95% glycerol.
Immunochemistry and TUNEL assays
Whole-mount immunochemistry staining for proliferation was performed with a rabbit polyclonal antibody against phosphorylated histone 3 (pH3) as previously described.33 Whole-mount TUNEL assay was performed using POD in situ Cell Death Detection Kit (Roche Diagnostics) according to the manufacturer's instructions. All results were imaged by stereomicroscope Nikon SMZ 1500 with 1× HR plan Apo objective and Version ACT-1 software.
E-ChIP
Embryos at 22 hpf were enzymatically dechorionated and then fixed in 1.85% formaldehyde. Embryonic chromatin immunoprecipitation (E-ChIP) with a polyclonal antibody against EGFP (Clontech) was performed as previously described.34 The 15 and 10 kb of potential promoter sequences upstream of the transcriptional start site (TSS) of murine and zebrafish bmi1 gene, respectively, were analyzed with rVista software (http://genome.lbl.gov/vista/rvista/submit.shtml). Potential C/EBP binding motifs were identified (http://www.genome.jp/tools/motif). Primer sequences are available in supplemental Table 1.
Results
Establishment and characterization of C/ebpαDN transgenic zebrafish lines
The zebrafish C/ebpαDN (previously designated as zD420 and termed C/ebpαDN hereafter) carries a deletion at the evolutionarily conserved cytosine, which is also found in human patient with M2-subtype acute myeloid leukemia (previously designated as hD395).7,18 The C/ebpαDN deletion mutation prevents expression of the full-length protein, allowing the expression of truncated isoforms from internal translational initiation sites, and has similar DN activities as its human counterpart hD395.7 To investigate the function of C/ebpαDN in the developing HSPCs, we took use of a previously established Tg(lmo2:Cre) transgenic line (Figure 1A)27 and first established 2 novel transgenic lines, Tg(lmo2:LDL-EGFP) (Figure 1B) and Tg(lmo2:LDL-EGFP-C/ebpαDN) (Figure 1C), in which a floxed DsRed gene (loxP-DsRed-loxP, abbreviated as LDL) followed by either a downstream EGFP or an in-frame fused EGFP-C/ebpαDN gene, are expressed under the lmo2 promoter,28 respectively. Because the lmo2 transcripts are richly detected in the developing HSPCs and bipotential EMPs in the ICM and PBI regions,2,11 the combinational use of the Tg(lmo2:Cre) and the Tg(lmo2:LDL-EGFP-C/ebpαDN) transgenic lines provides an invaluable tool to investigate the function of the leukemogenic C/ebpαDN in the short-lived HSPCs in a physiologic context.
The control Tg(lmo2:LDL-EGFP) and the Tg(lmo2:LDL-EGFP-C/ebpαDN) heterozygous transgenic adults were first mated to either wild-type or Tg(lmo2:Cre) heterozygote to evaluate the Cre recombinase activity in vivo through examining the shift of red fluorescence to green fluorescence at 22 hpf. In the LDL-EGFP+/−;wild-type embryos, only red fluorescence was detected in the head vasculature, ICM, and PBI (Figure 1D; red filter), without any detectable EGFP fluorescence (Figure 1D; green filter). In the double-transgenic LDL-EGFP+/−;Cre+/− embryos, however, only residual red fluorescence was retained (Figure 1E stars; red filter), with robust EGFP fluorescence being observed in the head vasculature, ICM, and PBI (Figure 1E; green filter). Highly similar fluorescent expression and shift patterns were also observed in the LDL-EGFP-C/ebpαDN+/−;wild-type and LDL-EGFP-C/EBPαDN+/−;Cre+/− embryos (Figure 1F-G), except that the EGFP fluorescence of LDL-EGFP-C/ebpαDN+/−;Cre+/− embryos was weaker than that of LDL-EGFP+/−;Cre+/− embryos (Figures 1E,G; green filters). The EGFP fluorescence was predominantly detected in the PBI of the LDL-EGFP-C/ebpαDN+/−;Cre+/− embryos (Figure 1G arrowhead; green filter), a site that hosts the short-lived bipotent hematopoietic progenitor (EMP) development and forms the CHT (equivalent of mammalian fetal liver) at later stages (48 hpf to 7 dpf).11,13,35 WISH analyses using a digoxigenin-labeled EGFP antisense probe verified these results (Figure 1D,G bottom panels). In addition, Western blot analyses confirmed the expression of the EGFP (27 kDa) and the EGFP-tagged C/ebpαDN proteins (54 kDa) in the lysates of LDL-EGFP+/−;Cre+/− and LDL-EGFP-C/ebpαDN+/−;Cre+/− embryos at 22 hpf, respectively (Figure 1H lanes E′ and G′). Furthermore, genomic PCR (primer locations indicated by black arrows in Figure 1C) and subsequent sequencing analyses confirmed that the Cre-mediated recombination occurred precisely at the LoxP sites (supplemental Figure 1; and data not shown).
Circulation congestion and erythropoietic dysplasia in the transgenic embryos expressing C/ebpαDN
Because the lmo2 promoter is activated during primitive hematopoiesis (12-26 hpf) and gradually wanes as definitive hematopoiesis initiates in the AGM at 26 hpf and expands in the CHT from 48 hpf onwards,2,28 the EGFP fluorescence in the EGFP-C/ebpαDN transgenic embryos was hardly detected after 24 hpf. Consistently, Western blot analyses showed that the expression of EGFP-tagged C/ebpαDN protein was undetected at 36 and 75 hpf but still weakly expressed at 26 hpf (supplemental Figure 2). We therefore picked out the EGFP-positive embryos at 22 hpf from the clutches of mating the Tg(lmo2:Cre) heterozygote to either Tg(lmo2:LDL-EGFP) or Tg(lmo2:LDL-EGFP-C/ebpαDN) heterozygote. These EGFP-positive embryos carrying either LDL-EGFP+/−;Cre+/− or LDL-EGFP-C/ebpαDN+/−;Cre+/− double transgenes were subject to intensive visual investigation, and no any detectable morphologic abnormalities were found before 72 hpf. Starting at 75 hpf, however, 98% of EGFP-C/ebpαDN-positive embryos (n = 65/66) demonstrated a congested circulation with blood cells accumulated within the caudal vein of CHT, which resulted in a deteriorating cardiac edema and embryonic lethality between 7 and 14 dpf (Figure 2A-B arrows; supplemental Videos 1 and 2; and data not shown). No obvious abnormalities were observed in the heart and vasculature development (data not shown). To confirm whether the accumulated blood cells had undergone Cre-mediated genomic recombination, we performed nested genomic PCR with 2 specific primer pairs (Figure 1C black and red arrows) to genotype the DNA extracted from the dissected tissue or the released blood cells in indicated anatomic sites (Figure 2B: I, tail tip; II, CHT; III, trunk muscle indicated by red boxes; and IV, CHT released blood cells indicated by curved arrow). As shown in Figure 2B and C, a 0.3-kb recombinant fragment was exclusively amplified from the CHT-released blood cells (IV), whereas only a 1.5-kb unrecombinant fragment was obtained from the tail tip (I). As expected, the CHT and trunk-derived muscle showed a stronger unrecombinant fragment and a weaker recombinant fragment probably because of the contamination of blood cells within these tissues (II and III). The Giemsa staining of the released cells from the congested CHT showed immature erythroid morphology with higher nucleus-cytoplasm ratio, basophilic staining, and cells undergoing mitotic division (Figure 2D-E arrow). Furthermore, a significantly increased number of cells positive for the mitotic pH3 marker (Figure 2F-G arrow) and the apoptotic TUNEL staining (Figure 2H-I arrows) was also observed in the congested CHT and segmental vessels of EGFP-C/ebpαDN-positive embryos.
Circulation congestion and erythropoietic dysplasia in the C/ebpαDN transgenic embryos. (A-B) External features of the Tg(lmo2:LDL-EGFP) (A) and Tg(lmo2:LDL-EGFP-C/ebpαDN) (B) embryos at 75 hpf. The circulation congestion is only detected in the C/ebpαDN-expressing embryos (B, arrowheads and supplemental Videos 1 and 2). Black boxes with dashed lines indicate the regions amplified at the bottom. Red boxes and curved arrow indicate the sites where tissue or circulating cells were isolated and genomic DNA were extracted for genotyping shown in panel C. DA indicates dorsal aorta; and CV, caudal vein. (C) Analyses of Cre-mediated genomic recombination by genomic-nested PCR using 2 pairs of primers indicated at the bottom of Figure 1C. The Roman numerals indicate the anatomic sites where tissues and cells are dissected (I-III) or released from the CHT (IV). (D-E) Wright-Giemsa stainings and morphologic characterizations of circulating blood cells released from the CHT of transgenic embryos at 75 hpf from Tg(lmo2:LDL-EGFP). (D) All 3 panels are adopted from the same slide and composited together and Tg(lmo2:LDL-EGFP-C/ebpαDN). (E) Both panels are adopted from the same slide. Arrow indicates the cell undergoing mitosis. Images were acquired with a 100× oil objective. (F-I) Proliferation and apoptosis analyses by pH3 immunochemistry stainings (F-G) and TUNEL assays (H-I) in the embryos expressing EGFP and EGFP-C/ebpαDN at 75 hpf as described in “Immunochemistry and TUNEL assays.”
Circulation congestion and erythropoietic dysplasia in the C/ebpαDN transgenic embryos. (A-B) External features of the Tg(lmo2:LDL-EGFP) (A) and Tg(lmo2:LDL-EGFP-C/ebpαDN) (B) embryos at 75 hpf. The circulation congestion is only detected in the C/ebpαDN-expressing embryos (B, arrowheads and supplemental Videos 1 and 2). Black boxes with dashed lines indicate the regions amplified at the bottom. Red boxes and curved arrow indicate the sites where tissue or circulating cells were isolated and genomic DNA were extracted for genotyping shown in panel C. DA indicates dorsal aorta; and CV, caudal vein. (C) Analyses of Cre-mediated genomic recombination by genomic-nested PCR using 2 pairs of primers indicated at the bottom of Figure 1C. The Roman numerals indicate the anatomic sites where tissues and cells are dissected (I-III) or released from the CHT (IV). (D-E) Wright-Giemsa stainings and morphologic characterizations of circulating blood cells released from the CHT of transgenic embryos at 75 hpf from Tg(lmo2:LDL-EGFP). (D) All 3 panels are adopted from the same slide and composited together and Tg(lmo2:LDL-EGFP-C/ebpαDN). (E) Both panels are adopted from the same slide. Arrow indicates the cell undergoing mitosis. Images were acquired with a 100× oil objective. (F-I) Proliferation and apoptosis analyses by pH3 immunochemistry stainings (F-G) and TUNEL assays (H-I) in the embryos expressing EGFP and EGFP-C/ebpαDN at 75 hpf as described in “Immunochemistry and TUNEL assays.”
To characterize the molecular nature of these dysplastic blood cells, we performed WISH analyses with hematopoietic marker genes (α-E1 and α-E3 hemoglobin for embryonic erythroid cells; gata-1 for erythroid progenitors; mpo and l-plastin for embryonic neutrophilic granulocytes and monocytes/macrophages, respectively; and rag-1 for thymic lymphocytes) at various developmental stages from 22 to 75 hpf. Although no obvious changes for the erythroid α-E1 and α-E3 expression were detected at 22 and 26 hpf (supplemental Figure 3A), a significantly increased number of α-E1– and α-E3–expressing erythroid cells was observed at 75 hpf: in the EGFP-positive control embryos at 75 hpf, α-E1- and α-E3–positive erythroid cells were mainly detected in the regions of heart and CHT (Figure 3A; supplemental Figure 3B, star and arrow); in the EGFP-C/ebpαDN-positive embryos, however, the globin-expressing cells abundantly filled in the cardiac region (star), the trunk vessels (red arrow), dorsal aorta (red arrowheads), cranial vasculature (red arrowheads), and segmental vessel (black arrowheads) (Figure 3B; supplemental Figure 3C). No changes in the levels of gata-1 transcripts were detected at 22, 36, and 75 hpf (supplemental Figure 4), suggesting that a Gata-1–independent mechanism contributed to the erythropoietic dysplasia. A slight increase, if any, was also observed for the expression of l-plastin and mpo at 75 hpf (supplemental Figure 5; and data not shown). In addition, a robust reduction or lack of rag-1 expression was also detected in the developing thymus, probably resulting from the disrupted circulation that is required for HSCs to colonize the thymus (supplemental Figure 6).13 Thus, the congested circulation observed in the CHT of EGFP-C/ebpαDN-positive embryos at 75 hpf resulted from a dysplasia of immature erythrocytes that had undergone Cre-mediated genomic recombination and were progeny of the C/ebpαDN-expressing HSPCs derived from ICM/PBI at 22 hpf.
Developmental mechanism and origin of dyserythropoiesis. (A-B) WISH analyses of αE1-globin in the indicated transgenic embryos at 75 hpf. The star, red arrow, and arrowheads indicate the increased number of erythrocytes expressing αE1-globin transcripts within the embryonic vasculature. (C-J) Time course of WISH analyses of scl in the transgenic embryos at indicated developmental stages. Red arrows and arrowheads indicate the increased scl+ HSPCs in the CHT and bilateral PDs. (K) Two-color WISH of EGFP (black color) and scl (red color) transcripts in the transgenic embryos at 32 hpf. (K-L) Fast red labeling of scl (RITC filter) is shown to the right. The red and black arrows indicate the colocalization of scl and EGFP.
Developmental mechanism and origin of dyserythropoiesis. (A-B) WISH analyses of αE1-globin in the indicated transgenic embryos at 75 hpf. The star, red arrow, and arrowheads indicate the increased number of erythrocytes expressing αE1-globin transcripts within the embryonic vasculature. (C-J) Time course of WISH analyses of scl in the transgenic embryos at indicated developmental stages. Red arrows and arrowheads indicate the increased scl+ HSPCs in the CHT and bilateral PDs. (K) Two-color WISH of EGFP (black color) and scl (red color) transcripts in the transgenic embryos at 32 hpf. (K-L) Fast red labeling of scl (RITC filter) is shown to the right. The red and black arrows indicate the colocalization of scl and EGFP.
Developmental origin and mechanism of erythropoietic dysplasia induced by C/ebpαDN
The PBI/ICM-derived HSPCs (including EMPs in the PBI region) occurred in the primitive hematopoiesis wave are thought to be transient and short-lived with limited self-renewal ability. As a result, they become hardly detected by WISH using scl or lmo2 as a marker after 24 hpf and do not contribute to the definitive hematopoiesis initiating in the AGM (between 26 and 72 hpf) and subsequently expanding in the CHT (between 48 hpf and 7 dpf).11 How could these short-lived PBI/ICM-derived primitive HSPCs contribute to the dyserythropoiesis occurrence at the definitive hematopoiesis stage at 75 hpf? One possible explanation is that the C/ebpαDN extends the life span and promotes the self-renewal of these short-lived HSPCs, therefore allowing them to colonize the definitive hematopoiesis sites, such as CHT for proliferative expansion and aberrant erythroid differentiation. Because these PBI/ICM-derived HSPCs express the stem cell maker gene scl,36 we first performed time-course WISH analyses with scl as a probe to trace the developmental origin of the erythropoietic dysplasia. Consistent with the hypothesis, a significantly increased number of scl+ HSPCs was readily detected in the CHT region of C/ebpαDN-positive embryos (n = 14 of 19) at 75 hpf, compared with the EGFP+ control embryos (Figure 3C-D). Unexpectedly, a 4-fold increase (3.3 vs 11.6) of the scl+ HSPCs was also detected in the bilateral PDs at 36 hpf (Figure 3E-F arrows) and 30 hpf (Figure 3G-H arrows), although no such change was detected at 22 hpf (Figure 3I-J). This finding is intriguing in that the PD was recently shown to serve as a migratory path for the HSPCs (c-myb+CD41+) to colonize the developing glomerulus.15
As expected, these scl+ PD-HSPCs also expressed EGFP-C/ebpαDN transcripts as determined by double WISH (Figure 3K-L, arrows), suggesting that these PD-HSPCs are progeny of PBI/ICM-derived HSPCs that expressed C/ebpαDN. Collectively, the results suggested a model that targeted expression of C/ebpαDN in these scl+ PBI/ICM-HSPCs at 22 hpf and resulted in an extended life span, allowing them to colonize the PD and the CHT, where they underwent proliferative expansion and Gata-1-independent dyserythropoiesis at 75 hpf.
Specific up-regulation of bmi1 transcripts in the PBI/ICM-located HSPCs
To explore the possible molecular mechanisms responsible for the C/ebpαDN-induced extension of HSPCs' life span, we screened a panel of zebrafish orthologs of mammalian epigenetic and developmental regulators that have been shown essential for self-renewal of HSCs and leukemia stem cells. These genes are bmi1,22 prdm3,37 prdm16,38 ezh2, rae28, notch1a, her6, cdx4, cdx1a, hox9a, hoxb4,39 and sonic hedgehog (shh).40 Interestingly, of 12 self-renewal genes screened, 4 genes (bmi1, rae28, prdm3, and prdm16) are specifically induced in the PBI and/or ICM regions (Figure 4 arrows) and no changes were detected for the other 8 genes (shh, notch1a, her6, ezh2, cdx4, cdx1a, hoxa9, and hoxb4; supplemental Figure 7). Of 4 induced genes, the significant induction of bmi1 transcripts specifically in the PBI and ICM (Figure 4A-B arrows) is particularly interesting because the Cre recombinase activity and EGFP-C/ebpαDN expression were also detected in these 2 regions (Figure 1G), suggesting that the Bmi1 may act downstream of C/ebpαDN signaling to mediate the expansion of the scl+ HSPCs and extension of their life span in the bilateral PD (30-36 hpf) and CHT (75 hpf), which in turn caused erythropoietic dysplasia.
Specific induction of epigenetic regulatory genes by C/ebpαDN. (A-H) WISH analyses of a panel of epigenetic regulators involved in the HSC maintenance and self-renewal at 22 hpf. The arrows indicate the specific induction of bmi1 (A-B), rae28 (C-D), prdm3 (E-F), and prdm16 (G-H) transcripts in the ICM or PBI region.
Specific induction of epigenetic regulatory genes by C/ebpαDN. (A-H) WISH analyses of a panel of epigenetic regulators involved in the HSC maintenance and self-renewal at 22 hpf. The arrows indicate the specific induction of bmi1 (A-B), rae28 (C-D), prdm3 (E-F), and prdm16 (G-H) transcripts in the ICM or PBI region.
Targeted overexpression of Bmi1 in the PBI/ICM-located HSPCs recapitulates phenotypes induced by C/ebpαDN
To test the hypothesis, we established another stable transgenic line Tg(lmo2:LDL-Bmi1-EGFP) to specifically express the EGFP-tagged Bmi1 protein in the HSPCs of PBI/ICM (Figure 5A). The zebrafish Bmi1 protein located on chromosome 24 shares 80% and 81% amino acid identities with its human and murine counterparts, respectively (data not shown). By mating the Tg(lmo2:LDL-Bmi1-EGFP) heterozygote to the Tg(lmo2:Cre) heterozygote, targeted expression of Bmi1-EGFP in the HSPCs of PBI/ICM was verified in both EGFP fluorescence and transcript levels at 22 hpf (Figure 5B, arrow and arrowhead). Time-course WISH analyses showed that the number of scl+ HSPCs was unchanged at 22 hpf (Figure 5C-D) but significantly increased along the bilateral PD in the LDL-Bmi1-EGFP+/−;Cre+/− embryos at 36 hpf (Figure 5E-H arrows and arrowheads in the inset). More importantly, the morphologic phenotypes characterized by circulation congestion (Figure 6A-B) and accumulation of hematopoietic cells positive for both pH3 (Figure 6C-D) and TUNEL staining (Figure 6E-F) in the CHT region, along with a massive increase of the α-E1-positive erythroid cells throughout the embryos (Figure 6G-H), were also observed in the LDL-Bmi1-EGFP+/−;Cre+/− embryos at 80 hpf. The results indicate that targeted expression of Bmi1 protein in the PBI/ICM-located HSPCs recapitulates most, if not all, of the morphologic and molecular abnormalities observed in the C/ebpαDN transgenic embryos and suggest that the self-renewal enhancement and life span elongation of the short-lived, scl+ HSPCs are conferred by transcriptional activation of bmi1 by C/ebpαDN.
Characterization of Bmi1 transgenic zebrafish line. (A) Schematic representation of lmo2:LDL-Bmi1-EGFP transgenic construct. pA indicates SV40 polyadenylation site. (B) Expression of Bmi1-EGFP fusion protein and transcripts in the progeny of mating Tg(lmo2:LDL-Bmi1-EGFP) with Tg (lmo2:Cre) line. (C-H) time-course WISH analyses of scl in the Bmi1-expressing transgenic embryos at indicated developmental stages. Red arrows and arrowheads indicate the increased scl+ HSPCs along the PD at 36 hpf.
Characterization of Bmi1 transgenic zebrafish line. (A) Schematic representation of lmo2:LDL-Bmi1-EGFP transgenic construct. pA indicates SV40 polyadenylation site. (B) Expression of Bmi1-EGFP fusion protein and transcripts in the progeny of mating Tg(lmo2:LDL-Bmi1-EGFP) with Tg (lmo2:Cre) line. (C-H) time-course WISH analyses of scl in the Bmi1-expressing transgenic embryos at indicated developmental stages. Red arrows and arrowheads indicate the increased scl+ HSPCs along the PD at 36 hpf.
Capitulation of C/ebpαDN-induced abnormalities in the Bmi1 transgenic embryos. (A-B) External features of the Tg(lmo2:LDL-EGFP) (A) and Tg(lmo2:LDL-Bmi1-EGFP) (B) embryos at 80 hpf. The circulation congestion is readily detected in the Bmi1-expressing embryos (B, arrowheads; and supplemental Videos 1 and 3). Black boxes with dashed lines indicate the regions amplified at the bottom. DA indicates dorsal aorta; and CV, caudal vein. (C-F) Proliferation and apoptosis analyses by pH3 immunochemistry stainings (C-D) and TUNEL assays (E-F) in the embryos expressing EGFP and Bmi1-EGFP at 80 hpf. (G-H) WISH analyses of αE1-globin in the transgenic embryos at 80 hpf. The star, red arrow, and arrowheads indicate the increased number of cells with expression of αE1-globin transcripts within the embryonic vasculatures.
Capitulation of C/ebpαDN-induced abnormalities in the Bmi1 transgenic embryos. (A-B) External features of the Tg(lmo2:LDL-EGFP) (A) and Tg(lmo2:LDL-Bmi1-EGFP) (B) embryos at 80 hpf. The circulation congestion is readily detected in the Bmi1-expressing embryos (B, arrowheads; and supplemental Videos 1 and 3). Black boxes with dashed lines indicate the regions amplified at the bottom. DA indicates dorsal aorta; and CV, caudal vein. (C-F) Proliferation and apoptosis analyses by pH3 immunochemistry stainings (C-D) and TUNEL assays (E-F) in the embryos expressing EGFP and Bmi1-EGFP at 80 hpf. (G-H) WISH analyses of αE1-globin in the transgenic embryos at 80 hpf. The star, red arrow, and arrowheads indicate the increased number of cells with expression of αE1-globin transcripts within the embryonic vasculatures.
Bmi1 is a direct downstream target of C/ebpαDN
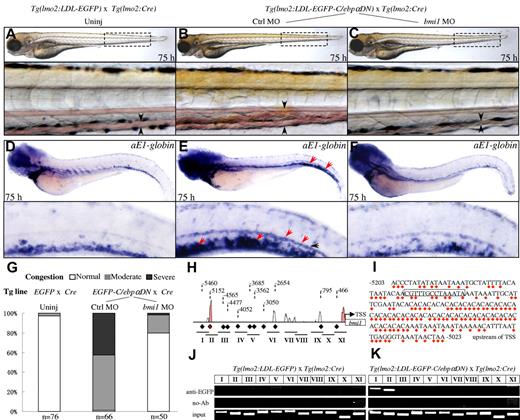
The specific induction of bmi1 in the Tg(lmo2:LDL-EGFP-C/ebpαDN) embryos and highly similar phenotypes between the Tg(lmo2:LDL-EGFP-C/ebpαDN) and Tg(lmo2:LDL-Bmi1-EGFP) transgenic lines suggest that the Bmi1 may act downstream of C/ebpαDN. Indeed, injection of a zebrafish bmi1-specific morpholino oligonucleotide (supplemental Figure 8) efficiently relieved the C/ebpαDN-mediated circulation congestion and erythropoietic dysplasia (Figure 7A-G). Of 50 C/ebpαDN-positive embryos microinjected with bmi1-specific morpholino oligonucleotides (0.15mM), 80% showed no detectable circulation congestion and α-E1 abnormalities (Figure 7C,F-G). In contrast, in the C/ebpαDN-positive embryos microinjected with an unrelated mismatch morpholino oligonucleotide (0.15mM), 98.5% (65 of 66) maintained these abnormal phenotypes induced by C/ebpαDN (Figure 7B,E,G). Moreover, WISH analyses showed that not only the number of the scl+ HSPCs in the PD region of C/ebpαDN embryos at 36 hpf was reduced to normal level (supplemental Figure 9), but the enhanced proliferation (supplemental Figure 10) and enhanced apoptosis (supplemental Figure 11) were also reversed by bmi1 knockdown. As a control, injection of either bmi1-specific or control morpholino into EGFP control embryos did not cause detectable morphologic and hematopoietic abnormalities (supplemental Figure 12). To further validate whether Bmi1 is a direct downstream target, we analyzed the 20-kb promoter region upstream of the TSS of zebrafish and murine bmi1 genes and found one evolutionarily conserved region containing a C/EBP binding motif at 5.2 kb upstream of the zebrafish bmi1 TSS (Figure 7H). The region shares 63% nucleotide acid identities with a region (−11 780 to −11 612 bp) located at the upstream of the murine Bmi1 TSS (Figure 7I, red dots). We therefore tested whether the EGFP-tagged C/ebpαDN was able to directly bind to this C/EBP motif as well as additional predicted C/EBP motifs indicated by the diamonds in Figure 7H (bottom) by whole E-ChIP assays. Chromatin fragments were extracted from the EGFP+/−;Cre+/− and EGFP-C/ebpαDN+/−;Cre+/− transgenic embryos at 22 hpf, when the specific up-regulation of bmi1 transcripts has been readily detected (Figure 4A-B), and were immunoprecipitated with a polyclonal anti-EGFP antibody. DNA from the immunoprecipitates was amplified by PCR using primers located to the indicated genomic regions (I-XI, Figure 7H horizontal lines). The results showed that EGFP-tagged C/ebpαDN specifically interacted with the distant C/EBP motif II (Figure 7J-K). A slightly increased binding activity was also detected for the C/EBP motif I (Figure 7J-K). No binding activity was detected for other regions (Figure 7J-K). The results indicated that Bmi1 is a direct downstream target of C/ebpαDN signaling to promote the self-renewal of these short-lived, scl+ and PBI/ICM-derived HSPCs, allowing them to colonize the bilateral PD and CHT, where they initiate an abnormal proliferation and differentiation program toward erythroid lineage.
Bmi1 is a direct downstream target of C/ebpαDN. (A-C) External features of the LDL-EGFP+/−;Cre+/− transgenic embryos without injection (A, uninj), and the LDL- EGFP-C/ebpα+/−;Cre+/− transgenic embryos injected with either unrelated 5-mismatch (B) or bmi1-specifcic morpholino oligonucleotides (C). The circulation congestion is readily detected in the C/ebpαDN-expressing embryos injected with control morpholino (B, arrowheads) but largely rescued by bmi1 knockdown (C, arrowheads). Boxes with dashed lines indicate the regions amplified and shown at the bottom. (D-F) WISH analyses of αE1-globin in the indicated transgenic embryos at 80 hpf. The red arrowheads indicate that the increased number of erythrocytes with expression of αE1-globin transcripts can be corrected to normal level in the LDL-EGFP-C/ebpα+/−;Cre+/− embryos injected with the bmi1-specifcic morpholino oligonucleotide (F). (G) Statistics of circulation congestion in the transgenic embryos shown in panels A to C. Normal, Moderate, and Severe indicate the extent of circulation congestion at the CHT region. (H) Conservation of the zebrafish bmi1 promoter region 5.5 kb upstream of the TSS with murine Bmi1 promoter region by rVista software. Diamonds and corresponding dashed lines with Arabic numerals represent the positions of 11 predicted C/EBP binding motifs. Horizontal lines with Roman numerals I to XI indicate the locations of primers used in the E-ChIP analyses. (I) The nucleotide acid sequence of the zebrafish bmi1 promoter region between −5203 and −5023 bp, corresponding to the conserved region indicated by the red diamond in panel H. Red dots indicate the nucleotide acid identities compared with murine bmi1 promoter region between −11 780 and −11 612 bp upstream of TSS. The predicated C/EBP binding motif is indicated by a box. (J-K) E-ChIP analyses of chromatins extracted from the LDL-EGFP+/−;Cre+/− and LDL-EGFP-C/ebpα+/−;Cre+/− embryos at 22 hpf. PCR reactions were performed using the primers located to the indicated promoter region (I-XI) in panel H. The EGFP-tagged C/ebpα specifically interacts with the C/EBP binding motifs II and I. The results were repeated 3 times with separate clutches of embryos and batches of chromatin preparations.
Bmi1 is a direct downstream target of C/ebpαDN. (A-C) External features of the LDL-EGFP+/−;Cre+/− transgenic embryos without injection (A, uninj), and the LDL- EGFP-C/ebpα+/−;Cre+/− transgenic embryos injected with either unrelated 5-mismatch (B) or bmi1-specifcic morpholino oligonucleotides (C). The circulation congestion is readily detected in the C/ebpαDN-expressing embryos injected with control morpholino (B, arrowheads) but largely rescued by bmi1 knockdown (C, arrowheads). Boxes with dashed lines indicate the regions amplified and shown at the bottom. (D-F) WISH analyses of αE1-globin in the indicated transgenic embryos at 80 hpf. The red arrowheads indicate that the increased number of erythrocytes with expression of αE1-globin transcripts can be corrected to normal level in the LDL-EGFP-C/ebpα+/−;Cre+/− embryos injected with the bmi1-specifcic morpholino oligonucleotide (F). (G) Statistics of circulation congestion in the transgenic embryos shown in panels A to C. Normal, Moderate, and Severe indicate the extent of circulation congestion at the CHT region. (H) Conservation of the zebrafish bmi1 promoter region 5.5 kb upstream of the TSS with murine Bmi1 promoter region by rVista software. Diamonds and corresponding dashed lines with Arabic numerals represent the positions of 11 predicted C/EBP binding motifs. Horizontal lines with Roman numerals I to XI indicate the locations of primers used in the E-ChIP analyses. (I) The nucleotide acid sequence of the zebrafish bmi1 promoter region between −5203 and −5023 bp, corresponding to the conserved region indicated by the red diamond in panel H. Red dots indicate the nucleotide acid identities compared with murine bmi1 promoter region between −11 780 and −11 612 bp upstream of TSS. The predicated C/EBP binding motif is indicated by a box. (J-K) E-ChIP analyses of chromatins extracted from the LDL-EGFP+/−;Cre+/− and LDL-EGFP-C/ebpα+/−;Cre+/− embryos at 22 hpf. PCR reactions were performed using the primers located to the indicated promoter region (I-XI) in panel H. The EGFP-tagged C/ebpα specifically interacts with the C/EBP binding motifs II and I. The results were repeated 3 times with separate clutches of embryos and batches of chromatin preparations.
Discussion
In this study, we show, for the first time, a novel genetic and mechanistic link between the leukemogenic mutation C/ebpαDN and stem cell self-renewal regulator Bmi1, and how the C/ebpαDN-Bmi1 signaling axis acts as an intrinsic factor to extend the life span of the short-lived HSPCs with limited self-renewal potential during primitive hematopoietic wave. Once these transient HSPCs are reprogrammed into “long-lived,” they are able to survive into definitive hematopoiesis stage and colonize the definitive hematopoietic sites, where they undergo proliferative expansion resulting in severe dyserythropoiesis and embryonic lethality because of circulation congestion and cardiac edema.
Although the human and zebrafish C/ebpαDN were shown to inhibit the function of wild-type full-length C/ebpα in a DN fashion, the possibility of C/ebpαDN gain of function has not been formally excluded. Recently, we and others showed that the C/EBPαDN is able to bind to the promoters of α-catenin,21 MMP11, p84N5, and SMYD220 to suppress their transcription through recruiting transcription repressive complex PRC2 and subsequent trimethylation of H3K27,21 suggesting that C/EBPαDN acquires a novel function other than its ability to inactivate wild-type C/EBPα. Intriguingly, the current study also indicates that the C/ebpαDN can also act as a transactivator to directly or indirectly induce the expression of a panel of important epigenetic regulatory genes, including bmi1 and rae28 and set domain-containing genes prdm3 and prdm16, all of which have been reported to be critical for HSC self-renewal and/or leukemia transformation.32,37,41-43 In addition, our results did not exclude the possibility that the C/ebpαDN interacts with wild-type C/ebpα as a heterodimer at the promoter region of bmi1 gene to reverse the transcription suppressive effect endowed by full-length C/ebpα. In support of this model, the wild-type c/ebpα transcripts are detected in both HSPC-containing ICM and EMP-containing PBI regions (supplemental Figure 13). In addition, an increased level of Bmi1 has been observed in C/ebpα-deficient murine HSCs.16 To clarify the issue, a zebrafish C/ebpα antibody needs to be generated to test whether the wild-type C/ebpα also binds to the bmi1 promoter in a physiologic context. In any case, the current studies suggest that the C/ebpαDN is an important upstream signaling of epigenetic regulators to maintain HSC life span and probable stemness or self-renewal.
In addition to the demonstration of the C/ebpαDN-Bmi1 signaling as a regulator of HSPC activity, the study also uncovers a novel genetic evidence for the involvement of C/ebpαDN-Bmi1 in erythroid proliferative disorders. Although increased numbers of erythroid progenitors and erythroid cells in C/EBPα−/− fetal liver have been documented,44 and the C/ebpα deficiency fails to induce a bcr/abl-mediated murine myeloid leukemia but contributes to a fatal erythroleukemia instead,45 the underlying mechanism has not been defined. The Bmi1 is originally cloned from a human erythroleukemia cell line K562,46 and its role in erythropoiesis remains to be elucidated. It is probable that the increased erythropoiesis by Bmi1 is only a consequence of both short-lived HSPC expansion and differentiation in a microenvironment or “niche ” in the CHT region favoring erythroid fate. However, the possibility of direct involvement of Bmi1 in erythroid commitment and differentiation through epigenetic regulation of other erythroid-specific signaling transduction pathways rather than Gata-1–associated signaling needs to be excluded. In any case, the possible involvement of C/EBPαDN-Bmi1 axis in human erythroid disorders associated with dysplasia, such as polycythemia and erythroleukemia, warrants further investigation.
The bilateral PD was recently demonstrated to be a novel pathway for HSPC migration anteriorly to colonize the developing kidney marrow, the adult definitive hematopoietic organ in zebrafish. Although time-lapse analyses showed that the PD-located HSPCs are derived from the dorsal AGM, where the definitive HSCs originally develop, other sites contributing to the PD-HSPCs are possible. Indeed, an increased number of scl+ HSPCs detected in the PD of C/ebpαDN embryos raises the possibility of both PBI-derived EMPs and ICM-derived HSPCs as another source of PD-HSPCs. In addition, we also detected a specific up-regulation of CD41, a marker for PBI-derived EMPs, but not for ICM-derived HSPCs by C/ebpαDN at 22 hpf (supplemental Figure 14), suggesting that the PBI-derived EMPs are probably the major cell source of PD-HSPCs and cause of erythropoietic dysplasia.
In conclusion, it should be pointed out that a unique characteristic in the current study is the use of the lmo2 promoter to drive the expression of C/ebpαDN and Bmi1. The lmo2 promoter activity is transient, peaking at 22 hpf and hardly detected by WISH and Western blot analyses after 24 hpf, which results in the rapid wane of primitive hematopoietic wave. This feature provides a unique opportunity to investigate the fate and consequence of the short-lived HSPCs once reprogrammed by “pulsed” or instantaneous expression of either C/ebpαDN or Bmi1. However, a potential disadvantage of using the lmo2 promoter is that the progeny of these reprogrammed HSPCs in the CHT region ∼ 75 hpf can no longer be followed up by EGFP fluorescence because of the lack of the sustained expression of EGFP-tagged proteins. Replacing the lmo2 promoter with a ubiquitously and sustainably activated promoter such as β-actin promoter, in combination with the Tg(lmo2:Cre) transgenic line, should complement the technical limitation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the anonymous reviewer I for insightful comments, and Dr Li-Ping Su and all the members of our laboratory for helpful discussions.
This work was supported in part by the National Basic Research Program of China (2007CB947003 and 2011CB964803), the National Natural Science Foundation of China (30525019, 30830047), Science and Technology Commission of Shanghai Municipality (09XD1404700), and the Strategic Priority Research Program of the Chinese Academy of Science (XDA 01010106).
Authorship
Contribution: T.Z., L.W., and K.-Y.Z. performed experiments and analyzed data; M. Dong, P.-F.X., Y.C., and M. Deng assisted with experiments; M. Deng, S.-J.C., Z.C., and T.X.L. designed the research plan; and T.Z. and T.X.L. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ting Xi Liu, Laboratory of Development and Diseases, Institute of Health Sciences, Room 408, Building 1, 225 South Chong Qing Road, Shanghai, People's Republic of China 200025; e-mail: txliu@sibs.ac.cn.