Abstract
Blood flow has long been thought to be important for vessel development and function, but its role in HSC development is not yet fully understood. Here, we take advantage of zebrafish embryos with circulation defects that retain relatively normal early development to illustrate the combinatorial roles of genetic and hemodynamic forces in HSC development. We show that blood flow is not required for initiation of HSC gene expression, but instead is indispensable for its maintenance. Knockdown of klf2a mimics the silent heart (sih/tnnt2a) phenotype while overexpression of klf2a in tnnt2a morphant embryos can rescue HSC defects, suggesting that klf2a is a downstream mediator of blood flow. Furthermore, the expression of NO synthase (nos) was reduced in klf2a knockdown embryos, and ChIP analysis showed that endogenous Klf2a is bound to the promoters of nos genes in vivo, indicating direct gene regulation. Finally, administration of the NO agonist S-nitroso N-acetylpenicillamine (SNAP) can restore HSC development in tnnt2a and klf2a morphants, suggesting that NO signaling is downstream of Klf2a which is induced by hemodynamic forces. Taken together, we have demonstrated that blood flow is essential for HSC development and is mediated by a klf2a-NO signaling cascade in zebrafish.
Introduction
The origin and migratory path of definitive HSCs has been delineated in zebrafish embryos, and is highly conserved in higher vertebrates.1-5 In zebrafish, HSCs are derived from the ventral wall of the dorsal aorta via an endothelial-hematopoietic transition (EHT), migrate to the caudal hematopoietic tissue (CHT), and finally home to definitive hematopoiesis sites in the thymus and kidney to produce T cells and all other blood lineages during adulthood.3,6-9 The specification and maintenance of the HSC program during early embryogenesis are tightly controlled by a complex of extrinsic and intrinsic signaling.3,6 However, the interplay between intrinsic genetic factors and local hemodynamic cues has not been fully understood. For example, it remains to be addressed how hemodynamic factors contribute significantly to the remodeling of artery and vein and then HSC development, and how physiologic factors are integrated with genetic factors. Most relevant knockout mice die of circulation defects before HSC emergence in the aorta-gonad-mesonephros (AGM).10 Therefore, it is difficult to separate the roles played by genetic and physiologic factors during artery-vein specification and then HSC development.
Recent evidence suggests that NO is responsible for HSC development in zebrafish and in mice. By chemical screening, North et al found that a group of chemical blood flow modulators can regulate HSC induction in zebrafish, mainly via flow-induced NO signaling, which is consistent with the finding reported in mice.11,12 However, how blood flow exerts its effect on the endothelial NO signaling is still obscure. Interestingly, the endothelial gene klf2, an immediate early responder to blood flow, has been demonstrated to be involved in blood vessel remodeling and heart valve development in zebrafish.13-15 Thus, an attractive hypothesis would be that flow-induced klf2 expression regulates NO signaling16 and thereby HSC development.
Here, we took advantage of available zebrafish blood flow–deficient embryos silent heart/tnnt2a, heart-of-glass/heg, and santa/krit1 combined with chemical treatment, to test this hypothesis in a comprehensive manner. Our findings show that blood flow is not required for the initiation of HSC gene expression, but is essential for its maintenance in zebrafish. Blood flow–induced expression of klf2a, a zinc-finger transcription factor, directly regulates the NO signaling pathway, and thereby controls artery/vein maturation and continued HSC programming.
Methods
Fish strains and embryos
Zebrafish embryos were obtained by natural spawning of adult AB strain zebrafish. Embryos were raised and maintained at 28.5°C in system water and staged as described.17 fli1a:GFP transgenic line was kindly provided by Steve Wilson (King's College London, London, United Kingdom). The flk1GFP/gata1:dsRed transgenic line was kindly provided by Stefan Schulte-Merker (Hubrecht Institute, Utrecht, The Netherlands). fli1a:nGFP line and moonshine were kindly provided by Anming Meng (Tsinghua University, Beijing, China). This study was approved by the Faculty of Clinical Medicine, Local Ethical Review Committee, University of Oxford, and the Ethical Review Committee in the Institute of Zoology, Chinese Academy of Sciences, China.
Morpholinos, mRNA synthesis, and microinjection
Standard morpholinos (MOs) and antisense MOs were purchased from GeneTools and prepared as 1mM stock solutions using dH2O. Standard MOs were used as negative controls in all MO injection experiments alongside with gene-specific MOs. The sequences of gene-specific MOs are described in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Capped fish klf2a full-length mRNA for injection was synthesized from NotI-digested pCS2 expression plasmid using the mMessage mMachine SP6 kit as instructed by the manufacturer (Ambion). For fish embryo injections, MOs (2-10 ng) and capped mRNA were injected alone or in combination, into 1-2 cell-stage zebrafish embryos at the yolk/blastomere boundary.
Chemical treatment
NO staining was performed by incubating zebrafish embryos with 5μM cell-permeant DAF-FM-DA (diamino-fluorophore 4-amino-5-methylamino-2′-7′-difluoro-fluorescein diacetate; Molecular Probes) for 2 hours in the dark at 28°C.18 Zebrafish embryos were placed in the NO-donor SNAP (S-nitroso N-acetylpenicillamine; Sigma-Aldrich) from bud stage at a concentration of 10μM in 0.1% DMSO. During the next 24 hours, the embryos were examined periodically to ensure that the pharmacologic effect remained constant over time.19
In situ hybridization
Whole-mount in situ hybridization (WISH) for zebrafish embryos was performed as described previously with probes including scl, gata1, runx1, cmyb, dll4, dlc, gata1, klf2a, nos1, hif2α, and rag1.20
Transverse sectioning
The embryos were mounted with the JB-4 Embedding Kit (Polysciences) before sectioning with a Leica RM2255 vibratome into 5- to 10-μm thick slices.
Quantitative RT-PCR
Total RNA from the trunk region of control and MO-injected embryos (20-40 embryos pooled for each sample) at 24 and 36 hours postfertilization (hpf) was extracted by TriReagent and was reversely transcribed. The cDNAs were all diluted 5 times to be used as templates. The PCR primers used and the expected product length are summarized in supplemental Table 2. The GoTaq qPCR Master Mix (Promega) was used in quantitative PCR on the Bio-Rad CFX96 Real-Time PCR system. All the experiments were repeated 3 times with triplicates. We analyzed the data using the ΔΔCt method. Data were represented as mean ± SD and t test was performed for comparison between control and experimental groups.21 P < .05 indicates significant difference.
TUNEL assay
Embryos were dechorionated and fixed in 4% paraformaldehyde overnight at 4°C then washed with PBS buffer twice and permeabilized with 0.1% sodium citrate and 0.1% Triton X for 2 minutes on ice. After washing 3 times with PBST for 5 minutes each and incubated with 90 μL of labeling solution plus 10 μL of enzyme solution (In Situ Cell Death Detection Kit, Fluorescein; Roche) at 37°C for 4 hours or at 4°C overnight. They were washed 3 times with PBST for 5 minutes each and the images were examined by confocal microscopy.
ChIP assay
ChIP analysis was carried out with wild-type embryos of 36 hpf. Embryos were lyzed using lysis buffer (10mM Tris-HCl pH 8.0, 10mM NaCl 0.5% NP-40) and Klf2a polyclonal Ab (Millipore) were used for immunoprecipitation with rabbit serum as negative control. After extensive washing, the DNA was eluted using elution buffer (25mM Tris-HCl, 10mM EDTA, 0.5% SDS) and the eluted DNA was assayed by PCR. The primers specific and unspecific to the Klf2 binding site in the upstream regions of nos1, nos2a, and nos2b were designed by searching for the conserved CACCC box and the primers unspecific to the Klf2-binding sites were included as negative controls. At least 3 pairs of specific primers and one pair of control primers for each gene were used in ChIP PCR. The primers and the expected product length were summarized in supplemental Table 3. The PCR program was as follows: 94°C, 5 minutes; 94°C, 30 seconds; 55°C, 30 seconds; 72°C, 15 seconds, 35 circles (nos2b-3F/R with 28 circles); 72°C, 10 minutes. All the PCR products were ∼ 200 bp and were evaluated on a 2% agarose gel.
Western blot analysis
Zebrafish embryos at 36 hpf were washed with fish water and the trunk region was dissected out after manual dechorionation, then homogenized with 1-mL syringe and needle in lysis buffer (10mM Tris-HCl pH 8.0, 10mM NaCl 0.5% NP-40) containing protease inhibitor (Roche) at 1× concentration (1 tablet in 2 mL of redistilled water as 25× concentration) for 30 seconds. Lysate was centrifuged at 12 000g for 2 minutes at 4°C and the resulting supernatant was loaded as protein samples. After 12% SDS-PAGE separation, proteins were transferred to nitrocellulose membranes. After blocking with 5% nonfat milk for 3 hours, nitrocellulose membranes were incubated with Klf2a polyclonal Ab (1:1000; Millipore) or β-actin Ab (1:1000; Cell Signaling) overnight. Membranes were washed in 1× TBST (Tris buffer solution plus Tween 20: 136.8mM NaCl, 24.8mM Tris, 0.5% [v/v] Tween 20, pH7.4) 3 times with each for 15 minutes, and incubated for 3 hours with anti–rabbit secondary Ab (1:5000; Jackson ImmunoResearch Laboratories). After extensive washing membranes were detected by a chemiluminescent HRP substrate (Millipore).
Confocal microscopy
Confocal images were acquired with a Zeiss LSM 510 META confocal laser microscope and 3-dimensional (3D) projections were generated using Zeiss LSM software (Carl Zeiss Inc).20
Results
Primitive hematopoiesis is normal in tnnt2a morphants
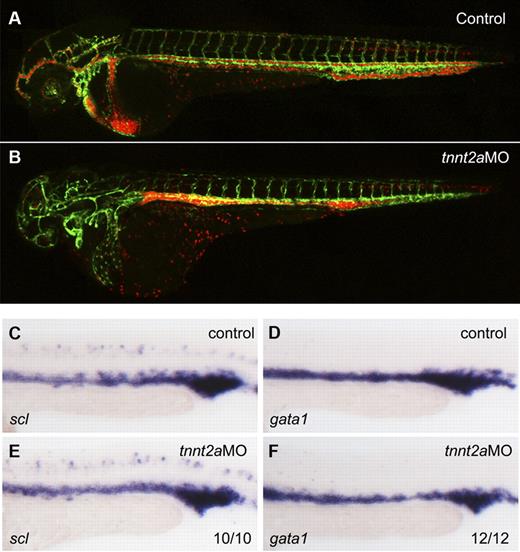
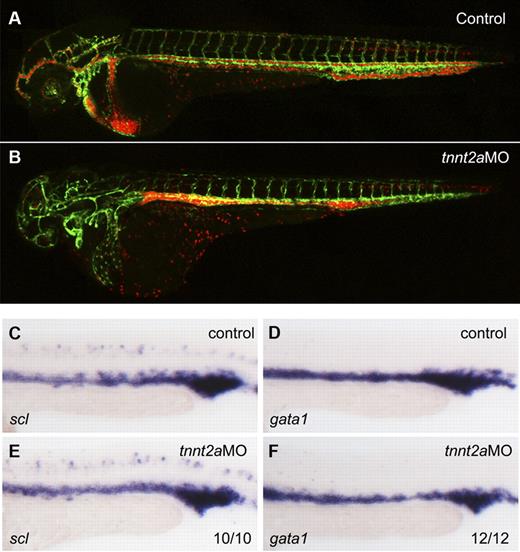
To address whether primitive hematopoiesis is affected in the absence of flow, we used 2 different methods in silent heart (sih/tnnt2a) embryos which have no heartbeat and therefore no circulation.22 First, we applied a flk1:GFP/gata1:dsRed double transgenic line. flk1:GFP marks all endothelial cells in blood vessels, while gata1:dsRed marks erythroid lineage derived from primitive wave of hematopoiesis during embryogenesis. As shown in Figure 1B, both endothelial cells and RBCs developed normally in tnnt2a morphants, albeit the latter were accumulated in the trunk region because of lack of heartbeat. Second, we examined expression of primitive hematopoietic markers, scl (stem cell leukemia), a primitive hematopoietic progenitor marker, gata1, an erythroid marker, by WISH. Expression of scl and gata1 in the intermediate cell mass (ICM), the primitive hematopoiesis site in zebrafish embryo, was relatively normal at 24 hpf in tnnt2a morphants, compared with that in controls (Figure 1C-F). Taken together, these experiments demonstrate that primitive hematopoiesis is not altered in flow-deficient tnnt2a morphants.
Primitive hematopoiesis is normal in tnnt2a morphants. (A-B) Confocal images of control MO (A) and tnnt2aMO (B) injected flk1:GFP/gata1:dsRed transgenic embryos at 2 dpf. (C-F) scl and gata1 expression in the intermediate cell mass (ICM) was normal in tnnt2a morphants at 24 hpf. Anterior is to the left and dorsal is up.
Primitive hematopoiesis is normal in tnnt2a morphants. (A-B) Confocal images of control MO (A) and tnnt2aMO (B) injected flk1:GFP/gata1:dsRed transgenic embryos at 2 dpf. (C-F) scl and gata1 expression in the intermediate cell mass (ICM) was normal in tnnt2a morphants at 24 hpf. Anterior is to the left and dorsal is up.
HSC programming is initiated but not maintained in tnnt2a morphants
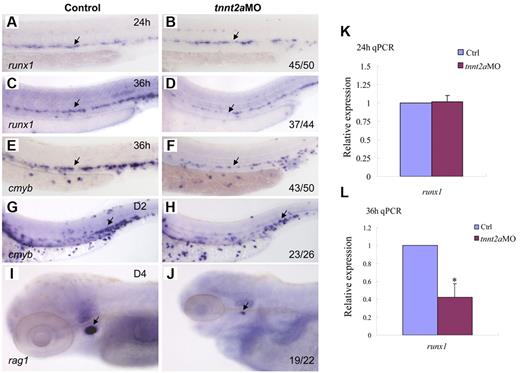
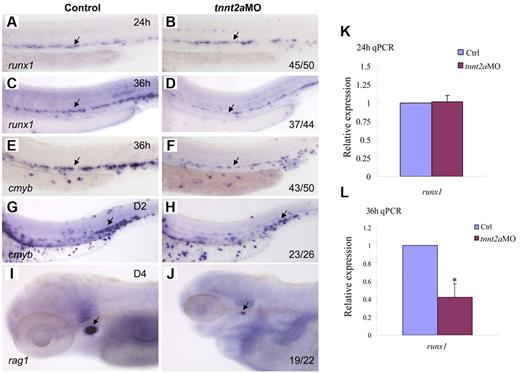
To determine whether blood flow is required for HSC development, we examined runx1 expression in silent heart (sih/tnnt2a) embryos (Figure 2A-D).22 At 24 hours, runx1 expression in the dorsal aorta region was normal in tnnt2aMO-injected embryos, compared with that in the controls, suggesting that the HSC program was initiated normally (Figure 2B). However, from 36 hpf onward, runx1 expression was not maintained in tnnt2a morphants (Figure 2D). Quantitative RT-PCR analysis showed that runx1 expression in the trunk region was comparable between control and tnnt2aMO embryos at 24 hpf (Figure 2K), then subsequently decreased at 36 hpf in tnnt2aMO embryos (Figure 2L), further supporting the WISH results. TUNEL assay showed that there were more apoptotic cells in the tnnt2a morphants (supplemental Figure 1), potentially at least partly explaining the down-regulation of HSC genes.
Blood flow is not required for the initiation of HSC gene expression, but is essential for its maintenance in zebrafish. Expression of runx1 and cmyb at 24 and 36 hpf in control MO (A,C,E), tnnt2aMO (B,D,F) injected embryos. Arrows mark runx1 and cmyb expression in HSCs. (G-J) The maintenance and homing of HSCs were affected in tnnt2a morphants. Expression of cmyb in CHT in control MO (G) and tnnt2aMO (H) injected embryos at 2 dpf. Expression of rag1 in thymus in control MO (I) and tnnt2aMO (J) injected embryos at 4 dpf. (G-H) Arrows indicate CHT. (I-J) Arrows indicate thymus. Anterior is to the left and dorsal is up. CHT, caudal hematopoietic tissue. (K,L) Quantitative RT-PCR analysis of runx1 and cmyb expression in the dissected trunk region of control and tnnt2aMO embryos at 24 and 36 hpf (mean ± SD, t test, *P < .05, n = 3).
Blood flow is not required for the initiation of HSC gene expression, but is essential for its maintenance in zebrafish. Expression of runx1 and cmyb at 24 and 36 hpf in control MO (A,C,E), tnnt2aMO (B,D,F) injected embryos. Arrows mark runx1 and cmyb expression in HSCs. (G-J) The maintenance and homing of HSCs were affected in tnnt2a morphants. Expression of cmyb in CHT in control MO (G) and tnnt2aMO (H) injected embryos at 2 dpf. Expression of rag1 in thymus in control MO (I) and tnnt2aMO (J) injected embryos at 4 dpf. (G-H) Arrows indicate CHT. (I-J) Arrows indicate thymus. Anterior is to the left and dorsal is up. CHT, caudal hematopoietic tissue. (K,L) Quantitative RT-PCR analysis of runx1 and cmyb expression in the dissected trunk region of control and tnnt2aMO embryos at 24 and 36 hpf (mean ± SD, t test, *P < .05, n = 3).
To examine whether HSCs or their derivatives appear in the CHT or thymus/kidney, we performed WISH with, cmyb, a conserved HSC marker, and rag1, a T-cell marker, in tnnt2a morphants (Figure 2E-J). Similar to runx1, cmyb expression in the dorsal aorta was also down-regulated in tnnt2aMO-injected embryos (Figure 2F). Expression of cmyb was significantly decreased in the CHT at 2 days postfertilization (dpf), when HSC derivatives can first be detected there,1,2 and onward (Figure 2H arrow; data not shown). Expression of the thymic T-cell marker, rag1, was also nearly abolished (Figure 2J). These results show that the loss of HSCs in the dorsal aorta seen in the absence of blood flow is permanent. Taken together, these data indicate that blood flow in the trunk is critical for maintenance of HSC programming during zebrafish embryogenesis.
Two other heartbeat defective morphants mimic the phenotype of tnnt2aMO-injected embryos
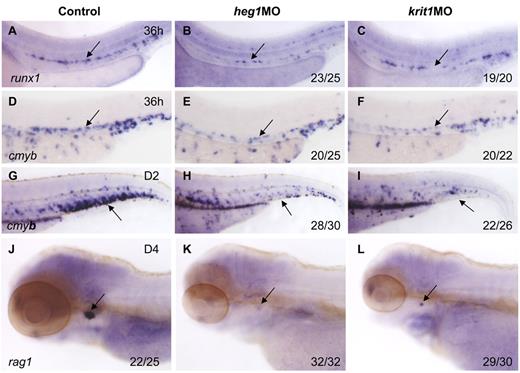
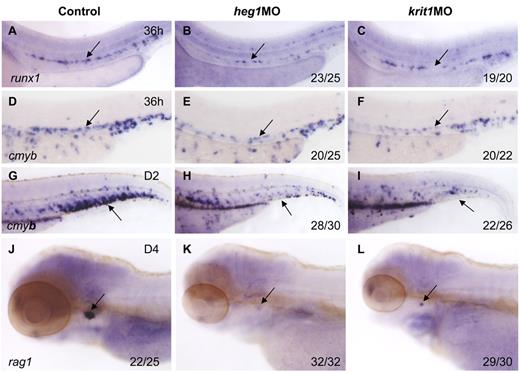
To confirm the role of blood flow in HSC development, we applied alternative methods to block heartbeat and confer circulation defects. heart of glass/heg1 and santa/krit1 encode 2 proteins that are vital for patterning concentric growth of the heart.23,24 Loss of function of these 2 proteins individually, by mutations or MO injection, leads to heartbeat defects and virtually no circulation in the body. In heg1 and krit1 morphants, expression of runx1 and cmyb was relatively normal at 24 hpf (supplemental Figure 2), but significantly decreased in the AGM region at 36 hpf (Figure 3B,C,E,F). In addition, cmyb expression in the CHT and rag1 expression in the thymus were also down-regulated or even abolished in these 2 morphants (Figure 3H,I,K,L), compared with controls. We therefore conclude that when circulation was inhibited by 3 different perturbations, that is, tnnt2a, heg1, and krit1 morphants, the HSC program was not maintained in the DA in zebrafish.
Heart of glass/heg1 and santa/krit1 embryos display similar HSC phenotypes to silent heartembryos. Expression of cmyb (A-C) or runx1 (D-F) in HSCs in the AGM region (A-F) or CHT (G-I) in control MO (A,D,G), heg1MO (B,E,H), and krit1MO (C,F,I) injected embryos. Expression of rag1 in the thymus in control MO (J), heg1MO (K), krit1MO (L) injected embryos. (A-F) Arrows mark AGM HSCs, 36 hpf. (G-I) Arrows mark CHT, 2 dpf. (J-L) Arrows mark T cells, 4 dpf. Anterior is to the left and dorsal is up.
Heart of glass/heg1 and santa/krit1 embryos display similar HSC phenotypes to silent heartembryos. Expression of cmyb (A-C) or runx1 (D-F) in HSCs in the AGM region (A-F) or CHT (G-I) in control MO (A,D,G), heg1MO (B,E,H), and krit1MO (C,F,I) injected embryos. Expression of rag1 in the thymus in control MO (J), heg1MO (K), krit1MO (L) injected embryos. (A-F) Arrows mark AGM HSCs, 36 hpf. (G-I) Arrows mark CHT, 2 dpf. (J-L) Arrows mark T cells, 4 dpf. Anterior is to the left and dorsal is up.
Arterial development is disrupted in tnnt2a-deficient embryos
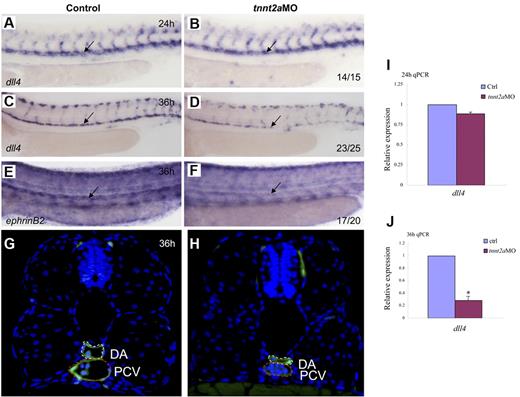
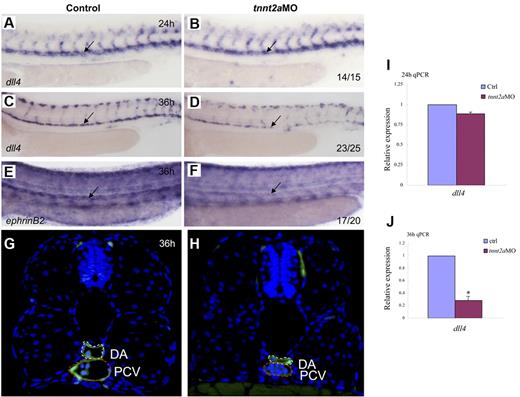
Because the dorsal aorta is thought to be the source of the first definitive HSCs,7,8 we assessed arterial development in tnnt2a-deficient embryos. Time-course studies showed that the lumen was formed at 24 hpf in tnnt2aMO-injected embryos, but shrunk after 30 hpf (supplemental Figure 3; Figure 4G-H), which coincides with simultaneously down-regulated expression of 2 arterial markers, that is, early specification marker, dll4 and late differentiation marker, ephrinB2 (Figure 4A-F). Quantitative RT-PCR analysis (Figure 4I-J) with the dissected trunk region of control and tnnt2aMO embryos at 24 and 36 hpf, respectively, further supports that dll4 expression in the dorsal aorta was specifically decreased, as shown by WISH results. Thus, the smaller lumen did not result merely from lack of inflation but apparently from a reduced number of endothelial cells in the DA (supplemental Figures 3-4). In mammals, HSCs are kept quiescent in the hypoxic niche, the endosteal zone of the BM.25 To test whether the heartbeat stop causes hypoxia in tnnt2aMO injected embryos, we examined hif2α expression but did not observe any alteration, suggesting no involvement of hypoxia (data not shown). Taken together, these data support that blood flow affects arterial cell maturation as well as HSC formation in zebrafish.
Artery maturation is attenuated in tnnt2a morphants. Expression of dll4 (A-D) or ephrinB2 (E,F) in the dorsal aorta in control MO and tnnt2aMO-injected embryos at 24 and 36 hpf, respectively. Anterior is to the left and dorsal is up. (G-H) Transverse section of control MO (G) and tnnt2aMO (H) injected fli1a:GFP embryos at 36 hpf. White dotted line denotes dorsal aorta; red dotted line denotes cardinal vein. (I-J) Quantitative RT-PCR analysis of dll4 expression in the dissected trunk region of control and tnnt2aMO embryos at 24 and 36 hpf, respectively (mean ± SD, t test, *P < .05, n = 3).
Artery maturation is attenuated in tnnt2a morphants. Expression of dll4 (A-D) or ephrinB2 (E,F) in the dorsal aorta in control MO and tnnt2aMO-injected embryos at 24 and 36 hpf, respectively. Anterior is to the left and dorsal is up. (G-H) Transverse section of control MO (G) and tnnt2aMO (H) injected fli1a:GFP embryos at 36 hpf. White dotted line denotes dorsal aorta; red dotted line denotes cardinal vein. (I-J) Quantitative RT-PCR analysis of dll4 expression in the dissected trunk region of control and tnnt2aMO embryos at 24 and 36 hpf, respectively (mean ± SD, t test, *P < .05, n = 3).
klf2a is a mediator of blood flow in the control of artery and HSC development
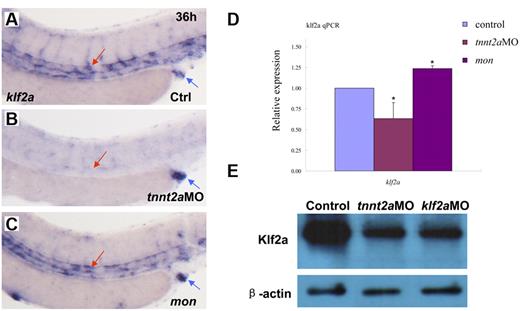
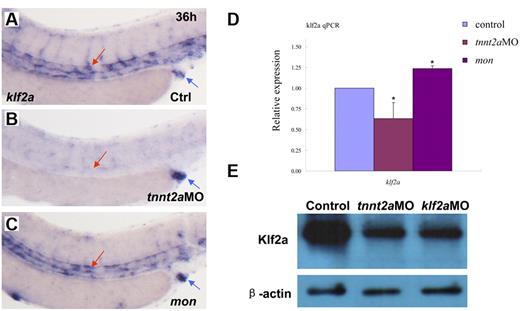
In vitro cell-culture studies showed that a kruppel-like transcription factor, klf2, is an immediate mediator of hemodynamic forces created by blood flow.14,26-29 Klf2 regulates flow-dependent endothelial phenotypes in cultured endothelial cells from zebrafish and mice.14,30 Mice lacking klf2 die during embryogenesis, defective in assembly of the vessel wall, which leads to massive hemorrhaging.31 Zebrafish klf2a is expressed in trunk vessels and pronephric ducts during embryogenesis (supplemental Figure 5).32 Zebrafish moonshine (mon) mutant has no red blood cells during embryogenesis.33 klf2a expression in mon embryos is similar to that in controls, further suggesting its specific expression in endothelial cells (Figure 5C).
klf2a expression is reduced in tnnt2a morphants, but maintained in moonshine mutants. (A-C) WISH showed that klf2a expression in the trunk region was reduced in the tnnt2aMO-injected embryos, but maintained in moonshine mutants. (D) Quantitative RT-PCR analysis of klf2a expression in the dissected trunk region of control and tnnt2aMO embryos at 24 and 36 hpf, respectively (mean ± SD, t test, *P < .05, n = 3). (E) Western blot showed Klf2a protein was significantly down-regulated in tnnt2a and klf2aMO-injected embryos.
klf2a expression is reduced in tnnt2a morphants, but maintained in moonshine mutants. (A-C) WISH showed that klf2a expression in the trunk region was reduced in the tnnt2aMO-injected embryos, but maintained in moonshine mutants. (D) Quantitative RT-PCR analysis of klf2a expression in the dissected trunk region of control and tnnt2aMO embryos at 24 and 36 hpf, respectively (mean ± SD, t test, *P < .05, n = 3). (E) Western blot showed Klf2a protein was significantly down-regulated in tnnt2a and klf2aMO-injected embryos.
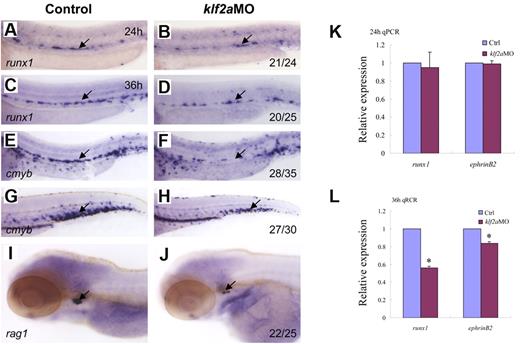
To determine whether zebrafish klf2a expression is affected by blood flow, we monitored klf2a expression in tnnt2aMO-injected embryos. As shown in Figure 5B, klf2a expression specifically in the vessel was significantly decreased in tnnt2aMO-injected embryos (red arrow), while the strong klf2a expression in the cloaca was not altered (Figure 5B, blue arrow). This observation was further supported by quantitative RT-PCR and Western blot of dissected trunk regions (Figure 5D-E), in which klf2a expression in the trunk region was down-regulated in tnnt2aMO-injected embryos compared with controls. These data strongly suggest that klf2a may be a downstream effector of blood flow. To address whether klf2a is involved in HSC development, we examined runx1 and cmyb expression in the AGM and/or the CHT in klf2a-deficient embryos generated by antisense MO knockdown. Similar to tnnt2a knockdown, runx1 expression in the AGM was only modestly changed at 24 hpf (Figure 6B) but was significantly reduced at 36 hpf (Figure 6D) in klf2a-deficient embryos. cmyb expression in the AGM and CHT was also attenuated, respectively (Figure 6F,H). Quantitative RT-PCR analysis of runx1 and ephrinB2 expression in the dorsal aorta of control and klf2aMO embryos at 24 and 36 hpf was consistent with the WISH data (Figure 6K-L, supplemental Figure 6). To further confirm that HSC numbers were reduced, we monitored expression of the thymic T-cell marker, rag1, and found that it was nearly abolished in klf2aMO-injected embryos (Figure 6J). Moreover, overexpression of klf2a mRNA partially rescued runx1 and dll4 expression in the DA region in tnnt2a MO embryos (Figure 7I; supplemental Figures 7D, 8), strongly supporting that klf2a is a major player downstream of blood flow. These data demonstrate that flow-induced klf2a expression is required for artery maturation and maintenance of HSC programming in zebrafish.
klf2a is downstream of tnnt2a in the regulation of artery maturation and maintenance of HSC programming. Expression of runx1 (A-D) and cmyb (E-H) in HSCs in control MO (A,C,E,G) and klf2aMO (B,D,F,H) embryos at 24, 36, and 48 hpf, respectively. (I-J) Expression of rag1 in thymus in control MO (I) and klf2aMO (J) injected embryos at 4dpf. (K-L) Quantitative RT-PCR analysis of runx1 and ephrinB2 expression in the dissected trunk region of control and klf2aMO embryos at 24 and 36 hpf, respectively (mean ± SD, t test, *P < .05, n = 3).
klf2a is downstream of tnnt2a in the regulation of artery maturation and maintenance of HSC programming. Expression of runx1 (A-D) and cmyb (E-H) in HSCs in control MO (A,C,E,G) and klf2aMO (B,D,F,H) embryos at 24, 36, and 48 hpf, respectively. (I-J) Expression of rag1 in thymus in control MO (I) and klf2aMO (J) injected embryos at 4dpf. (K-L) Quantitative RT-PCR analysis of runx1 and ephrinB2 expression in the dissected trunk region of control and klf2aMO embryos at 24 and 36 hpf, respectively (mean ± SD, t test, *P < .05, n = 3).
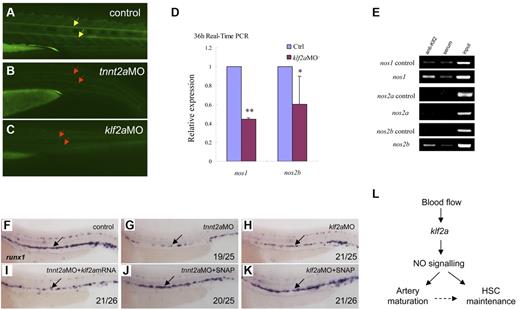
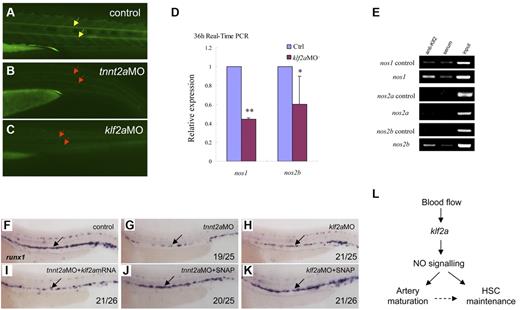
NO signaling is directly regulated by klf2a in the control of artery maturation and maintenance of HSC programming in zebrafish. (A-C) DAF-FM imaging showed reduced NO production in tnnt2a- and klf2aMO-injected embryos at 36 hpf. Yellow arrows mark normal NO production in the notochord area; red arrows mark reduced NO production. (D) Quantitative RT-PCR analysis of nos1 and nos2 expression in the dissected trunk region of control and klf2aMO embryos at 36 hpf (mean ± SD, t test, *P < .05, **P < .01, n = 3). (E) ChIP assay showing that Klf2a directly binds to nos1 and nos2b promoter region. (F-K) Rescue of runx1 expression in tnnt2a- and klf2aMO-injected embryos by klf2a mRNA or SNAP treatment. (F,G,I) Expression of runx1 in HSCs of tnnt2aMO-injected embryos (G) was partially rescued by coinjection of klf2a mRNA (I) at 36 hpf. (J-K) SNAP treatment at tailbud stage can rescue runx1 expression in tnnt2aMO (G,J) and klf2aMO (H,K) injected embryos. (L) Model of blood flow-induced klf2a-NO signaling in HSC maintenance.
NO signaling is directly regulated by klf2a in the control of artery maturation and maintenance of HSC programming in zebrafish. (A-C) DAF-FM imaging showed reduced NO production in tnnt2a- and klf2aMO-injected embryos at 36 hpf. Yellow arrows mark normal NO production in the notochord area; red arrows mark reduced NO production. (D) Quantitative RT-PCR analysis of nos1 and nos2 expression in the dissected trunk region of control and klf2aMO embryos at 36 hpf (mean ± SD, t test, *P < .05, **P < .01, n = 3). (E) ChIP assay showing that Klf2a directly binds to nos1 and nos2b promoter region. (F-K) Rescue of runx1 expression in tnnt2a- and klf2aMO-injected embryos by klf2a mRNA or SNAP treatment. (F,G,I) Expression of runx1 in HSCs of tnnt2aMO-injected embryos (G) was partially rescued by coinjection of klf2a mRNA (I) at 36 hpf. (J-K) SNAP treatment at tailbud stage can rescue runx1 expression in tnnt2aMO (G,J) and klf2aMO (H,K) injected embryos. (L) Model of blood flow-induced klf2a-NO signaling in HSC maintenance.
NO signaling is directly regulated by Klf2a during HSC development
Several studies have shown that klf2 activates the expression of vascular genes including endothelial NO synthase (eNOS).16,30,34 NO is involved in several physiologic and pathologic events. In particular, NO produced by the eNOS is a fundamental determinant of cardiovascular homeostasis, including systemic blood pressure, vascular remodeling, and angiogenesis.35 In cultured endothelial cells, shear stress and other factors have been shown to increase eNOS expression.36 To determine whether NO signaling was altered in tnnt2a- and klf2a-deficient embryos, we first used NO imaging to visualize NO production in live embryos. DAF-FM staining showed that NO production was reduced in the tnnt2a- and klf2a-deficient embryos (Figure 7B-C red arrows), compared with that in controls. To further test whether NO signaling was involved in the blood flow response required for HSC development, we examined nos1 and nos2a/2b expression in klf2a-deficient embryos. nos1 and nos2b expression was significantly decreased in klf2a morphants at 36 hpf (Figure 7D). To test whether nos1 and nos2a/2b were directly regulated by Klf2a, ChIP analysis was carried out. nos1 and nos2b but not nos2a were specifically enriched in the Klf2a Ab but not in serum-incubated samples (Figure 7E), suggesting that endogenous Klf2a bound to the promoters of nos1 and nos2b in vivo. This strongly indicates that klf2a regulates NO signaling directly. Finally, addition of a NO agonist, SNAP, slightly increased runx1 expression in wild-type embryos (supplemental Figure 8E) and rescued runx1 and dll4 expression in the AGM of tnnt2a (Figure 6G,J; supplemental Figures 8B,E) and klf2a-deficient embryos (Figure 7H,K, supplemental Figure 7C,F), suggesting that NO signaling acts downstream of blood flow controlling artery maturation and definitive HSC development. Taken together, these results demonstrate that blood flow induces expression of klf2a, which in turn, directly activates NO signaling to transduce the flow-dependent response to endothelial cells necessary for HSC development and artery maturation in zebrafish (Figure 7L).
Discussion
In the present study, we provide evidence for a novel klf2a-NO signaling cascade in integrating blood flow with HSC development in zebrafish embryos. This conclusion is based on the following findings: First, blood flow promotes continued HSC program development, but not its initiation. Second, HSC maintenance defects in the blood flow–deficient embryos were closely associated with artery maintenance defects. Third, Klf2a is the immediate downstream effector of blood flow in regulating dorsal aorta and HSC development. Fourth, Klf2a directly regulates nos1 and nos2, and thereby promotes the role of NO signaling in HSC development. These data lead us to propose a model in which blood flow controls dorsal aorta and HSC program maintenance through the klf2a-NO signaling cascade in zebrafish (Figure 7L).
The importance of mechanical forces in cardiovascular development has been largely neglected. However, recent data have identified fluid blood flow as a dominant modifier of cardiogenesis, vessel remodeling, vessel integrity, and HSC development.11,13,15,35,37,38 However, some questions remained concerning the role of blood flow in HSC development. Previously, North et al and Lam et al showed that blood flow is required for HSC development in zebrafish embryos.9,11 They looked at runx1/cmyb expression in 28- or 36-hpf zebrafish embryos, 6 or 14 hours after the start of HSC gene expression. The earliest expression of runx1 in HSC progenitors is detected at 22-24 hpf in zebrafish.39,40 In another 2 studies, Jin et al and Murayama et al showed relatively normal cmyb expression in HSCs at 30 hpf and also later stages in tnnt2aMO-injected embryos.1,2 To reconcile the obvious discrepancy, we have systematically checked several HSC markers, including runx1, cmyb, and ikaros at various stages, by using in situ hybridization and real time RT-PCR (Figure 2 and data not shown). Our data show that runx1 in fact initiated normally at 24 hpf, but it was not maintained after 36 hpf (Figure 2). This indicates that blood flow is not required for the initiation of HSC gene expression in the DA, but is essential for its continued expression, therefore providing a more precise description of the role of blood flow in HSC development. Our findings, together with those from others, add HSC development to a growing list of developmental processes requiring fluid dynamic inputs for proper growth and patterning, and further demonstrate that physiologic cues are part of the embryonic developmental program.
The kruppel-like factor (KLF) family of transcription factors is widely involved in vessel development in vertebrates. Specifically, klf2, responds to blood flow immediately, then exerts its function in angiogenesis and heart valve formation in zebrafish and in human vascular endothelial cells.13,15,26,27,30 Previous studies showed that induction of klf2 by fluid shear stress was mediated by several factors, including PCAF (P300/cAMP-response element-binding protein-associated factor), PI3K, Histone acetylation and MEF2 (myocyte enhancer factor-2) in HUVECs.29 Recently, Nicoli et al showed that in zebrafish, hemodynamic force-induced klf2a acts upstream of miR-126 which in turn represses spred1, the repressor of Vegf signaling, to promote aortic arch angiogenesis.15 Recent Xenopus studies showed that klf2 regulates expression of the VEGF receptor flk1 directly in endothelial cells.41 Together, these suggest Vegf signaling might also be involved in flow-dependent HSC development. Therefore, although our data show that in zebrafish, the klf2a-NO signaling cascade controls continued HSC development as well as dorsal aorta maturation, we cannot rule out a role for klf2a-induced Vegf signaling during this process.
HSC emergence has been tightly associated with the dorsal aorta in the AGM region in vertebrates. Recent evidence has demonstrated that HSCs are directly derived from hemogenic endothelium through an endothelial-hematopoietic transition in zebrafish embryos in vivo and in mouse AGM ex vivo.7,8,42 Thus, the dorsal aorta acts as the requisite de novo site or microenvironment for definitive hematopoiesis to occur. Our data shows that blood flow-dependent klf2a-NO signaling affects both dorsal aorta maturation and HSC program maintenance, but not its initiation. Whether this blood flow-induced signaling cascade regulates both dorsal aorta and HSC maintenance programs, or primarily controls dorsal aorta maturation which in turn affects HSC maintenance still needs to be clarified.
Taken together, our studies demonstrate a requirement for blood flow in blood stem cell program maintenance during zebrafish embryogenesis. Because the signaling cascade comprising the components of NO signaling and klf2 are highly conserved across the vertebrates, they might be considered as candidates for disease genes contributing to later HSC (BM) failure in mammals.43
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Maggie Walmsley for critical reading of the manuscript, and Anming Meng and Qiang Wang for help with fish facilities.
This work was supported by National Basic Research Program of China grants 2010CB945300 and 2011CB943900, National Natural Science Foundation of China (NSFC) grant 30971678, and the Chinese Academy of Sciences (all to F.L.), and the Medical Research Council (R.P.).
Authorship
Contribution: L.W., P.Z., Y.W., and Y.G. performed experiments; R.P. revised the manuscript; and F.L. designed and performed the experiments, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Roger Patient, Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, University of Oxford, Oxford, United Kingdom; e-mail: roger.patient@imm.ox.ac.uk; or Feng Liu, State Key Laboratory of Biomembrane and Membrane Biotechnology, Key Laboratory of Stem Cell Development, Institute of Zoology, Chinese Academy of Sciences, Beijing, 100101, China; e-mail: liuf@ioz.ac.cn.