Abstract
Plasmacytoid dendritic cells (pDCs) reside in bone marrrow and lymphoid organs in homeostatic conditions and typically secrete abundant quantities of type I interferons (IFNs) on Toll-like receptor triggering. Recently, a pDC population was identified within Peyer patches (PPs) of the gut that is distinguished by its lack of IFN production; however, the relationship of PP pDCs to pDCs in other organs has been unclear. We report that PP pDCs are derived from common DC progenitors and accumulate in response to Fms-like tyrosine kinase 3 ligand, yet appear divergent in transcription factor profile and surface marker phenotype, including reduced E2-2 and CCR9 expression. Type I IFN signaling via STAT1 has a cell-autonomous role in accrual of PP pDCs in vivo. Moreover, IFN-α enhances pDC generation from DC progenitors by a STAT1-dependent mechanism. pDCs that have been developed in the presence of IFN-α resemble PP pDCs, produce inflammatory cytokines, stimulate Th17 cell generation, and fail to secrete IFN-α on Toll-like receptor engagement. These results indicate that IFN-α influences the development and function of pDCs by inducing emergence of an inflammatory (Th17-inducing) antigen-presenting subset, and simultaneously regulating accumulation of pDCs in the intestinal microenvironment.
Introduction
Plasmacytoid dendritic cells (pDCs) were originally defined by their plasma cell-like morphology, surface marker profile, and ability to produce massive amounts of type I interferons (IFNs) in response to Toll-like receptor (TLR) triggering during viral infection.1-3 IFNs regulate antiviral genes and activate effector cells to initiate adaptive immunity4 ; thus, pDCs have been considered crucial to the host antiviral response. After TLR stimulation, pDCs mature into antigen-presenting cells (APCs) with up-regulated MHC class II and costimulatory molecules.5 pDCs reside in bone marrow (BM) and lymphoid organs and regulate adaptive immunity by modulating T helper (Th) cell polarization (eg, induction of Th1 and Th2 and suppression of Th17 generation), activating CD8+ cytotoxic T cells, and inducing regulatory T cell (Treg) function.6-8 Aberrant pDC activity is linked to autoimmune and inflammatory diseases, including systemic lupus erythromatosus, psoriasis, and diabetes.9,10 Moreover, IFN-α can be toxic in high concentrations and appears to contribute to autoimmunity when administered to humans11,12 ; however, whether type I IFNs influence pDC function and/or development has been unclear.
Recently, a pDC population was found in the subepithelial and interfollicular regions of the Peyer patches (PPs),13 lymphoid organs adjacent to the intestine, suggesting potential contact with T lymphocytes infiltrating the gut. The PP pDC subset is distinguished from pDCs in other tissues by its inability to secrete abundant type I IFN in response to the TLR agonist CpG. Conditioning by factors that are highly expressed in mucosal tissues, including TGF-β, IL-10, and prostaglandin E2, repressed IFN production from splenic pDCs,13 suggesting that the microenvironment of the gut regulates PP pDC function. Although the developmental origin of PP pDCs and their relationship to pDCs found in BM and other lymphoid organs has remained unclear, these results suggest the potential for localized extracellular signals to regulate pDC function.
Infection and other physiologic stresses stimulate cytokine output regionally and systemically. Certain cytokines, such as Fms-like tyrosine kinase 3 ligand (Flt3L) and GM-CSF, enhance proliferation of BM progenitor cells and support immune cell development, including pDCs and conventional DCs (cDCs).14,15 Cytokines elicit cellular responses by activating members of the STAT transcription factor family through receptor-Jak tyrosine kinase signaling cascades.16 STATs are critical mediators of emergency hematopoiesis and immune cell generation, with important functions in processes such as Th differentiation, DC development, and inflammation.17-19 Although IFN-α has classically been considered an antiproliferative factor, it was found to stimulate growth and survival of activated T cells20 and to promote the entry of dormant hematopoietic stem cells (HSCs) into the cell cycle via a STAT1-dependent pathway.21 These data suggest that IFN-α may enhance the generation of specific blood cell lineages.
pDC and cDC development initiates within the lin− Flt3+ progenitor population in the BM,22,23 proceeding sequentially through a macrophage-DC progenitor and a common DC progenitor (CDP).24-26 Under homeostatic conditions, cDC precursors exit the BM and seed lymphoid tissues before completing terminal stages of cDC maturation.27,28 Inflammation or high-dose GM-CSF supplements cDC numbers in peripheral organs by driving generation of “inflammatory cDCs.”29,30 By contrast, development of pDCs occurs in BM, with differentiated cells released to blood for dispersion to lymphoid tissues.24,25 Whether separate mechanisms regulate the generation of PP pDCs and pDCs found in peripheral lymphoid organs or BM has not been investigated.
We report here that PP pDCs are derived from CDPs and are induced by Flt3L, indicating similar developmental cues as pDCs localized in other tissues. Moreover, type I IFN receptor (IFNAR) and STAT1 have cell-autonomous roles in accrual of PP pDCs, and IFN-α enhances pDC production in BM and spleen via a STAT1-dependent mechanism, suggesting regulation of pDCs by type I IFN signals. pDCs generated in IFN-α conditions produce IL-6, IL-23, and TNF-α and elicit robust Th17 differentiation from naive CD4+ T cells, yet fail to secrete type I IFNs on TLR triggering, distinguishing them functionally from Flt3L-generated pDCs or pDCs present in steady state in BM or spleen. Furthermore, pDCs generated in response to IFN-α resemble PP pDCs, with each population demonstrating comparable Irf8 and Irf7 expression and reduced Tcf4 mRNA, relative to pDCs isolated from BM or spleen under homeostatic conditions, or pDCs generated ex vivo in Flt3L. Collectively, our results indicate that type I IFNs control mucosal pDC abundance in steady state, up-regulate pDC production from BM progenitors, and influence pDC function, rendering the emergence of an inflammatory (Th17-inducing) subset.
Methods
Mice
C57BL/6, CD45.1+ congenic B6.SJL, Rag1−/−, OT-II TCR transgenic, 129Sv/Ev, and Stat1−/− mice were purchased from NCI, The Jackson Laboratory, or Taconic Farms. Hematopoietic STAT3-deficient mice [ie, Tg(Tek-cre)12Flv, Stat3f/Δ] were obtained by breeding Tg(Tek-cre)12Flv transgenic mice and Stat3f/f mice.31 Ifnar−/− mice were provided by Dr Tadatsugu Taniguchi. All animals were housed in an SPF barrier facility at University of Texas M. D. Anderson Cancer Center. Experimental procedures were approved by the Institutional Animal Care and Use Committee (University of Texas M. D. Anderson Cancer Center).
Cytokine delivery by HGT
Mice were injected intravenously with pORF vector or pORF encoding Flt3L (10 μg), IFN-α (3 μg), or GM-CSF (5 μg) by hydrodynamic gene transfer (HGT).32 At day 4 after HGT, mice were sacrificed and BM, spleens, and PPs were collected for analysis.
Flow cytometry, cell isolation, adoptive transfer, and CFSE staining
Single-cell suspensions were prepared from BM and spleens after RBC lysis, or from PPs of the mice of indicated genotypes; cells were stained with fluorescently labeled antibodies (BD Biosciences PharMingen or eBioscience) and analyzed by FACS. Lin− Flt3+ progenitors, pDCs, and cDCs were isolated by FACS after magnetic bead depletion. In some experiments, purified CDPs (BM lin−Flt3+ CD115+ c-kitint cells) were transferred to Ifnar−/− and Stat1−/− mice by intravenous injection. CFSE labeling was performed according to the manufacturer's protocol (Invitrogen).
Cytokine stimulation and immunoblotting
Lin− Flt3+ progenitors were cultured in RPMI medium supplemented with 10% FCS, 1mM sodium pyruvate, and 5 × 10−5M β-mercaptoethanol. Murine cytokines Flt3L, GM-CSF, or IFN-α were added to cultures, and cells, were collected for FACS, RNA isolation, or whole cell lysate preparation. Immunoblotting was performed with antibodies specific for phospho-STAT1, phospho-STAT5 (Cell Signaling), total STAT1, or total STAT5 (Santa Cruz Biotechnology). RNA isolation and quantitative PCR were performed as described.33 Threshold cycle values for each gene were normalized to RPL13A threshold cycle values, and arbitrary expression levels were calculated as described.33
ELISAs and Th differentiation assays
FACS-purified pDCs were stimulated with 5 μM of CpGA overnight; cytokine levels in culture supernatants were determined by ELISA. For in vitro Th differentiation assays, pDCs and IFN-α–induced pDCs were pretreated with or without CpGA for 4 hours and then cultured with CD4+ CD25− CD62L+ CD44− naive T cells from OT-II mice and OVA peptide 323-339. In some cultures, the medium was supplemented with TGF-β, TGF-β + IFN-α, Th-polarizing cytokines, antibody cocktail,34 or IL-6–blocking antibodies. Th differentiation was determined at day 4 by intracellular staining after restimulation with phorbol myristate acetate and ionomycin in the presence of BD Golgi-Stop. For in vivo assays, pDCs or IFN-α–induced pDCs were pulsed with OVA peptide 323-339 and CpGA for 4 hours, then transferred subcutaneously into the tail base of Rag1−/− mice that received naive OT-II CD4+ T cells intravenously. Inguinal lymph nodes were collected at day 7 and Th induction was determined.
Reporter assays, ChIP, and EMSAs
The murine DC cell line D2SC/1 was transiently transfected with pGL4.12 containing an Irf8 promoter fragment (−351 to +52) with an intact or mutated STAT element,33 pMNC-mSTAT1, pMNC-mSTAT2, and phRL-TK (Promega) via Lipofectamine2000 (Invitrogen) for 36 hours. Cells were treated with or without IFN-α (300 U/mL) for 2 hours, and luciferase activity was measured by the Dual-Luciferase Reporter Assay System (Promega). For EMSAs, D2SC/1 cells were stimulated with IFN-α for 30 minutes; nuclear extracts (10 μg) were incubated with 32P-labeled oligonucleotides corresponding to the STAT consensus site in the murine Irf8 promoter or a STAT1-binding oligonucleotide (Santa Cruz Biotechnology) with or without unlabeled competitor oligonucleotides or STAT1 antibody (Santa Cruz Biotechnology). ChIPs were performed with a ChIP Assay kit (Upstate Biotechnology) using a mixture of antibodies for STAT1 and p-STAT1 (Santa Cruz Biotechnology; Cell Signaling).
Statistical analysis
Data are presented as mean ± SEM. The statistical significance between groups was calculated by 2-tailed t test using GraphPad Prism Version 4 software (www.graphpad.com).
Results
Developmental regulation of PP pDCs
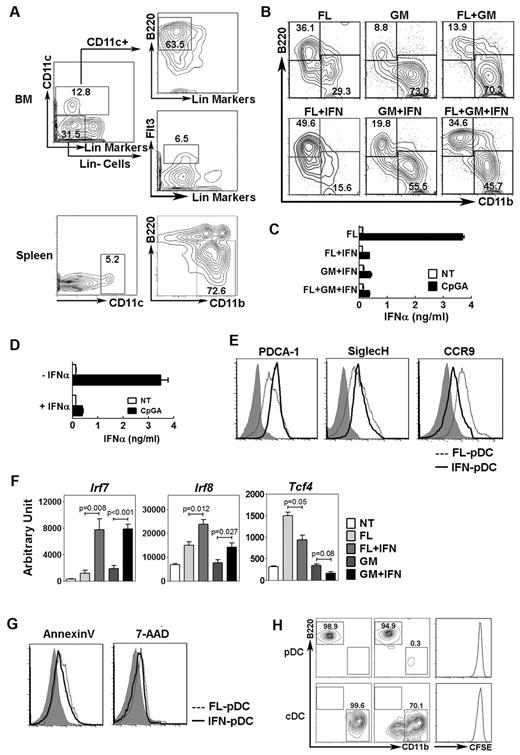
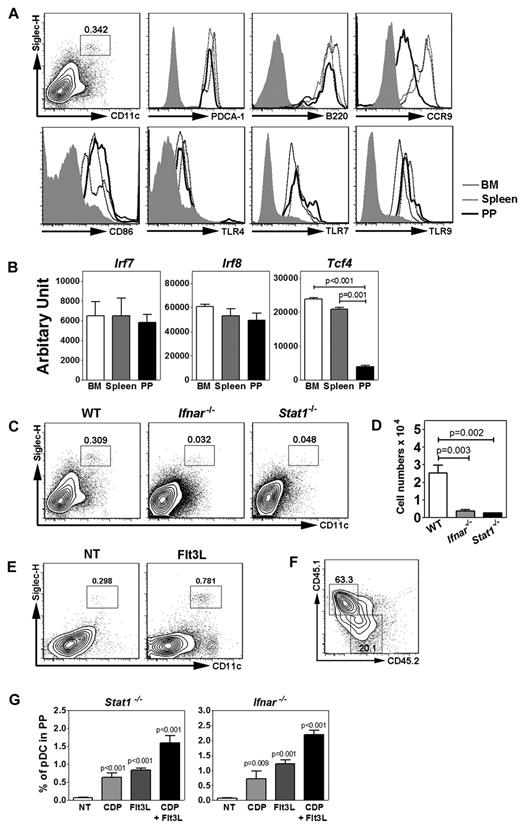
To investigate the development of PP pDCs, we identified a discrete CD11c+ Siglec H+ population that coexpressed PDCA-1 and B220, similar to PP pDCs described previously13 and pDCs from BM or spleen (Figure 1A). Expression of the mucosal migratory receptor CCR9 was reduced on PP pDCs relative to pDCs isolated from BM or spleen under homeostatic conditions; however, PP pDCs expressed similar, if not higher, amounts of cell surface CD86 and TLR4 as well as intracellular TLR7 and TLR9 compared with BM or splenic pDCs (Figure 1A). Moreover, PP pDCs had reduced amounts of Tcf4 (E2-2) mRNA yet showed similar Irf7 and Irf8 expression compared with pDCs from other tissues (Figure 1B). These data suggest that, although PP pDCs are phenotypically similar to pDCs found in BM or spleen, they may have undergone distinct conditioning events to affect expression of pDC-related genes.
Characterization of PP pDCs and their requirement for IFN-STAT1 signaling. (A) Gating strategy of PP pDCs. Surface expression of PDCA-1, B220, CCR9, CD86, and TLR4 and intracellular TLR7 and TLR9 expression in pDCs from BM, spleen, and PP. Shaded area represents isotype control staining. (B) Gene expression of Irf7, Irf8, and Tcf4 in pDCs from BM, spleen, and PP, determined by quantitative PCR. (C-D) Proportion (C) and absolute number (D) of PP pDCs from Stat1−/−, Ifnar−/−, and wild-type (WT) controls. Error bars represent SEM of results from 4-8 mice. (E) PP pDC proportions in mice receiving HGT with pORF vector (NT) or pORF encoding Flt3L (10 μg), 4 days after treatment. (F) PP pDC proportions in Stat1−/− mice at 8 days after transfer of 105 congenic CD45.1+ DC progenitors (BM lin− Flt3+ CD115+ c-kitint cells) intravenously. Similar results were obtained after transfer into Ifnar1−/− mice (not shown). (G) PP pDC proportions in Stat1−/− or Ifnar1−/− mice 8 days after transfer of 105 wild-type DC progenitors (BM lin− Flt3+ CD115+ c-kitint cells) intravenously (CDP) or in animals left untreated (NT), as indicated. Some mice received Flt3L HGT 2 days before DC progenitor transfer (CDP + Flt3L) or Flt3L HGT alone (Flt3L), as shown. (B-G) Data represent 2 or 3 independent experiments. N = 4-10.
Characterization of PP pDCs and their requirement for IFN-STAT1 signaling. (A) Gating strategy of PP pDCs. Surface expression of PDCA-1, B220, CCR9, CD86, and TLR4 and intracellular TLR7 and TLR9 expression in pDCs from BM, spleen, and PP. Shaded area represents isotype control staining. (B) Gene expression of Irf7, Irf8, and Tcf4 in pDCs from BM, spleen, and PP, determined by quantitative PCR. (C-D) Proportion (C) and absolute number (D) of PP pDCs from Stat1−/−, Ifnar−/−, and wild-type (WT) controls. Error bars represent SEM of results from 4-8 mice. (E) PP pDC proportions in mice receiving HGT with pORF vector (NT) or pORF encoding Flt3L (10 μg), 4 days after treatment. (F) PP pDC proportions in Stat1−/− mice at 8 days after transfer of 105 congenic CD45.1+ DC progenitors (BM lin− Flt3+ CD115+ c-kitint cells) intravenously. Similar results were obtained after transfer into Ifnar1−/− mice (not shown). (G) PP pDC proportions in Stat1−/− or Ifnar1−/− mice 8 days after transfer of 105 wild-type DC progenitors (BM lin− Flt3+ CD115+ c-kitint cells) intravenously (CDP) or in animals left untreated (NT), as indicated. Some mice received Flt3L HGT 2 days before DC progenitor transfer (CDP + Flt3L) or Flt3L HGT alone (Flt3L), as shown. (B-G) Data represent 2 or 3 independent experiments. N = 4-10.
To understand the origin of PP pDCs, we first investigated whether they were controlled by STAT3, which has been reported to regulate abundance of pDCs as well as cDC subsets.35 Mice deficient in STAT3 expression in hematopoietic tissues (Tg(Tek-cre)12Flv, Stat3f/Δ mice; termed herein Tie2cre+Stat3f/Δ)31 showed a severe reduction in PPs with some animals lacking PPs altogether (not shown). We therefore extended our analysis to Stat1−/− mice, since the role for STAT1 in DC development has not been determined previously. Although Stat1−/− mice contain normal amounts of pDCs in BM and spleen (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), they show a significant reduction in PP pDCs (Figure 1C-D). To determine potential STAT1-activating signals that control PP pDC accumulation, we measured PP pDC amounts in Ifnar−/− mice. These animals also showed a reduction in PP pDCs relative to wild-type controls, whereas pDC abundance was normal in BM or spleen (Figure 1C-D; supplemental Figure 1), suggesting that type I IFN signals via STAT1 are necessary for controlling PP pDC accumulation.
To determine whether PP pDCs were regulated by similar mechanisms as BM or splenic pDCs, we assessed their ability to respond to Flt3L overexpression mediated by HGT. This method induces circulating Flt3L from basal levels of 500 ng/mL to 2 μg/mL by day 4 of treatment, and significantly stimulates pDC amounts in PPs by day 4, indicating the PP pDC subset is responsive to Flt3L (Figure 1E; and data not shown). By contrast, IFN-α HGT did not induce PP pDC amounts significantly, suggesting that systemic IFN-α has little effect on PP pDC abundance (not shown). Purified CDPs from CD45.1+ congenic mice were able to reconstitute PP pDCs in Stat1−/− or Ifnar−/− animals (Figure 1F; and data not shown). Flt3L HGT expanded PP pDC proportions and absolute numbers in Stat1−/− or Ifnar−/− mice (Figure 1G; and data not shown). Moreover, purified CDPs transferred into Stat1−/− or Ifnar−/− animals appeared to respond to Flt3L overexpression, as judged by increased PP pDC amounts in Stat1−/− or Ifnar−/− mice treated with Flt3L and CDP transfer versus either condition alone (Figure 1G). These data suggest PP pDCs have a similar developmental origin as BM and splenic pDCs yet use cell-intrinsic type I IFN-STAT1 signals and potentially localized IFN cues for accrual in PPs.
IFN-α induces pDC generation from DC progenitors
Because type I IFN signals were important in regulating PP pDCs, we asked whether IFN-α affected the development of pDCs from hematopoietic progenitor cells in response to Flt3L or the pDC suppressive cytokine GM-CSF. These studies showed that IFN-α enhanced production of pDCs yet inhibited CD11c+ CD11b+ cDC generation from DC progenitors in Flt3L cultures (Figure 2A-B; supplemental Table 1). Furthermore, IFN-α partially overcame the suppressive effect of GM-CSF on pDC formation (Figure 2B; supplemental Table 1). Our gating strategy also detected a minor CD11c+ CD11b+ B220+ population that did not seem to be strongly influenced by IFN-α (Figure 2B); the identity of this population remains to be determined. pDCs generated in the presence of IFN-α demonstrated a diminished capacity to secrete IFN-α in response to the TLR ligand CpGA or influenza in vitro, relative to Flt3L-induced pDCs (Figure 2C; supplemental Figure 2). Moreover, exposure of BM pDCs purified from mice in steady-state conditions to IFN-α suppressed their ability to secrete IFN-α after TLR ligation (Figure 2D). pDCs generated in Flt3L + IFN-α cultures expressed PDCA-1, Siglec-H, CCR7, and modest amounts of CD8α, similar to Flt3L-generated pDCs, whereas neither pDC population expressed the migratory cDC marker CD103 or the B-cell marker CD19 (Figure 2E; supplemental Figure 3). pDCs exposed to IFN-α showed reduced CCR9 similar to PP pDCs yet retained the pDC ultrastructural morphology with well-developed endoplasmic reticulum (Figure 2E; and data not shown),1 consistent with their identity as a pDC population.
Effects of IFN-α on pDC development and function. (A) FACS purification strategy of DC progenitor cells (lin− CD11c− B220− Flt3+ cells) and pDCs (CD11c+ CD11b− B220+ cells) from BM, as well as cDCs (CD11c+ CD11b+ B220− cells) from spleen. (B) Proportion of pDCs (top left quadrant) and cDCs (bottom right quadrant) after 4 days of culture of DC progenitors in Flt3L (FL, 50 ng/mL), GM-CSF (GM, 25 ng/mL), IFN-α (IFN, 300 U/mL), or the indicated combinations. Results were gated on the CD11c+ population (not shown); B220 and CD11b analysis is shown. Data represent 3 independent experiments. (C) pDCs were generated from total BM cells cultured in Flt3L, Flt3L + IFN-α, GM-CSF + IFN-α, or Flt3L + GM-CSF + IFN-α as indicated in panel B for 4 days. pDCs were then purified by FACS and stimulated with or without 5 μM CpGA overnight, as indicated. Culture supernatants were assayed by ELISA. Error bars represent SEM of values obtained from triplicate wells. (D) pDCs were purified from total BM and treated with or without IFN-α for 24 hours in the presence of Flt3L. pDCs were then stimulated with CpGA and analyzed for IFN-α secretion as indicated in panel C. (E) Representative surface staining of pDCs purified from Flt3L alone (dashed line) or Flt3L + IFN-α cultures (solid line) with antibodies to PDCA-1, Siglec-H, or CCR9. Shaded area represents isotype control staining. Data represent 3 independent experiments. (F) Gene expression in DC progenitors stimulated with Flt3L, Flt3L + IFN-α, GM-CSF, or GM-CSF + IFN-α for 4 days, as indicated, or immediately after purification (NT). Data are the mean ± SEM of 3-5 independent experiments. (G) Annexin V and 7-amino-actinomycin D (7-AAD) staining of DC progenitors cultured 4 days with Flt3L (dashed line) or Flt3L + IFN-α (solid line). Shaded area represents isotype control staining. Data represent 3 independent experiments. (H) Phenotype of FACS-purified BM pDCs and splenic cDCs. After purification, pDCs and cDCs were cultured 4 days in the presence of Flt3L + IFN-α and analyzed by flow cytometry (middle panels) or analyzed immediately (left panels); pDC and cDC frequencies within the CD11c+ population are shown. (Right panels) Purified DCs were stained with CFSE, cultured 4 days in Flt3L + IFN-α, and analyzed by flow cytometry.
Effects of IFN-α on pDC development and function. (A) FACS purification strategy of DC progenitor cells (lin− CD11c− B220− Flt3+ cells) and pDCs (CD11c+ CD11b− B220+ cells) from BM, as well as cDCs (CD11c+ CD11b+ B220− cells) from spleen. (B) Proportion of pDCs (top left quadrant) and cDCs (bottom right quadrant) after 4 days of culture of DC progenitors in Flt3L (FL, 50 ng/mL), GM-CSF (GM, 25 ng/mL), IFN-α (IFN, 300 U/mL), or the indicated combinations. Results were gated on the CD11c+ population (not shown); B220 and CD11b analysis is shown. Data represent 3 independent experiments. (C) pDCs were generated from total BM cells cultured in Flt3L, Flt3L + IFN-α, GM-CSF + IFN-α, or Flt3L + GM-CSF + IFN-α as indicated in panel B for 4 days. pDCs were then purified by FACS and stimulated with or without 5 μM CpGA overnight, as indicated. Culture supernatants were assayed by ELISA. Error bars represent SEM of values obtained from triplicate wells. (D) pDCs were purified from total BM and treated with or without IFN-α for 24 hours in the presence of Flt3L. pDCs were then stimulated with CpGA and analyzed for IFN-α secretion as indicated in panel C. (E) Representative surface staining of pDCs purified from Flt3L alone (dashed line) or Flt3L + IFN-α cultures (solid line) with antibodies to PDCA-1, Siglec-H, or CCR9. Shaded area represents isotype control staining. Data represent 3 independent experiments. (F) Gene expression in DC progenitors stimulated with Flt3L, Flt3L + IFN-α, GM-CSF, or GM-CSF + IFN-α for 4 days, as indicated, or immediately after purification (NT). Data are the mean ± SEM of 3-5 independent experiments. (G) Annexin V and 7-amino-actinomycin D (7-AAD) staining of DC progenitors cultured 4 days with Flt3L (dashed line) or Flt3L + IFN-α (solid line). Shaded area represents isotype control staining. Data represent 3 independent experiments. (H) Phenotype of FACS-purified BM pDCs and splenic cDCs. After purification, pDCs and cDCs were cultured 4 days in the presence of Flt3L + IFN-α and analyzed by flow cytometry (middle panels) or analyzed immediately (left panels); pDC and cDC frequencies within the CD11c+ population are shown. (Right panels) Purified DCs were stained with CFSE, cultured 4 days in Flt3L + IFN-α, and analyzed by flow cytometry.
The expression of DC transcription factors in developing progenitors was modulated by IFN-α exposure. Irf8, Sfpi1 (PU.1), Spib and Tcf4 (E2-2) and, to a lesser extent, Irf7, were induced in DC progenitors by Flt3L, whereas addition of IFN-α significantly up-regulated Irf7 and Irf8, maintained Sfpi1 and Spib, and repressed Tcf4 mRNA amounts (Figure 2F; and data not shown). By contrast, exposure to GM-CSF either modestly increased or had no effect on Irf7, Irf8, and Tcf4 yet stimulated Irf4, Sfpi1, and Spib expression; IFN-α addition significantly enhanced Irf7 and Irf8 expression and inhibited Tcf4 (Figure 2F; and data not shown). We also observed reduced Tcf4 expression in FACS-purified pDCs isolated from animals treated with IFN-α HGT relative to pDCs from steady state (not shown). IFN-α did not induce pDC apoptosis or stimulate phenotypic conversion between pDCs and cDCs; however, a subset of cDCs lost CD11b expression with IFN-α exposure (Figure 2G-H). CFSE labeling suggested that IFN-α had minimal effects on proliferation of DCs or DC progenitors in vitro (Figure 2H; supplemental Figure 4). Expression of the type I IFN receptor was detected on DC progenitor cells (supplemental Figure 5). Thus, our results suggest that IFN-α signaling in DC progenitors enhances pDC development; moreover, IFN-α conditions pDCs to down-regulate Tcf4 and CCR9 and suppress TLR-responsive IFN secretion.
IFN-α augments pDC production in vivo
To investigate the role of IFN-α in DC generation in vivo, mice were treated by HGT, which induces circulating IFN-α to amounts similar as those observed in infection with rhabdo- or orthomyxovirus family members VSV and Thogoto virus (Figure 3A; and data not shown).36 GM-CSF HGT was also performed to determine whether it suppressed pDCs in vivo and what effect IFN-α might have on GM-CSF-mediated pDC inhibition. Mice treated with the HGT pORF vector had similar amounts of pDCs and cDCs in BM and spleen compared with untreated controls (not shown), indicating that HGT does not affect DC homeostasis. IFN-α treatment caused a substantial reduction (∼ 40%) in total BM cellularity (not shown) and suppressed cDC numbers and proportions, in agreement with previous reports.37 By contrast, pDC absolute numbers were increased significantly in BM and spleen of IFN-α–treated mice relative to controls. pDC proportions were also increased, a change that reflects both the increase in pDC absolute numbers as well as the reduction in cDCs (Figure 3B-C).
Effects of IFN-α on pDCs in vivo. C57BL/6 mice were injected intravenously with pORF vector or pORF encoding GM-CSF (GM, 5 μg), IFN-α (IFN, 3 μg), or a combination of both plasmids (GM + IFN, 8 μg) by HGT, or left untreated (NT). Four days later, BM and spleens were collected for analysis. (A) ELISA results on sera samples collected at day 4 from mice treated by HGT, as indicated. Data are the mean ± SEM of 8-10 individual samples. (B) Proportion of pDCs (top left quadrant) and cDCs (botom right quadrant) in BM and spleen of mice treated by HGT, as indicated. Results were gated on the CD11c+ population (not shown); B220 and CD11b analysis is shown. Data represent 3 independent experiments. (C) Absolute pDC and cDC numbers in BM (2 femurs + 2 tibias) and spleen of mice treated by HGT, as indicated. n = 8 to 10 (3 independent experiments). Error bars represent SEM. (D) IFN-α production from pDCs sorted by FACS from BM of HGT-treated mice on CpGA stimulation. Error bars represent SEM of 8 to 10 individual samples. (E) Numbers of DC progenitors (lin− Flt3+ cells) in mice treated by HGT, as indicated. n = 6 (2 independent experiments). Error bars represent SEM.
Effects of IFN-α on pDCs in vivo. C57BL/6 mice were injected intravenously with pORF vector or pORF encoding GM-CSF (GM, 5 μg), IFN-α (IFN, 3 μg), or a combination of both plasmids (GM + IFN, 8 μg) by HGT, or left untreated (NT). Four days later, BM and spleens were collected for analysis. (A) ELISA results on sera samples collected at day 4 from mice treated by HGT, as indicated. Data are the mean ± SEM of 8-10 individual samples. (B) Proportion of pDCs (top left quadrant) and cDCs (botom right quadrant) in BM and spleen of mice treated by HGT, as indicated. Results were gated on the CD11c+ population (not shown); B220 and CD11b analysis is shown. Data represent 3 independent experiments. (C) Absolute pDC and cDC numbers in BM (2 femurs + 2 tibias) and spleen of mice treated by HGT, as indicated. n = 8 to 10 (3 independent experiments). Error bars represent SEM. (D) IFN-α production from pDCs sorted by FACS from BM of HGT-treated mice on CpGA stimulation. Error bars represent SEM of 8 to 10 individual samples. (E) Numbers of DC progenitors (lin− Flt3+ cells) in mice treated by HGT, as indicated. n = 6 (2 independent experiments). Error bars represent SEM.
GM-CSF administration markedly suppressed the number and frequency of BM and spleen pDCs, whereas increased cDCs were observed, compared with pORF-treated mice (Figure 3B-C). GM-CSF also induced splenomegaly (ie, a 3- to 5-fold enlargement in spleen weight) in recipient mice (not shown), possibly resulting from accumulation of DCs and other cells. When GM-CSF and IFN-α were codelivered, IFN-α partially restored pDC amounts (Figure 3B-C), consistent with its ability to stimulate pDC production in the presence of GM-CSF ex vivo (Figure 2). pDCs generated in the presence of IFN-α in vivo failed to secrete a significant amount of IFN-α on CpGA stimulation and showed reduced CCR9 surface amounts (Figure 3D; supplemental Figure 6); however, Siglec-H and PDCA-1 were expressed at comparable, if not higher, amounts compared with pDCs isolated under steady-state conditions (not shown). Furthermore, DC progenitor numbers increased 2- to 2.5-fold in IFN-α–treated mice, compared with pORF-treated controls (Figure 3E). Our data thus suggest that IFN-α not only promotes pDC generation from DC progenitors but also drives the formation of DC progenitors from HSCs, and conditions pDCs in vivo to a state in which they are unable to produce type I IFN. These results suggest that IFN-α affects both the developmental pathway and function of pDCs in vivo.
IFN-α effects on pDC development are independent of STAT3
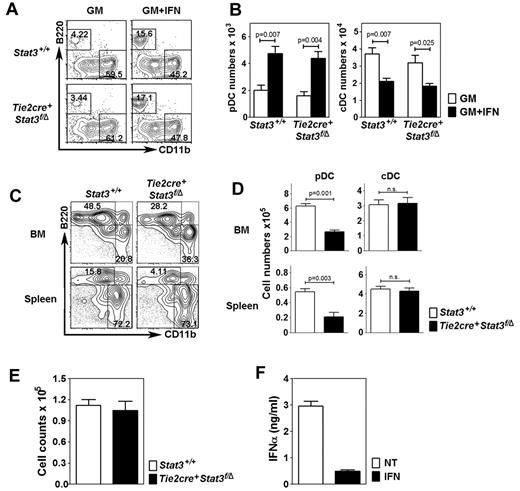
IFN-α stimulated STAT3 tyrosine phosphorylation in DC progenitor cells and pDCs (not shown); therefore, we tested whether STAT3 mediated IFN-α–induced pDC development ex vivo using BM cells from Tie2cre+Stat3f/Δ mice. Because Flt3L depends on STAT3, we used GM-CSF, which supports pDC generation in the presence of IFN-α (Figures 2–3). Similar pDC amounts were generated from Tie2cre+Stat3f/Δ and littermate control Stat3+/+ BM in cultures with GM-CSF + IFN-α, indicating that STAT3 is not required for IFN-α–driven pDC development (Figure 4A-B). Increased pDC numbers and proportions were found in GM-CSF + IFN-α cultures versus GM-CSF alone (Figure 4A-B), in agreement with the ability of IFN-α to partially restore pDC formation in GM-CSF (Figures 2B, 3B-C). Tie2cre+Stat3f/Δ mice produced pDCs in vivo yet showed reduced proportions and absolute numbers in BM and spleen (Figure 4C-D). By contrast, cDC numbers were suppressed by IFN-α similarly in Tie2cre+Stat3f/Δ and Stat3+/+ BM samples (Figure 4B), yet not significantly affected by STAT3 deficiency in vivo (Figure 4C-D). These results indicate that regulation of cDCs by IFN-α or in homeostatic conditions in vivo is STAT3-independent. Furthermore, DC progenitor numbers in BM are comparable between Tie2cre+Stat3f/Δ mice and Stat3+/+ mice (Figure 4E). As we reported previously, STAT3-deficient pDCs secrete abundant amounts of IFN-α on CpGA stimulation, similar to wild-type pDCs33 ; this capability was diminished after IFN-α treatment (Figure 4F; and data not shown). These results are consistent with an established role for STAT3 in pDC development,33,35 without affecting DC progenitor numbers. However, STAT3-independent pathways contribute to pDC generation under homeostatic conditions, as well as IFN-α–driven pDC production and IFN-α–dependent repression of pDC IFN-secretory responses.
STAT3-independent development of pDCs by IFN-α. (A) Proportion of pDCs (top left quadrant) and cDCs (bottom right quadrant) in total BM cultures (105 cells) from Tie2cre+Stat3f/Δ mice or littermate control (Stat3+/+) samples, after 4 days in GM-CSF or GM-CSF + IFN-α. Results were gated on the CD11c+ population (not shown); B220 and CD11b analysis is shown. Data represent 3 independent experiments. (B) Absolute number of pDCs and cDCs in total BM cultures shown in panel A. Error bars represent SEM of 3 independent experiments. (C) Proportion of pDCs and cDCs in BM and spleen of Tie2cre+Stat3f/Δ mice and Stat3+/+ mice, as indicated. Results were analyzed as indicated in panel A. Data represent 3 independent experiments. (D) Absolute number of pDCs and cDCs in BM and spleen of Tie2cre+Stat3f/Δ mice and Stat3+/+ mice, as indicated. Data are the mean ± SEM from 4-8 mice. (E) Absolute number of DC progenitors (lin− Flt3+ cells) in Tie2cre+Stat3f/Δ mice and Stat3+/+ mice, as indicated. Data are the mean ± SEM from 4-8 mice. (F) CpGA-induced IFN-α production from pDCs sorted from BM of Tie2cre+Stat3f/Δ mice before or after IFN-α treatment. Error bars represent SEM of 4 to 8 individual samples.
STAT3-independent development of pDCs by IFN-α. (A) Proportion of pDCs (top left quadrant) and cDCs (bottom right quadrant) in total BM cultures (105 cells) from Tie2cre+Stat3f/Δ mice or littermate control (Stat3+/+) samples, after 4 days in GM-CSF or GM-CSF + IFN-α. Results were gated on the CD11c+ population (not shown); B220 and CD11b analysis is shown. Data represent 3 independent experiments. (B) Absolute number of pDCs and cDCs in total BM cultures shown in panel A. Error bars represent SEM of 3 independent experiments. (C) Proportion of pDCs and cDCs in BM and spleen of Tie2cre+Stat3f/Δ mice and Stat3+/+ mice, as indicated. Results were analyzed as indicated in panel A. Data represent 3 independent experiments. (D) Absolute number of pDCs and cDCs in BM and spleen of Tie2cre+Stat3f/Δ mice and Stat3+/+ mice, as indicated. Data are the mean ± SEM from 4-8 mice. (E) Absolute number of DC progenitors (lin− Flt3+ cells) in Tie2cre+Stat3f/Δ mice and Stat3+/+ mice, as indicated. Data are the mean ± SEM from 4-8 mice. (F) CpGA-induced IFN-α production from pDCs sorted from BM of Tie2cre+Stat3f/Δ mice before or after IFN-α treatment. Error bars represent SEM of 4 to 8 individual samples.
STAT1 mediates pDC generation in response to IFN-α
To study the signaling pathways involved in IFN-α–enhanced pDC generation, we measured STAT1 and STAT5 tyrosine phosphorylation in cytokine-treated DC progenitors. We found that IFN-α strongly activated STAT1 but very little STAT5, GM-CSF stimulated STAT5, and modest amounts of STAT1, whereas GM-CSF + IFN-α activated both STAT1 and STAT5 (Figure 5A). The preferred activation of STAT1 by IFN-α suggested that it might be involved in pDC production. To test this, we examined the ability of Stat1−/− BM cells to generate pDCs ex vivo. These experiments showed that the IFN-α–mediated increase in pDC production was abrogated in Stat1−/− cells, whereas GM-CSF–responsive cDC generation was observed (Figure 5B). Stat1−/− mice have comparable pDC and cDC amounts in BM and spleen under steady-state conditions compared with Stat1+/+ animals, yet IFN-α HGT failed to promote pDC induction in these organs in Stat1−/− mice (Figure 5C-D; supplemental Figure 1). Moreover, suppression of cDCs by IFN-α appeared to require STAT1 (Figure 5C-D). Thus, our results indicate that STAT1 is necessary for enhanced pDC generation by IFN-α in vitro and in vivo, but not for homeostatic pDC or cDC production in BM and spleen.
Role for STAT1 in IFN-α–mediated pDC development. (A) STAT activation and expression in IFN-α–, GM-CSF-, or IFN-α + GM-CSF-treated (30 minutes) DC progenitors (lin− Flt3+ cells), determined by immunoblotting, as indicated. (B) Proportion of pDCs (top left quadrant) and cDCs (bottom right quadrant) in total BM cultures from Stat1−/− or wild-type mice in GM-CSF or GM-CSF + IFN-α for 4 days. Results were gated on the CD11c+ population (not shown); B220 and CD11b analysis is shown. n = 5. (C-D) Proportion (C) and absolute numbers (D) of pDCs and cDCs in BM and spleen of Stat1−/− or wild-type mice treated by HGT, as indicated. Results were analyzed as indicated in panel B. n = 5. Error bars represent SEM. (E) Gene expression in D2SC/1 cells stimulated with GM-CSF, IFN-α, or both for 2 hours or left unstimulated, as indicated. Error bars represent SEM. (F) EMSAs with nuclear extracts from D2SC/1 cells stimulated with or without IFN-α for 30 minutes. Some samples contained STAT1 competitor antibody or competitor oligonucleotides, as indicated. d indicates Irf8 STAT site; m, mutated Irf8 STAT site; and s, STAT1-consensus oligonucleotide. (G) Luciferase assays in IFN-α–stimulated (IFN) or untreated (NT) D2SC/1 cells transfected with empty vector (pGL4.12) or an Irf8 reporter (pGL4.12/IRF8) with an intact (WT) or mutated (MU) STAT element, plus pMNC/mSTAT1, pMNC/mSTAT2, and phRL-TK plasmids. Error bars represent SEM. (H) ChIPs from D2SC/1 cells with or without IFN-α stimulation (1 hour) with STAT1 antibodies or IgG controls, as indicated. PCR reactions were performed with total cell lysates (input) or immunoprecipitated samples, as shown. (A-H) Results represent 3 independent experiments.
Role for STAT1 in IFN-α–mediated pDC development. (A) STAT activation and expression in IFN-α–, GM-CSF-, or IFN-α + GM-CSF-treated (30 minutes) DC progenitors (lin− Flt3+ cells), determined by immunoblotting, as indicated. (B) Proportion of pDCs (top left quadrant) and cDCs (bottom right quadrant) in total BM cultures from Stat1−/− or wild-type mice in GM-CSF or GM-CSF + IFN-α for 4 days. Results were gated on the CD11c+ population (not shown); B220 and CD11b analysis is shown. n = 5. (C-D) Proportion (C) and absolute numbers (D) of pDCs and cDCs in BM and spleen of Stat1−/− or wild-type mice treated by HGT, as indicated. Results were analyzed as indicated in panel B. n = 5. Error bars represent SEM. (E) Gene expression in D2SC/1 cells stimulated with GM-CSF, IFN-α, or both for 2 hours or left unstimulated, as indicated. Error bars represent SEM. (F) EMSAs with nuclear extracts from D2SC/1 cells stimulated with or without IFN-α for 30 minutes. Some samples contained STAT1 competitor antibody or competitor oligonucleotides, as indicated. d indicates Irf8 STAT site; m, mutated Irf8 STAT site; and s, STAT1-consensus oligonucleotide. (G) Luciferase assays in IFN-α–stimulated (IFN) or untreated (NT) D2SC/1 cells transfected with empty vector (pGL4.12) or an Irf8 reporter (pGL4.12/IRF8) with an intact (WT) or mutated (MU) STAT element, plus pMNC/mSTAT1, pMNC/mSTAT2, and phRL-TK plasmids. Error bars represent SEM. (H) ChIPs from D2SC/1 cells with or without IFN-α stimulation (1 hour) with STAT1 antibodies or IgG controls, as indicated. PCR reactions were performed with total cell lysates (input) or immunoprecipitated samples, as shown. (A-H) Results represent 3 independent experiments.
We previously reported that GM-CSF inhibits pDC generation by suppressing Irf8 transcription via STAT5.33 We therefore examined whether IFN-α–responsive STAT1 signaling mediated Irf8 transcription. The murine DC cell line D2SC/1 recapitulated IFN-α–dependent induction of Irf7 and Irf8 (Figure 5E), confirming its ability to model DC Irf8 transcriptional regulation.33 EMSAs showed that IFN-α activated STAT1 binding to the Irf8 promoter STAT consensus site, as revealed by competition with a STAT1 consensus oligonucleotide or STAT1 blocking antibodies (Figure 5F). Moreover, IFN-α–stimulated transcription of an Irf8 reporter construct was dependent on an intact STAT element (Figure 5G). ChIP experiments demonstrated that STAT1 was recruited to the Irf8 promoter in vivo on IFN-α stimulation (Figure 5H). Thus, we conclude that IFN-α–induced STAT1 promotes Irf8 transcription by direct interaction at the endogenous Irf8 promoter. STAT1-responsive induction of Irf8 may contribute toward the generation of pDCs during IFN-α exposure, even in the presence of suppressive signals from GM-CSF–induced STAT5 and reduced Tcf4 expression.
pDCs generated in the presence of IFN-α preferentially stimulate Th17 differentiation
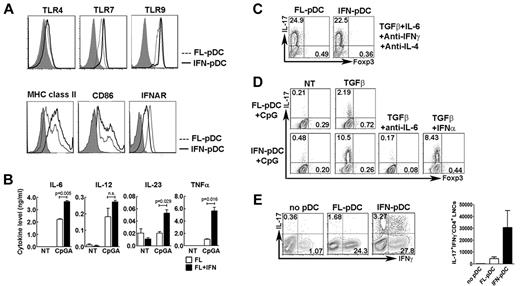
pDCs reside in T-cell rich areas in lymph nodes, suggesting potential regulation of T-cell responses. To examine whether IFN-α conditioning of pDCs affected functions involved in T lymphocyte activation, we compared the expression of pattern recognition receptors, costimulatory molecules, and cytokine production of pDCs derived from Flt3L cultures with pDCs generated in Flt3L + IFN-α. TLR4, TLR7, and TLR9 expression was similar in pDCs generated in Flt3L or Flt3L + IFN-α conditions, whereas modestly elevated amounts of surface MHC class II and CD86 and suppressed IFNAR amounts were detected on pDCs from IFN-α cultures (Figure 6A). Cytokine induction by the TLR agonist CpGA showed the most significant differences, as pDCs generated in IFN-α cultures produced increased amounts of the inflammatory cytokines IL-6, IL-23, and TNF-α compared with Flt3L-driven pDCs (Figure 6B). By contrast, IL-12 p70 induction was similar and IL-1β production negligible in both populations (not shown). Because IL-6, IL-23, and TNF-α are involved in the generation of inflammatory IL-17–secreting CD4+ T cells (Th17 cells),38 these data suggest that pDCs conditioned in the presence of IFN-α may have a preferential ability to elicit Th17 cells compared with pDCs generated by Flt3L.
Th17 generation by IFN-α–conditioned pDCs. (A) Intracellular TLR7 and TLR9 expression and surface expression of TLR4, MHC class II, CD86, and IFNAR in FACS-purified pDCs from Flt3L (FL-pDC; dashed line) or Flt3L + IFN-α cultures (IFN-pDC; solid line). Shaded area represents isotype control staining. (B) Cytokine secretion from FACS-purified, CpGA-stimulated pDCs from Flt3L (FL) or Flt3L + IFN-α cultures (FL + IFN), determined by ELISA. Error bars represent SEM of values obtained from triplicate wells. (C) Th differentiation stimulated by FACS-purified pDCs from Flt3L or Flt3L + IFN-α cultures in the presence of optimal Th17-inducing conditions and absence of CpGA. pDCs were cocultured with CD4+ CD25− CD62L+ CD44− naive T cells from OT-II mice and OVA peptide 323-339 in the presence of the indicated cytokines and antibodies. Results show intracellular staining for IL-17 and Foxp3 at 4 days, after restimulation with phorbol myristate acetate and ionomycin. (D) Th differentiation stimulated by FACS-purified CpGA-activated pDCs from Flt3L (FL-pDC) or Flt3L + IFN-α cultures (IFN-pDC) with or without exogenous TGF-β, as indicated. pDCs were cocultured with naive T cells from OT-II mice and OVA peptide 323-339 in the presence of TGF-β (1 ng/mL) with or without exogenous IFN-α (3 ng/mL) or IL-6 blocking antibodies (10 μg/mL), as indicated. Th differentiation was determined as indicated in panel C. (E) Th induction in vivo by CpGA-activated pDCs from Flt3L (FL-pDC) or Flt3L + IFN-α cultures (IFN-pDC). Rag1−/− mice received naive OT-II CD4+ T cells intravenously and CpGA-stimulated, OVA peptide-pulsed pDCs subcutaneously. Rag1−/− mice injected with only T cells were included as controls (NT). Inguinal lymph nodes were collected at 7 days, and Th induction was determined as indicated in the left panel by intracellular staining of IL-17 and IFN-γ. Absolute number of IL-17+ IFN-γ− CD4+ lymph node cells is shown in the right panel. n = 6. (A-E) Results represent 3 independent experiments.
Th17 generation by IFN-α–conditioned pDCs. (A) Intracellular TLR7 and TLR9 expression and surface expression of TLR4, MHC class II, CD86, and IFNAR in FACS-purified pDCs from Flt3L (FL-pDC; dashed line) or Flt3L + IFN-α cultures (IFN-pDC; solid line). Shaded area represents isotype control staining. (B) Cytokine secretion from FACS-purified, CpGA-stimulated pDCs from Flt3L (FL) or Flt3L + IFN-α cultures (FL + IFN), determined by ELISA. Error bars represent SEM of values obtained from triplicate wells. (C) Th differentiation stimulated by FACS-purified pDCs from Flt3L or Flt3L + IFN-α cultures in the presence of optimal Th17-inducing conditions and absence of CpGA. pDCs were cocultured with CD4+ CD25− CD62L+ CD44− naive T cells from OT-II mice and OVA peptide 323-339 in the presence of the indicated cytokines and antibodies. Results show intracellular staining for IL-17 and Foxp3 at 4 days, after restimulation with phorbol myristate acetate and ionomycin. (D) Th differentiation stimulated by FACS-purified CpGA-activated pDCs from Flt3L (FL-pDC) or Flt3L + IFN-α cultures (IFN-pDC) with or without exogenous TGF-β, as indicated. pDCs were cocultured with naive T cells from OT-II mice and OVA peptide 323-339 in the presence of TGF-β (1 ng/mL) with or without exogenous IFN-α (3 ng/mL) or IL-6 blocking antibodies (10 μg/mL), as indicated. Th differentiation was determined as indicated in panel C. (E) Th induction in vivo by CpGA-activated pDCs from Flt3L (FL-pDC) or Flt3L + IFN-α cultures (IFN-pDC). Rag1−/− mice received naive OT-II CD4+ T cells intravenously and CpGA-stimulated, OVA peptide-pulsed pDCs subcutaneously. Rag1−/− mice injected with only T cells were included as controls (NT). Inguinal lymph nodes were collected at 7 days, and Th induction was determined as indicated in the left panel by intracellular staining of IL-17 and IFN-γ. Absolute number of IL-17+ IFN-γ− CD4+ lymph node cells is shown in the right panel. n = 6. (A-E) Results represent 3 independent experiments.
To examine CD4+ Th subset generation, we used in vitro cocultures with FACS-purified pDCs and naive CD4+ T cells from OT-II transgenic mice, carrying a T-cell receptor transgene that recognizes ovalbumin. Cocultures were performed in the presence or absence of CpGA or Th-polarizing cytokines ex vivo, plus OVA peptide 323-339, to examine intrinsic Th-generating activity with or without exogenous stimuli. In the presence of cytokines that are known to elicit Th polarization, unactivated pDCs from Flt3L + IFN-α cultures (ie, without CpGA stimulation) were as efficient as Flt3L-derived pDCs in driving Th1, Th2, Th17, and inducible Treg differentiation (Figure 6C; supplemental Figure 7). These data indicate both pDC subsets act similarly as APCs under optimal Th-polarizing conditions.
By contrast, in cocultures that lacked all exogenously added cytokines but TGF-β, a serum factor that regulates Th17 and Treg development, CpGA-stimulated pDCs from Flt3L + IFN-α cultures promoted significantly enhanced development of the inflammatory Th17 subset, compared with CpGA-stimulated Flt3L-derived pDCs (Figure 6D). Th17 generation by CpGA-stimulated IFN-α–conditioned pDCs required IL-6, as revealed by interference with IL-6–blocking antibodies (Figure 6D), in agreement with the essential role for this cytokine in Th17 polarization.38 Type I IFNs have been reported to inhibit Th17 cells39,40 yet preferential Th17 generation by IFN-α–conditioned pDCs was not the result of their lack of IFN-α production, as exogenous IFN-α did not abrogate Th17 development (Figure 6D).
To examine whether IFN-α–conditioned pDCs showed functional differences from Flt3L-generated pDCs in vivo, we used an adoptive transfer model to compare Th responses. We transferred CpGA-activated, OVA peptide-pulsed pDCs and naive OT-II CD4+ T cells into Rag1−/− mice, which lack endogenous T and B lymphocytes. At 7 days after transfer, we collected inguinal lymph nodes and assessed Th induction by intracellular cytokine staining. IFN-α–conditioned pDCs showed an enhanced ability to elicit Th17 generation in vivo, as judged by increased Th17 frequency and absolute numbers, relative to Flt3L-generated pDCs (Figure 6E). These results suggest that IFN-α exposure during development influences pDC function, conditioning pDCs to an inflammatory Th17-inducing APC state.
Discussion
Here, we establish that IFN-α regulates both the development and function of pDCs, and has a crucial role in PP pDC accrual in vivo. Moreover, we find a new role for STAT1 in controlling PP pDC accumulation as well as IFN-α–mediated induction of pDCs ex vivo or in vivo. IFN-α–STAT1 signaling is required in the hematopoietic compartment to control PP pDC abundance; however, systemic IFN-α delivery does not induce PP pDCs, suggesting that type I IFNs expressed within the gut affect the developmental pathway and/or migratory responses of pDCs or their progenitors. This idea is consistent with the ability of purified wild-type CDPs to generate PP pDCs in Ifnar−/− or Stat1−/− mice after adoptive transfer, as well as IFNAR expression on DC progenitors and IFN-α–dependent accumulation of the DC progenitor subset in BM, results indicating that DC progenitors respond to type I IFN cues. pDCs that have been exposed to IFN-α during or after development elicit inflammatory Th17 generation in the presence of TGF-β and TLR triggering by CpGA ex vivo, or on CpGA activation followed by adoptive transfer in vivo yet fail to secrete abundant amounts of type I IFN. Reduced IFN secretion has also been observed in PP pDCs,13 suggesting similarity with IFN-α–induced pDCs derived ex vivo. Collectively, our results indicate that pDCs conditioned by IFN exposure either during or after development serve as an APC subset that stimulates adaptive Th17 responses.
IFN-α is traditionally considered an inflammatory, antiviral, and antiproliferative factor because of its well-documented effects, including induction of host genes that interfere with viral replication and cellular proliferation.4 IFN-α was recently shown to stimulate HSC proliferation through both direct and indirect mechanisms involving STAT1,21 a response that may contribute to the accumulation of DC progenitors and pDCs in IFN-α–treated mice. In ex vivo conditions, we found that a second cytokine (eg, Flt3L or GM-CSF) was required to stimulate optimal generation of pDCs by IFN-α, and IFN-α did not enhance DC progenitor proliferation in response to Flt3L or GM-CSF. These results suggest that IFN-α influences specification, maturation, or survival of the pDC lineage, whereas other factors regulate proliferative expansion of pDC precursors. IFN-α activates STAT3 and STAT1, yet pDC development induced by IFN-α does not require functional STAT3 and depends instead on STAT1, similar to IFN-α–mediated HSC regulation.
Tie2cre+Stat3f/Δ mice showed reduced pDC amounts within BM and spleen, relative to STAT3-sufficient controls, in agreement with the requirement for STAT3 in Flt3L-mediated DC progenitor proliferation.33 We considered that type I IFNs and STAT1 may provide compensatory signals; however, results from Tie2cre+Stat3f/ΔIfnar−/− animals indicate that additional factors control residual pDC amounts in Tie2cre+Stat3f/Δ mice (H.S.L. and S.S.W., data not shown). Moreover, Tie2cre+Stat3f/Δ animals have CD11b+ CD11c+ cDCs in BM and spleen in near-normal abundance. These findings contrast with a previous report35 yet concur with a recent study of a DC-specific STAT3 deletion (CD11c-cre).41 Collectively, these studies indicate the importance of STAT3 in pDCs, as well as STAT3-independent pathways for pDC and cDC development in vivo.
The mechanisms regulating IFN-α–enhanced pDC production from DC progenitors and/or the distinct activity of IFN-α–conditioned pDCs may involve modulation of transcription factors that control maturation and function of pDCs in steady state. We found that IFN-α induced Irf8, Irf7, Sfpi1, and Spib, consistent with their role in pDCs18 ; however, a notable exception was Tcf4, which appears to be repressed by IFN-α signaling in vitro or in vivo. Furthermore, PP pDCs express similar amounts of Irf7 and Irf8 yet reduced Tcf4 compared with pDCs from BM or spleen in homeostatic conditions, in agreement with the concept that PP pDCs may receive type I IFN conditioning signals during accrual in the gut. Although we determined that Irf8 is directly induced by IFN-α via STAT1, the mechanism of Tcf4 down-regulation remains unclear. IFN-α did not affect TLR or CD86 costimulatory molecule expression dramatically on pDCs generated in vitro. PP pDCs and BM pDCs also showed similar TLR and costimulatory molecule amounts. Thus, Tcf4 reduction in PP pDCs and/or IFN-α–conditioned pDCs may contribute to their deficiency in IFN-α production, in agreement with the finding that E2–2/Tcf4 haploinsufficiency is accompanied by impaired pDC IFN secretion,42 whereas IFN-α–STAT1 induction of Irf8 may be involved in promoting pDC development.
By contrast with IFN-α, we found that GM-CSF suppresses BM and splenic pDCs in vivo, concurring with our report that GM-CSF abrogates Flt3L-driven pDC development from DC progenitors.33 pDC inhibition by GM-CSF is offset by an increase in CD11c+ CD11b+ cDCs, which may include GM-CSF–driven inflammatory cDCs.43 The outcome of GM-CSF signaling is a decreased pDC/cDC ratio in vivo. Because the balance of pDCs to cDCs affects viral immunity,44 pDC inhibition by GM-CSF might increase susceptibility to viruses; however, patients receiving GM-CSF therapy do not appear to have more frequent or potent viral infections.45 Although the purpose of GM-CSF–mediated pDC suppression remains unclear, we suggest that it may dampen IFN-α production, which inhibits cDCs37 (results herein). Hence, GM-CSF–dependent induction of inflammatory cDCs and pDC/IFN-α suppression might ensure sufficient generation of APCs during infection.
In conclusion, our data are consistent with a model in which local IFN cues regulate migration or retention, and potentially activity, of pDCs within PPs while systemic IFN-α delivery stimulates production and affects function of pDCs in other organs. Intestinal immune cells provide a crucial front-line defense to constrain gut microbes and induce tolerance; IFNs and DCs are central components of protective gut responses.46-49 By contrast, prolonged clinical (systemic) IFN-α treatment, which may condition pDCs to elicit pathogenic Th17 responses, is associated with a propensity to develop autoimmunity.11,12 In agreement, pDC depletion in murine encephalomyelitis suppressed Th17 production and disease severity.50 Therefore, IFN-α conditioning of pDCs in the gut may contribute to protective responses, whereas sustained systemic exposure may shape the APC repertoire and adaptive immunity toward a more inflammatory response that may be detrimental.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Chen Dong, Wei Cao, and Michel Gilliet for helpful discussions and K. Ramirez and Z. He for assistance with FACS.
This work was supported by the National Institutes of Health (grant CA128913, W.W.O.; grant AI059718, Y.-J.L.; and grants AI073587 and AR059010, S.S.W.), The Netherlands Organization for Scientific Research (VENI grant 916.046.014, W.W.O.), and the National Cancer Institute (Core grant P30CA16672). A.G. was supported by the National Research Service Award (Training grant T32DC007367). G.J.M. is a Fellow of the Schissler Foundation. H.N.-J. was supported by the National Institutes of Health Training Program in Cancer Immunobiology (T32-CA-09598-16) at the University of Texas M. D. Anderson Cancer Center.
National Institutes of Health
Authorship
Contribution: H.S.L. designed and performed experiments, analyzed data, and wrote the paper; A.G., G.J.M., and E.E. designed and performed experiments; H.Z. and H.N.-J. provided the STAT3-deficient mice; Y.-J.L. and W.W.O. supervised the project, analyzed data, and communicated unpublished results; and S.S.W. supervised the project, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stephanie S. Watowich, Department of Immunology and Center for Inflammation and Cancer, University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: swatowic@mdanderson.org.