Abstract
M-CSF–driven differentiation of peripheral blood monocytes is one of the sources of tissue macrophages. In humans and mice, the differentiation process involves the activation of caspases that cleave a limited number of proteins. One of these proteins is nucleophosmin (NPM1), a multifunctional and ubiquitous protein. Here, we show that caspases activated in monocytes exposed to M-CSF cleave NPM1 at D213 to generate a 30-kDa N-terminal fragment. The protein is further cleaved into a 20-kDa fragment, which involves cathepsin B. NPM1 fragments contribute to the limited motility, migration, and phagocytosis capabilities of resting macrophages. Their activation with lipopolysaccharides inhibits proteolytic processes and restores expression of the full-length protein that negatively regulates the transcription of genes encoding inflammatory cytokines (eg, NPM1 is recruited with NF-κB on the MCP1 gene promoter to decrease its transcription). In mice with heterozygous npm gene deletion, cytokine production in response to lipopolysaccharides, including CXCL1 (KC), MCP1, and MIP2, is dramatically enhanced. These results indicate a dual function of NPM1 in M-CSF–differentiated macrophages. Proteolysis of the protein participates in the establishment of a mature macrophage phenotype. In response to inflammatory stimuli, the full-length protein negatively regulates inflammatory cytokine production.
Introduction
The mononuclear phagocytic system is composed of monocytes, macrophages, and dendritic cells that form a network of phagocytic cells throughout most tissues and play a major role in development, inflammation, antipathogen defenses, and scavenging. These cells have a remarkable heterogeneity related to their origin, phenotype, tissue localization, and function.1 Circulating blood monocytes develop from bone marrow progenitor cells and enter tissues to further differentiate, mainly into resident tissue macrophages and dendritic cells. The later steps of these pathways can be reproduced ex vivo by incubating blood monocytes with macrophage–colony-stimulating factor (M-CSF) to generate macrophages or with granulocyte macrophage–colony-stimulating factor (GM-CSF) and IL-4 to generate dendritic cells.1 M-CSF interaction with its receptor at the surface of blood monocytes provokes the oscillatory activation of phosphatidylinositol-3-kinase and the kinase AKT, which within 2-3 days leads to the formation of a multimolecular platform that includes the adaptor Fas-associated death domain (FADD), the serine-threonine kinase RIP1, the long and short isoforms of FLIP, and procaspase-8.2,3 Caspase-8 activation in this platform is required for M-CSF–driven macrophage formation.4 This protease provokes a limited activation of several downstream caspases that cleave intracellular proteins.5 These proteolytic events cannot be detected in monocytes undergoing GM-CSF plus IL-4–induced differentiation into dendritic cells. Their contribution to the M-CSF–driven monocyte-to-macrophage differentiation remains poorly understood.
Nucleophosmin (NPM1, B23, numatrin, or NO38) is one of the caspase targets in the M-CSF–driven differentiation of monocytes into macrophages.5 NPM1 is a ubiquitously and abundantly expressed 38-kDa phosphoprotein that constantly shuttles between the granular region of the nucleolus and the cytoplasm, especially during the S phase of the cell cycle. This multifunctional protein is involved in centrosome duplication,6 maintenance of genome integrity,7 transport of preribosomal particles,8 and ribosome biogenesis.9 NPM1 also exerts a chaperone activity for both proteins and nucleic acids.10,11 In addition, the protein is part of a basic complex for NF-κB transcriptional activity12 and regulates gene transcription, either as a coactivator or a corepressor.13-15 NPM1 is also a caspase substrate in cells undergoing apoptosis16 and a substrate of the cytotoxic granule protease granzyme M during natural killer cell–mediated killing.17
Here, we demonstrate that NPM1 is cleaved by caspases and cathepsins in monocytes undergoing differentiation into macrophages on exposure to M-CSF. The 30- and 20-kDa N-terminal fragments generated by the sequential proteolysis of the native protein down-regulate phagocytosis, motility, and transmigration in resting macrophages. TLR4-mediated activation of these cells inhibits the proteolytic processes and restores expression of the full-length protein that negatively regulates the transcription of a series of cytokine genes to control the amplitude of the inflammatory response. Analysis of lipopolysaccharide (LPS)–treated Npm+/− mice further demonstrates the importance of fine-tuning NPM1 proteolysis in macrophages generated by M-CSF stimulation of monocytes.
Methods
Reagents and antibodies
Human M-CSF, GM-CSF, and IL-4 and murine M-CSF were purchased from Miltenyi Biotec; MG132 from the Peptide Institute; clasto-lactacystin β-lactone from Cayman Chemical; cathepsin B inhibitor CA-O74-Me, cathepsin K inhibitor II, cathepsin inhibitor III, and cathepsin B substrate III from Calbiochem; Q-VD-OPH from MBL International; Suc-LLVY-AMC from Enzo Life Sciences; cathepsin K activity assay kit from Abcam; LPS, flagellin, and Gardiquimod from Sigma-Aldrich; poly I:C (polyinosinic:polycytidylic acid) from Invitrogen; and Pam from EMC Microcollections. We used polyclonal rabbit anti-NPM1, cleaved caspase-3, caspase-7, and phospho-IκB-α antibodies from Cell Signaling Technology; mouse monoclonal anti–caspase-8 from MBL or Santa Cruz Biotechnology; mouse monoclonal anti–cathepsin B from Sigma-Aldrich; and mouse monoclonal anti-HSC70 and goat polyclonal anti–lamin B and HSP60 antibodies from Santa Cruz Biotechnology. HRP-conjugated goat anti–mouse and anti–rabbit antibodies were purchased from Jackson Immunoresearch, and HRP-conjugated rabbit anti–goat antibody was purchased from Dako. allophycocyarin-conjugated anti-CD71, and anti-CD16, PE-conjugated anti-CD163 and anti-CD14, and isotype-matched controls were from BD.
Monocyte culture and differentiation
Human peripheral blood monocytes were obtained from healthy donors with informed consent according to recommendations of an independent scientific review board. Cells were enriched with a monocyte isolation kit with an autoMACS Separator (Miltenyi Biotec) according to the manufacturer's instructions, grown in RPMI 1640 medium with glutaMAX-I (Lonza) supplemented with 10% (vol/vol) FBS (Lonza) in standard conditions, and exposed to 100 ng/mL M-CSF. Macrophage differentiation (adhesion to culture flasks and fibroblast-like shape) was visualized with standard optics (Zeiss). Wild-type and npm+/− C57BL6 mice (kindly provided by P. P. Pandolfi, Harvard Medical School, Boston, MA) were bred in our animal facility. Experiments were performed with the approval of the ethics committee of the University of Burgundy. Bone marrow cells were extracted from tibias and femurs of wild-type, npm+/−, casp3−/−, casp7−/−, or cathB−/− C57/BL6 mice; incubated with FITC-labeled anti-CD49b, -CD45R, -CD3ϵ, and -ter119 antibodies (Miltenyi Biotec); and washed and incubated with anti-FITC microbeads (Miltenyi Biotec). Cells were then separated with autoMACS (Miltenyi Biotec), and the negative fraction that contained purified monocytes was treated with murine M-CSF (100 ng/mL). When indicated, LPS (0.5 mg/kg) was injected in the tail vein of wild-type, npm+/−, and tlr4+/− mice, and blood samples were collected intraorbitally at different times to measure plasma IL-6 by CBA (BD Biosciences). Peritoneal macrophages were obtained from wild-type mice, by peritoneal washes. Cells were incubated for 4 hours at 37°C, and supernatant cells were recovered. Cytokines were analyzed in human macrophage culture supernatant or mice plasma by use of cytokine arrays (R&D Systems). Human macrophages (2 × 106) were transfected with small interfering RNA (siRNA, 100 pmol/L; Applied Biosystems; sequences in supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) or plasmids (250 ng) using JetPEI-Macrophage from Polyplus Transfection according to the manufacturer's instructions.
Flow cytometry
Macrophage differentiation was studied by cytometry. Cells were washed with ice-cold PBS; incubated at 4°C for 1 hour in PBS/BSA (BSA 0.1%) with anti-CD71, -CD163, -CD14, and -CD16 antibodies or an isotype control; and washed and fixed in 2% paraformaldehyde. Fluorescence was measured with an LSRII flow cytometer (BD Biosciences). To detect caspase activity, we used FAM-DEVD-fmk and FAM-IETD-fmk detection kits (PromoCell).
Immunoblot assays
Cells were lysed for 15 minutes at 4°C in lysis buffer (1% SDS, 0.4mM Na3VO4, 10mM Tris, pH 7.4). Lysates were sonicated (12 seconds, 5% amplitude, on ice), and 30 μg of proteins was separated and transferred according to standard protocols before analysis with a chemiluminescence detection kit (Santa Cruz Biotechnology).
Real-time PCR and mutagenesis
Total RNA was isolated with TRIzol (Invitrogen) and reverse transcribed by Moloney murine leukemia virus reverse transcriptase (Promega) with random hexamers (Promega). Real-time PCR was performed with AmpliTaq Gold polymerase in an Applied Biosystems 7500 Fast thermocycler using the standard SyBr Green detection protocol. Primer sequences are available on request. Mutagenesis was performed by PCRs. Briefly, 2 PCRs were performed on normal npm cDNA with different couples of primers: sense NPM/antisense MUT and sense MUT/antisense NPM with PfuUltra polymerase (Stratagene), which does not add an A in 3′. Sequences are provided in supplemental Table 2. The PCR products were purified (Promega Wizard kit) and mixed, and the 2 fragments were annealed before they were amplified with AccuPrime polymerase (Invitrogen) using NPM forward and reverse primers. PCR product was then purified and cloned into pcDNA3.1V5/His-TOPO (Invitrogen) according to the manufacturer's instructions. The NPM-p30 and NPM-p20 fragments were also cloned in pcDNA3.1V5 His-TOPO with primers mentioned in supplemental Table 2.
In vitro production and cleavages
NPM wild-type or mutant proteins were produced in vitro with TNT Quick Coupled Transcription/Translation Systems using Transcend tRNA according to the manufacturer's instructions (Promega). Cleavages were assessed with 5 μL of the TNT production in caspase buffer (25mM HEPES, 10mM DTT, 1% 3 [(3-chloamidopropyl)dimethylammonio]-1 propanesulfonic acid) and the different recombinant caspases: 2, 3, 7, 8, and 10 from R&D Systems, 6 and 9 from PromoKine. Cathepsin B was activated before the assay in buffer that contained 25mM MES, 5mM DTT, pH 5 for 15 minutes at room temperature. The assay was performed in 25mM MES, pH 5 for 2 hours at 37°C with 5 μL of the TNT reaction. Cathepsin S was activated in 50mM NaOAc, 5mM DTT, 250mM NaCl, pH 4.5 for 2 hours at room temperature, and the assay was performed in the same buffer for 2 hours at 37°C. Cathepsin L was activated in 50mM MES, 10mM DTT, pH 6 for 15 minutes at 4°C, and the assay was performed in the same buffer for 2 hours at 37°C. Human recombinant cathepsins were purchased from R&D Systems.
Functional assays
A wound-healing assay was performed by implanting cells with high density the day before the experiment, then scraping the culture with a 1-10-μL tip before washing and after recolonization by video microscopy during a 30-hour period. Apoptotic bodies were collected from CFSE-stained VAL cells (10μM for 30 minutes at 37°C) treated overnight with Fas ligand, which induced > 80% cell death. Macrophages were implanted in 24-well plates and incubated for 30 minutes with Alexa Fluor 488 Escherichia coli BioParticles conjugate (Invitrogen) or for 1 hour with VAL apoptotic bodies at 37°C and washed in cold PBS. Extracellular fluorochromes were quenched with Trypan blue, and fluorescence was measured by flow cytometry. Engulfment of bacteria and apoptotic bodies was inspected by microscopy. Transmigration assay was performed by counting the cells that remained in the upper chamber of a cell culture insert (Millicell; Millipore) 24 hours after 2 × 105 cells had been seeded in this chamber. All of these experiments were performed in quadruplicate.
Statistical analysis
Statistical analyses were performed with a Mann-Whitney test.
Chromatin immunoprecipitation and immunoprecipitation
ChIP experiments were performed according to the manufacturer's instructions (Millipore). Briefly, cells were cross-linked with paraformaldehyde and then with glycine, washed in PBS, lysed, and sonicated. After preclearance with protein G agarose, 5 μg of antibodies was added and incubated overnight (B23 antibody and FLAG [as IgG control] from Sigma-Aldrich). After different washes, DNA was eluted and purified by ethanol precipitation. ChIP-Re-ChIP experiments were performed with Re-ChIP-IT (Active Motif) according to the manufacturer's instructions. Briefly, chromatin immunoprecipitated by NPM (Sigma-Aldrich) or HDAC1 (Active Motif) antibodies was removed from the magnetic beads, the chromatin was desalted, and a second ChIP was performed with p65 (Santa Cruz Biotechnology), NPM (Sigma-Aldrich), or IgG control (Santa Cruz Biotechnology) antibodies. The cross-links of these sequentially immunoprecipitated protein-DNA complexes were then reversed, and the DNA was analyzed by real-time PCR for MCP1 promoter (forward: CGGGCCCAGTATCTGGAAT; reverse: AGGAGGCAGCTTTGG AAGTTC). Macrophages treated by LPS or not treated were harvested, washed twice in ice-cold PBS, and lysed. Equal amounts of protein extract were incubated with the indicated antibodies (2 μg) at 4°C overnight on rotation. The immune complex was captured by protein A/G PLUS agarose (Santa Cruz Biotechnology) at 4°C for 4 hours with rotation. Complexes were washed 4 times with lysis buffer and analyzed by immunoblotting.
Cytokine antibody blot array and cytokine quantitation
Monocytes were induced to differentiate into macrophages in the presence of recombinant human M-CSF, as described previously, in the presence or absence of LPS. Two hours later, cell culture supernatant was collected and stored at −80°C until analysis. The presence of chemokines/cytokines in macrophage supernatant was assessed with a human cytokine antibody array, panel A (R&D Systems) according to the manufacturer's instructions. Human MCP1 and IL-8 (CXCL8) were quantified in culture supernatants and murine KC in plasma with a bead-based FlowCytomix multiple-analyte detection system (eBioscience) according to the manufacturer's instructions. Data were analyzed with FlowCytomix software. Mice were injected with low-dose LPS (0.5 mg/kg) in the tail vein. Wild-type and npm+/− mouse behavior was videotaped during the first 30 minutes. Forward and spin-around movements were scored.
Results
Nucleophosmin is cleaved by caspases in M-CSF–treated monocytes
The differentiation of human blood monocytes into macrophages on exposure to M-CSF is associated with caspase-8 and -3 activation that occurs at day 2-3 after the beginning of cell treatment (Figure 1A; supplemental Figure 1A).3 Caspase activation is associated with a decrease in the expression of full-length NPM1 and the appearance of a 30-kDa peptide, then a 20-kDa peptide, which suggests NPM1 cleavage. Caspases are not activated, NPM1 remains uncleaved, and the full-length protein accumulates in monocytes exposed to GM-CSF alone (supplemental Figure 1B) or to GM-CSF and IL-4 (Figure 1A). The pan-caspase inhibitor qVD prevents caspase activation, the decrease in the expression of full-length NPM1, and the appearance of the 30- and 20-kDa peptides in M-CSF–treated cells (Figure 1B). This inhibitor also impairs the differentiation process, as exemplified by inhibition of characteristic fibroblast-like morphologic changes (Figure 1C) and the decreased expression of CD163 and CD16 on the cell surface (Figure 1D). A similar activation of caspases (not shown) and cleavage of NPM1 into p30 and p20 fragments (supplemental Figure 1C) are observed in bone marrow murine monocytes induced to differentiate into macrophages on exposure M-CSF, which can be prevented by qVD (data not shown). Human NPM1 produced in reticulocyte lysates is cleaved into a 30-kDa fragment by recombinant active caspase-3, -7, and -8, whereas caspase-2, -6, and -9 do not cleave the protein (Figure 1E). In addition to caspase-8 and -3, caspase-7 is activated in M-CSF–treated monocytes (Figure 1F), and NPM1 cleavage is conserved in M-CSF–treated monocytes from caspase-3−/− mice (Figure 1G-H) but is decreased in M-CSF–treated monocytes from caspase-7−/− mice (Figure 1H), which suggests that caspase-7 is a key enzyme in NPM1 cleavage, at least in mice. To determine NPM1 cleavage site by caspases, we deleted the acidic region enriched in aspartic residues (161-183) or changed aspartate residues into alanine (supplemental Figure 1D). Caspase-3 (Figure 1I) and caspase-7 (data not shown) cleaved all of these mutants except the D213A, which indicates that caspases cleave NPM at NGKD213 to generate the 30-kDa fragment. Of note, the 2 fragments of NPM1 were identified in murine macrophages collected from the peritoneum (Figure 1J). The caspase-mediated proteolysis of NPM may occur in the cytoplasm, because these enzymes are activated in this cell compartment (supplemental Figure 1E).
Nucleophosmin cleavage by caspases in M-CSF–treated monocytes. Human peripheral blood monocytes from healthy donors were cultured with M-CSF (100 ng/mL) or GM-CSF (100 ng/mL) and IL-4 (10 ng/mL). (A) Immunoblot analysis of caspase-8, caspase-3, and NPM1 in cells exposed for indicated times to cytokines. HSC70 was used as loading control. (B) Immunoblot analysis of caspase-8, caspase-3, and NPM1 in monocytes exposed for indicated times to M-CSF in the absence or presence of 50μM qVD-OPH. HSC70 was used as loading control. (C) Phase contrast examination of monocytes exposed for 4 days to M-CSF in the absence or presence of qVD. (D) Flow cytometric analysis of indicated markers at the surface of monocytes exposed for 4 days to M-CSF in the absence or presence of qVD. Each panel is representative of at least 4 independent experiments. (E) NPM1 was produced in reticulocyte lysates and incubated for 8 hours with indicated recombinant active caspases. EV indicates empty vector, and −, controls (no DNA). (F) Immunoblot analysis of caspase-7 in monocytes exposed for indicated times to M-CSF. (G) Immunoblot analysis of NPM1 in caspase-3−/− (C3−/−) and wild-type (WT) monocytes exposed for 4 days to M-CSF in the absence or presence of 50μM qVD-OPH. (H) Immunoblot analysis of NPM1 in C3−/−, C7−/−, and wild-type (WT) monocytes exposed for 4 days to M-CSF (3 independent WT and 2 independent C7−/− samples are shown). (I) NPM1 wild-type (WT) and mutants (Mut) were produced in reticulocyte lysates and incubated for 8 hours with recombinant active caspase-3. EV indicates empty vector; and −, controls (no DNA). (J) Immunoblot analysis of NPM1 in mouse peritoneal macrophages in the absence or presence of LPS. Each panel is representative of at least 3 independent experiments.
Nucleophosmin cleavage by caspases in M-CSF–treated monocytes. Human peripheral blood monocytes from healthy donors were cultured with M-CSF (100 ng/mL) or GM-CSF (100 ng/mL) and IL-4 (10 ng/mL). (A) Immunoblot analysis of caspase-8, caspase-3, and NPM1 in cells exposed for indicated times to cytokines. HSC70 was used as loading control. (B) Immunoblot analysis of caspase-8, caspase-3, and NPM1 in monocytes exposed for indicated times to M-CSF in the absence or presence of 50μM qVD-OPH. HSC70 was used as loading control. (C) Phase contrast examination of monocytes exposed for 4 days to M-CSF in the absence or presence of qVD. (D) Flow cytometric analysis of indicated markers at the surface of monocytes exposed for 4 days to M-CSF in the absence or presence of qVD. Each panel is representative of at least 4 independent experiments. (E) NPM1 was produced in reticulocyte lysates and incubated for 8 hours with indicated recombinant active caspases. EV indicates empty vector, and −, controls (no DNA). (F) Immunoblot analysis of caspase-7 in monocytes exposed for indicated times to M-CSF. (G) Immunoblot analysis of NPM1 in caspase-3−/− (C3−/−) and wild-type (WT) monocytes exposed for 4 days to M-CSF in the absence or presence of 50μM qVD-OPH. (H) Immunoblot analysis of NPM1 in C3−/−, C7−/−, and wild-type (WT) monocytes exposed for 4 days to M-CSF (3 independent WT and 2 independent C7−/− samples are shown). (I) NPM1 wild-type (WT) and mutants (Mut) were produced in reticulocyte lysates and incubated for 8 hours with recombinant active caspase-3. EV indicates empty vector; and −, controls (no DNA). (J) Immunoblot analysis of NPM1 in mouse peritoneal macrophages in the absence or presence of LPS. Each panel is representative of at least 3 independent experiments.
NPM p30 fragment is further cleaved by cathepsins
To explore how the 20-kDa N-terminal peptide was generated, we tested other protease inhibitors. Proteasome activity, as measured by cleavage of Suc-LLVY-AMC peptide, increases in monocytes undergoing M-CSF–driven differentiation (not shown). Exposure of M-CSF–treated cells for 4 hours to the proteasome inhibitors MG132 and clasto-lactacystin β-lactone induced accumulation of phospho-IκBα, a well-known proteasome target, whereas inhibition of proteasome activity was confirmed by use of a fluorescent peptide target. Lactacystin failed to prevent the appearance of the p20 fragment, whereas the tripeptide aldehyde MG132 (Z-Leu-Leu-Leu-H) completely inhibited this cleavage and increased the expression of the p30 fragment and, to a lesser extent, of full-length NPM (Figure 2A). MG132 is also a cathepsin K inhibitor, but the large-spectrum cathepsin inhibitor III (Z-Phe-Gly-NHO-Bz-pOMe, CI) strongly reduced NPM1 cleavage into p20 and induced accumulation of p30, whereas a more specific cathepsin K inhibitor (CKI) did not (Figure 2B). In vitro cleavage experiments with recombinant active enzymes at acidic pH demonstrated that cathepsin S did not affect NPM1 that was totally degraded by cathepsin L. Cathepsin B cleaved full-length NPM1 to generate primarily a 20-kDa fragment, although the protease could also generate a 30-kDa fragment (Figure 2C). Increased cathepsin B expression and activity is a well-known feature of M-CSF–driven monocyte differentiation into macrophages (Figure 2E-F).18-20 Both the cathepsin inhibitor III and CA-074Me, which preferentially inhibit cathepsin B, reduced the appearance of NPM-p20 and induced the accumulation of NPM-p30 (Figure 2D). The cleavage of NPM-p30 into NPM-p20 fragments was decreased strongly, although not completely suppressed, in cathepsin B−/− murine monocytes exposed to M-CSF (Figure 2G). Recombinant cathepsin B cleaved NPM1 into an N-terminal p20 fragment, which could not be prevented by mutagenesis of aspartate residues (not shown). The p30 fragment was identified in both the nucleus and the cytoplasm of M-CSF–treated monocytes, whereas the p20 fragment was detected only in the cytoplasm (supplemental Figure 1F).
Cathepsin B contributes to the formation of the NPM-p20 fragment. (A) Human monocytes were treated with M-CSF for 6 days. MG132 (30μM) or clasto-lactacystin β-lactone (10μM) was added 4 hours before cell lysis and immunoblotting with indicated antibodies or measurement of proteasome activity with Suc-LLVY-AMC substrate (mean ± SD of triplicates; ****P < .001). AU indicates arbitrary units. (B) Human monocytes were treated with M-CSF for 6 days. MG132 (30μM) or cathepsin inhibitor (5μM) or cathepsin K inhibitor (5μM) was added 4 hours before cell lysis and immunoblotting with indicated antibodies (HSC, loading control) or measurement of cathepsin K activity with Ac-LR-AFC (mean ± SD of triplicates; ****P < .001). (C) Full-length NPM1 was produced in reticulocyte lysates in the presence of biotinylated lysine, then exposed to indicated recombinant active cathepsins at acidic pH for 2 hours at 37°C. Peptides were identified with streptavidin-HRP and revealed by chemiluminescence. (D) Human monocytes were treated with M-CSF for 6 days. MG132 (30μM), cathepsin inhibitor (5μM), or CA-074Me cathepsin B inhibitor (5μM) was added 4 hours before cell lysis and immunoblotting with indicated antibodies (HSC70, loading control) or measurement of cathepsin B activity with Z-RR-AMC substrate (mean ± SD of triplicates; ****P < .001). (E) Measurement of cathepsin B activity with Z-RR-AMC substrate in monocytes treated with M-CSF for 0, 3, or 6 days. (F) Immunoblot analysis of cathepsin B in monocytes exposed for indicated times to M-CSF. HSC70 was used as loading control. (G) Monocytes were collected from the bone marrow of wild-type (WT) or cathepsin B−/− (CathB−/−) mice and induced to differentiate into macrophages on exposure to murine M-CSF (100 ng/mL) for indicated times before immunoblot analysis of NPM1 expression (HSC70, loading control). Each panel is representative of at least 3 independent experiments.
Cathepsin B contributes to the formation of the NPM-p20 fragment. (A) Human monocytes were treated with M-CSF for 6 days. MG132 (30μM) or clasto-lactacystin β-lactone (10μM) was added 4 hours before cell lysis and immunoblotting with indicated antibodies or measurement of proteasome activity with Suc-LLVY-AMC substrate (mean ± SD of triplicates; ****P < .001). AU indicates arbitrary units. (B) Human monocytes were treated with M-CSF for 6 days. MG132 (30μM) or cathepsin inhibitor (5μM) or cathepsin K inhibitor (5μM) was added 4 hours before cell lysis and immunoblotting with indicated antibodies (HSC, loading control) or measurement of cathepsin K activity with Ac-LR-AFC (mean ± SD of triplicates; ****P < .001). (C) Full-length NPM1 was produced in reticulocyte lysates in the presence of biotinylated lysine, then exposed to indicated recombinant active cathepsins at acidic pH for 2 hours at 37°C. Peptides were identified with streptavidin-HRP and revealed by chemiluminescence. (D) Human monocytes were treated with M-CSF for 6 days. MG132 (30μM), cathepsin inhibitor (5μM), or CA-074Me cathepsin B inhibitor (5μM) was added 4 hours before cell lysis and immunoblotting with indicated antibodies (HSC70, loading control) or measurement of cathepsin B activity with Z-RR-AMC substrate (mean ± SD of triplicates; ****P < .001). (E) Measurement of cathepsin B activity with Z-RR-AMC substrate in monocytes treated with M-CSF for 0, 3, or 6 days. (F) Immunoblot analysis of cathepsin B in monocytes exposed for indicated times to M-CSF. HSC70 was used as loading control. (G) Monocytes were collected from the bone marrow of wild-type (WT) or cathepsin B−/− (CathB−/−) mice and induced to differentiate into macrophages on exposure to murine M-CSF (100 ng/mL) for indicated times before immunoblot analysis of NPM1 expression (HSC70, loading control). Each panel is representative of at least 3 independent experiments.
NPM-p30 and NPM-p20 inhibit macrophage functions
To gain insight into the role of the proteolytic fragments of NPM1 that accumulate in monocytes undergoing M-CSF–induced differentiation into macrophages, we transfected either a noncleavable mutant of NPM1 (D213A; MUT) or NPM-p30 or NPM-p20 into monocytes that had been exposed to M-CSF for 4 days and checked the expression of the transfected genes by quantitative RT-PCR (Figure 3A). The overexpression of NPM-p30 and NPM-p20 reduced the ability of differentiating macrophages to phagocytose fluorescent E coli bioparticles, which is a specific property of activated macrophages, without affecting engulfment of apoptotic bodies, which is a property shared by quiescent and activated phagocytes (Figure 3A).21 The 2 fragments also significantly decreased macrophage transmigration measured in a transwell migration assay (Figure 3B). Lastly, overexpression of NPM-p30 inhibited macrophage motility in a wound-healing assay (Figure 3C), whereas the noncleavable, full-length NPM1 did not (not shown).
NPM-p30 and NPM-p20 participate in macrophage quiescence. Macrophages were obtained by M-CSF treatment of human peripheral blood monocytes for 4 days, transfected with an empty pcDNA3.1 vector, this vector expressing D213A-mutated NPM1 (MUT), the p30 N-terminal fragment of NPM1 (P30), or the p20 N-terminal fragment of NPM1 (P20). In all panels, NS indicates nonsignificant; **P < .025; ***P < .01; ****P < .001. (A) Left panel, reverse transcription–quantitative PCR of NPM1; control of overexpression. Middle panel, Phagocytosis of fluorescent E coli bioparticles was determined by flow cytometry (mean ± SD of mean fluorescence indexes, triplicates). Right panel, Phagocytosis of apoptotic bodies was determined by flow cytometry (mean ± SD of mean fluorescence indexes, triplicates) 24 hours after transfection. (B) Transwell migration assay; cells were counted in the top panel after 24 hours of migration. (C) Recolonization capabilities were monitored 24 hours later in a wound-healing assay. (Inset) NPM1 protein level was checked by immunoblotting 24 hours after transfection.
NPM-p30 and NPM-p20 participate in macrophage quiescence. Macrophages were obtained by M-CSF treatment of human peripheral blood monocytes for 4 days, transfected with an empty pcDNA3.1 vector, this vector expressing D213A-mutated NPM1 (MUT), the p30 N-terminal fragment of NPM1 (P30), or the p20 N-terminal fragment of NPM1 (P20). In all panels, NS indicates nonsignificant; **P < .025; ***P < .01; ****P < .001. (A) Left panel, reverse transcription–quantitative PCR of NPM1; control of overexpression. Middle panel, Phagocytosis of fluorescent E coli bioparticles was determined by flow cytometry (mean ± SD of mean fluorescence indexes, triplicates). Right panel, Phagocytosis of apoptotic bodies was determined by flow cytometry (mean ± SD of mean fluorescence indexes, triplicates) 24 hours after transfection. (B) Transwell migration assay; cells were counted in the top panel after 24 hours of migration. (C) Recolonization capabilities were monitored 24 hours later in a wound-healing assay. (Inset) NPM1 protein level was checked by immunoblotting 24 hours after transfection.
NPM1 cleavage is abrogated on LPS-mediated activation
LPS is a macrophage activator through TLR4. Exposure of M-CSF–induced macrophages to LPS provoked the disappearance of p30 and p20 fragments and the accumulation of the full-length NPM1 (Figure 4A) within 2 hours (supplemental Figure 2A). LPS also decreased the NPM fragments in murine peritoneal macrophages (Figure 1J). This was associated with an inhibition of the proteolysis of caspase-8 and -3, as well as DEVD-FITC cleavage, which is known to be performed by caspases-3 and -7 (Figure 4A-B), and cathepsin B activity (Figure 4C). We did not detect any significant change in NPM1 mRNA level in LPS-activated compared with quiescent macrophages (not shown). These observations argued for an inhibition of the proteolytic process involving caspases and cathepsins in LPS-activated macrophages. Quiescent macrophages were exposed to other TLR ligands, including Pam, which binds TLR2; poly I:C, which binds TLR3; flagellin, which binds TLR5; and Gardiquimod, which binds TLR7/8. Of these ligands, Pam and flagellin also inhibited NPM1 proteolysis, whereas the 2 others did not (supplemental Figure 2B).
LPS-induced inhibition of NPM1 proteolysis. (A) Macrophages obtained by M-CSF treatment of human peripheral blood monocytes for 6 days were left untreated (−) or exposed to LPS (1 mg/mL) for 24 hours (day 7) or 48 hours (day 8) before cell lysis and immunoblot analysis of indicated proteins (HSC70, loading control). (B) Flow cytometric analysis of active caspase-3/7 in macrophages 48 hours after LPS treatment. (C) Cathepsin B activity was measured as in Figure 2D 48 hours after LPS treatment. Each panel is representative of at least 3 independent experiments in triplicate (****P < .001).
LPS-induced inhibition of NPM1 proteolysis. (A) Macrophages obtained by M-CSF treatment of human peripheral blood monocytes for 6 days were left untreated (−) or exposed to LPS (1 mg/mL) for 24 hours (day 7) or 48 hours (day 8) before cell lysis and immunoblot analysis of indicated proteins (HSC70, loading control). (B) Flow cytometric analysis of active caspase-3/7 in macrophages 48 hours after LPS treatment. (C) Cathepsin B activity was measured as in Figure 2D 48 hours after LPS treatment. Each panel is representative of at least 3 independent experiments in triplicate (****P < .001).
NPM1 negatively regulates cytokine production in LPS-treated macrophages
To explore the function of NPM1 in LPS-activated macrophages, we examined the effects of full-length NPM1 on cytokine gene expression. First, macrophages were transfected with a plasmid vector that encoded full-length NPM1 with a D213A mutation to prevent protein cleavage by activated caspases, 24 hours before exposure to LPS. Overexpressed full-length NPM1 almost completely abrogated the increase in MIP1α (CCL3), MCP1 (CCL2), MIP1β (CCL4), GROα/MIP2 (CXCL2), and IL-6 gene expression observed in LPS-treated macrophages (Figure 5). Conversely, the siRNA-mediated decrease in NPM expression induced a dramatic increase in the expression of MCP1and MIP1β genes when measured 6 hours after LPS treatment (Figure 5B). The expression of NPM1, mutated at the site of proteolysis to prevent any cleavage by caspases, in M-CSF–differentiated macrophages provoked a decrease in MCP-1, MIP-1α, IL-8 (CXCL8), Groα/KC (CXCL1), MIF, and IL-1rα protein level in the cell supernatants of macrophages treated for 6 hours with LPS (Figure 5C-D). The effects on IL-8 and MCP1 levels were confirmed by multiplex flow cytometry with a statistically significant decreased level of MCP1 and IL-8 in the supernatant of NPM-transfected cells exposed to LPS for 6 hours (Figure 5E).
NPM1 expression modulates chemokine gene expression on LPS exposure in human macrophages. Macrophages were obtained by M-CSF treatment of human peripheral blood monocytes for 4 days before transfection and 24 hours after transfection, were exposed to LPS for the indicated times. mRNA was collected for reverse transcription–quantitative PCR analyses of indicated chemokines. (A) Overexpression of a noncleavable mutant of NPM1 (D213A; NPM-MUT) in human cells. ■ indicates control vector; □, NPM-MUT–expressing vector. (Inset) NPM protein level in transfected cells. (B) Down-regulation of NPM1 with siRNA. Reverse transcription–quantitative PCR analyses were performed 3 hours after LPS treatment. Each panel is representative of at least 3 independent experiments. Scr indicates scrambled; and si1 and si2, two different siRNAs targeting NPM1. (C) The ratio between cytokine quantities detected in control and NPM-MUT–transfected macrophage supernatants with or without LPS exposure was quantified and normalized to control pcDNA-transfected cells. (D) Culture supernatant cytokine array analysis of macrophages treated with LPS for indicated times. AU indicates arbitrary units. (E) IL-8 and MCP1 chemokines were quantified with FlowCytomix in the supernatants of control and NPM-MUT–transfected macrophages treated or not treated with LPS and normalized to control pcDNA-transfected cells (3 independent experiments). Mean ± SD of triplicates; **P < .025; ***P < .01; ****P < .001.
NPM1 expression modulates chemokine gene expression on LPS exposure in human macrophages. Macrophages were obtained by M-CSF treatment of human peripheral blood monocytes for 4 days before transfection and 24 hours after transfection, were exposed to LPS for the indicated times. mRNA was collected for reverse transcription–quantitative PCR analyses of indicated chemokines. (A) Overexpression of a noncleavable mutant of NPM1 (D213A; NPM-MUT) in human cells. ■ indicates control vector; □, NPM-MUT–expressing vector. (Inset) NPM protein level in transfected cells. (B) Down-regulation of NPM1 with siRNA. Reverse transcription–quantitative PCR analyses were performed 3 hours after LPS treatment. Each panel is representative of at least 3 independent experiments. Scr indicates scrambled; and si1 and si2, two different siRNAs targeting NPM1. (C) The ratio between cytokine quantities detected in control and NPM-MUT–transfected macrophage supernatants with or without LPS exposure was quantified and normalized to control pcDNA-transfected cells. (D) Culture supernatant cytokine array analysis of macrophages treated with LPS for indicated times. AU indicates arbitrary units. (E) IL-8 and MCP1 chemokines were quantified with FlowCytomix in the supernatants of control and NPM-MUT–transfected macrophages treated or not treated with LPS and normalized to control pcDNA-transfected cells (3 independent experiments). Mean ± SD of triplicates; **P < .025; ***P < .01; ****P < .001.
Murine macrophages obtained by ex vivo differentiation of bone marrow monocytes on M-CSF treatment were also exposed to LPS. In murine Npm+/− macrophages, the cytokine response to LPS was amplified; that is, Mip1α, Mip1β, and Groα/Mip2 mRNA was strongly accumulated at 3 hours after LPS treatment, whereas MCP1 mRNA was increased 24 to 48 hours after LPS treatment (Figure 6A). Wild-type and Npm+/− mice were injected intravenously with LPS (0.5 mg/kg). Strikingly, shivers, shaking, and bristly hairs were observed within 15 minutes in all the Npm+/− mice compared with 30 minutes in wild-type animals (supplemental Figure 3A). We measured a significant increase in spin-around movements between 10 and 30 minutes after LPS injection in npm+/− compared with wild-type mice (Figure 6B), which was associated with an increase in plasma IL-6 level 30 minutes after LPS injection (Figure 6C-D). In addition, cytokine array analysis of mouse plasma collected 30 minutes after LPS injection showed an increase in IL-6, CXCL1 (KC), and CCL2 (MCP1) protein levels (Figure 6C-D; supplemental Figure 3B). Plasma levels of MIP2, M-CSF, C5a, sICAM1, IL-1rα, and IFN-γ measured 1 hour after LPS injection were also increased (Figure 6C), and the KC and IL-6 increase was confirmed by flow cytometry (Figure 6D). As anticipated, TLR4 mutation prevented any reaction to LPS, as demonstrated by measurement of IL-6 secretion (Figure 6E; supplemental Figure 3A).
NPM1 expression modulates chemokine secretion on LPS exposure in murine macrophages. (A) Mouse macrophages were obtained ex vivo from wild-type (WT) or npm+/− mice treated for 4 days by M-CSF before exposure to LPS and measurement of cytokine gene expression by reverse transcription–quantitative PCR. (B) Wild-type and npm+/− mice were injected intravenously with LPS (0.5 mg/mL). Ten minutes after injection, their behaviors were video recorded for 20 minutes, and spin-around movements were scored. (C) Plasma cytokine array analysis of wild-type and npm+/− mice injected with LPS at indicated time. (D) The ratio between cytokine quantities detected in control and npm+/− mouse plasma injected or not injected with LPS was quantified and normalized to untreated control mice. (E) Wild-type, npm+/−, and tlr4-mutated transgenic mice were injected intravenously with LPS (0.5 mg/mL). Their peripheral blood was collected after 30 minutes and examined by CBA for IL-6 level measurement and by FlowCytomix for KC fold increase measurement. Mean ± SD of 5 mice per group, 3 independent experiments; ****P < .001). Co indicates control.
NPM1 expression modulates chemokine secretion on LPS exposure in murine macrophages. (A) Mouse macrophages were obtained ex vivo from wild-type (WT) or npm+/− mice treated for 4 days by M-CSF before exposure to LPS and measurement of cytokine gene expression by reverse transcription–quantitative PCR. (B) Wild-type and npm+/− mice were injected intravenously with LPS (0.5 mg/mL). Ten minutes after injection, their behaviors were video recorded for 20 minutes, and spin-around movements were scored. (C) Plasma cytokine array analysis of wild-type and npm+/− mice injected with LPS at indicated time. (D) The ratio between cytokine quantities detected in control and npm+/− mouse plasma injected or not injected with LPS was quantified and normalized to untreated control mice. (E) Wild-type, npm+/−, and tlr4-mutated transgenic mice were injected intravenously with LPS (0.5 mg/mL). Their peripheral blood was collected after 30 minutes and examined by CBA for IL-6 level measurement and by FlowCytomix for KC fold increase measurement. Mean ± SD of 5 mice per group, 3 independent experiments; ****P < .001). Co indicates control.
NPM1 is recruited on MCP-1 promoter in LPS-treated macrophages
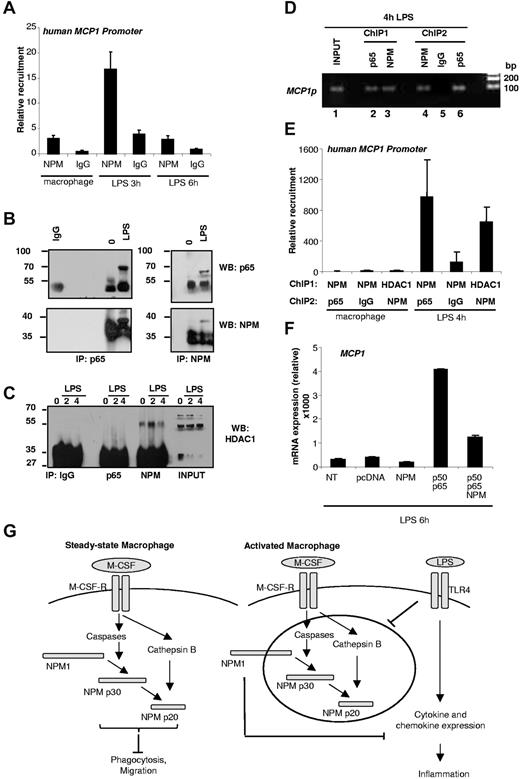
Using an anti-NPM1 antibody, we immunoprecipitated the chromatin of macrophages obtained by M-CSF–driven differentiation of human monocytes and exposed for 3 and 6 hours to LPS treatment. We detected a transient recruitment of NPM1 on the MCP-1 promoter after 3 hours of exposure to LPS. This recruitment occurred in the region of the κB-binding sites and was no longer detected after 6 hours of treatment (Figure 7A). LPS has been shown to activate the NF-κB transcription factor in human monocytes and macrophages.22 Coimmunoprecipitation experiments demonstrated that LPS treatment of macrophages for 4 hours provoked an interaction between endogenous NPM and the p65 subunit of NF-κB (Figure 7B). We also identified an interaction between endogenous NPM and the histone deacetylase HDAC1 that increased transiently after 2 hours of LPS exposure, whereas no interaction between p65 and HDAC1 could be detected (Figure 7C). ChIP and Re-ChIP experiments identified p65 interaction with NPM on MCP1 promoter (Figure 7D) and identified NPM and p65 or NPM and HDAC1 on this promoter (Figure 7E). Transient expression of p50 and p65 enhanced the expression of MCP1 mRNA, which could be prevented by the expression of NPM (Figure 7F). Altogether, NPM was a negative regulator of NF-κB–mediated induction of MCP1 gene expression in LPS-treated human macrophages.
NPM1 is recruited on MCP-1 promoter under LPS treatment. (A) ChIP experiment of monocyte-derived macrophages treated for indicated times with LPS using NPM1 antibody. One representative of 3 independent experiments is shown. (B) Monocyte-derived macrophages and 4-hour LPS–treated macrophage extracts were subjected to immunoprecipitation by control mouse IgG, p65 (left panel), or NPM (right panel) antibodies. The presence of p65 (top panel) or NPM (bottom panel) in the immunocomplexes was detected by immunoblotting. WB indicates Western blot. (C) Untreated macrophages and 2-hour or 4-hour LPS-treated macrophage extracts were subjected to immunoprecipitation by control mouse IgG, p65, or NPM antibodies. The presence of HDAC1 in the immunocomplexes was detected by immunoblotting. WB indicates Western blot. (D) Coexistence of p65 and NPM on MCP1 promoter was demonstrated by a sequential ChIP experiment. Chromatin was first immunoprecipitated by p65 (lane 2) or NPM (lane 3) antibody. A second round of ChIP was performed with NPM (lane 4) or IgG control (lane 5) for p65 or with p65 (lane 6) for NPM. Input is shown in lane 1. MCP1 promoter was analyzed by PCR. (E) Coexistence of NPM and p65 or HDAC1 and NPM on MCP1 promoter was demonstrated by a sequential ChIP experiment. Chromatin was first immunoprecipitated by NPM or HDAC1 antibody. A second round of ChIP was performed with p65 or IgG control for NPM or using NPM for HDAC1. MCP1 promoter was analyzed by quantitative PCR and normalized to input. (F) Primary human macrophages were transfected with the different indicated constructs. Six hours after LPS treatment, total RNA was isolated and analyzed for endogenous MCP1 mRNA expression. Absolute mRNA values were determined, normalized to L32 RNA, and reported as relative values. NT indicates nontransfected. (G) Proposed role of NPM1 in quiescent and activated macrophages. Ab differentiation into macrophages progresses, NPM1 is cleaved sequentially by caspases and cathepsin B. NPM-p30 or NPM-p20 overexpression inhibits phagocytosis and migration. On LPS activation, NPM1 proteolysis is abrogated, and NPM1 in turn inhibits cytokine expression.
NPM1 is recruited on MCP-1 promoter under LPS treatment. (A) ChIP experiment of monocyte-derived macrophages treated for indicated times with LPS using NPM1 antibody. One representative of 3 independent experiments is shown. (B) Monocyte-derived macrophages and 4-hour LPS–treated macrophage extracts were subjected to immunoprecipitation by control mouse IgG, p65 (left panel), or NPM (right panel) antibodies. The presence of p65 (top panel) or NPM (bottom panel) in the immunocomplexes was detected by immunoblotting. WB indicates Western blot. (C) Untreated macrophages and 2-hour or 4-hour LPS-treated macrophage extracts were subjected to immunoprecipitation by control mouse IgG, p65, or NPM antibodies. The presence of HDAC1 in the immunocomplexes was detected by immunoblotting. WB indicates Western blot. (D) Coexistence of p65 and NPM on MCP1 promoter was demonstrated by a sequential ChIP experiment. Chromatin was first immunoprecipitated by p65 (lane 2) or NPM (lane 3) antibody. A second round of ChIP was performed with NPM (lane 4) or IgG control (lane 5) for p65 or with p65 (lane 6) for NPM. Input is shown in lane 1. MCP1 promoter was analyzed by PCR. (E) Coexistence of NPM and p65 or HDAC1 and NPM on MCP1 promoter was demonstrated by a sequential ChIP experiment. Chromatin was first immunoprecipitated by NPM or HDAC1 antibody. A second round of ChIP was performed with p65 or IgG control for NPM or using NPM for HDAC1. MCP1 promoter was analyzed by quantitative PCR and normalized to input. (F) Primary human macrophages were transfected with the different indicated constructs. Six hours after LPS treatment, total RNA was isolated and analyzed for endogenous MCP1 mRNA expression. Absolute mRNA values were determined, normalized to L32 RNA, and reported as relative values. NT indicates nontransfected. (G) Proposed role of NPM1 in quiescent and activated macrophages. Ab differentiation into macrophages progresses, NPM1 is cleaved sequentially by caspases and cathepsin B. NPM-p30 or NPM-p20 overexpression inhibits phagocytosis and migration. On LPS activation, NPM1 proteolysis is abrogated, and NPM1 in turn inhibits cytokine expression.
Discussion
The catalytic activation of some caspases and cathepsins in human and mouse monocytes undergoing M-CSF–driven differentiation generates 2 NPM1 fragments that contribute to the differentiated phenotype by decreasing the migration, motility, and bacteria phagocytosis capabilities of the resting cells. As soon as the macrophage is activated by LPS, the proteolytic process is stopped, and the native NPM1 negatively regulates cytokine and chemokine gene expression and their secretion; that is, the full-length NPM1 is a transcriptional corepressor that maintains the right amount of cytokine gene expression.
Proteolysis of cellular components by cysteine proteases, including various combinations of caspases and cathepsins, has been associated with specific pathways of cell differentiation.23,24 Caspases and cathepsins are activated along M-CSF–driven differentiation of human peripheral blood monocytes.2,3,25 One of the protease targets in this differentiation context is NPM1,5 the proteolysis of which was also observed in resting mouse peritoneal macrophages. This protein was previously reported to be a caspase substrate in a cell-death setting,16 and a mass spectrometry approach identified a murine NPM1 cleavage site by caspase-7 at SVRD197.26 Using a mutagenesis approach, we show that recombinant caspase-3, -7, and -8 actually cleave the 38-kDa human protein at NGKD213 in vitro to generate a 30-kDa N-terminal fragment and that the proteolysis is not affected by mutation of D198, which corresponds to mouse D197. Analysis of caspase-3−/− and caspase-7−/− mouse monocytes exposed to M-CSF argues for a predominant function of caspase-7 in vivo. Whereas most of the NPM1 protein is cleaved in monocytes undergoing M-CSF–driven differentiation, the whole protein is hardly cleaved in vitro, which could indicate that NPM1 protein in the monocytes is in a conformation or cellular localization that is more accessible to caspases than NPM1 is in reticulocytes (eg, because of posttranslational modifications that include phosphorylation, sumoylation, and ubiquitylation).27-29 The 30-kDa N-terminal NPM1 peptide was identified in both the nucleus and the cytoplasm.
The cytotoxic granule protease granzyme M cleaves NPM1 at Leu-158 during natural killer cell–mediated killing, which was suggested to disable a death-protective function of the protein.17 In M-CSF–treated cells, we observed an additional proteolysis of the caspase-induced NPM-p30 fragment, which was mediated, at least in part, by cathepsin B (Figure 7B). Increased cathepsin B expression and activity has long been identified as a characteristic of M-CSF–driven monocyte differentiation.18-21 The cathepsin-mediated cleavage of the 30-kDa fragment may occur either in the lysosomes or phagolysosomes or in the cytosol, because the endopeptidase activity of some cathepsins could be conserved at neutral pH.30 The 20-kDa N-terminal fragment of NPM1 remains in the cytosol, which could indicate a cleavage site in the nuclear localization signal that is enriched in arginine and lysine residues.31 The reproducible observed slight residual proteolysis of NPM1 into p20 observed in cathepsin B−/− mouse monocytes exposed to M-CSF argues for an additional level of complexity of the proteolytic event.
In the absence of any inflammatory stimulus, the primary role of macrophages is to remove cellular debris such as apoptotic cells. When activated, macrophages eliminate invading microorganisms and trigger presentation of pathogen-derived peptide antigen.21 Here, we demonstrate that enforced expression of NPM-p30 and NPM-p20 during the differentiation process, for example, at day 4 of M-CSF exposure, decreases the capability of the cell to engulf bacteria and to migrate in a transwell or a wound-healing assay. These observations suggest that the caspase-mediated cleavage of NPM1 generates a fragment that participates in the resting stage of the unstimulated macrophage. Interestingly, enforced expression of NPM-p30 and NPM-p20 does not affect the cell's ability to engulf apoptotic bodies, which is a function exerted in normal tissues by resting macrophages. Future studies will address the mechanisms by which NPM-p30 and NPM-p20 fragments affect cellular motility and phagocytosis capabilities, which could involve the interaction of NPM1 fragments with Rock II and the ATPase domain of EG5.31,32 Recently, the cytoplasmic accumulation of NPM1, because of an AML-associated C-terminal frame-shift mutation of npm1, was shown to protect against death ligand–induced apoptosis by direct inhibition of caspase-8,33 which suggests that the NPM1 fragment could also prevent the amplification of the caspase cascade and subsequent cell death in M-CSF–treated monocytes.
Some of the mammalian TLRs, such as TLR2, TLR4, and TLR5, are expressed on the macrophage cell surface, whereas others, such as TLR3, TLR7, and TLR8, are in intracellular compartments. We show that agonists of the former group could inhibit NPM1 proteolysis in M-CSF–differentiated macrophages, whereas agonists of the latter could not. TLR signaling pathways share common molecules, such as MyD88, and demonstrate specific activities, for example, in the regulation of chemokine and cytokine secretion.34,35 Here, we demonstrate that the amount of full-length NPM1 in the macrophages determines the level of chemokine gene expression (namely, MIP1α, MIP1β, MIP2, and MCP1) on LPS stimulation. Overexpressed NPM1 completely abrogates the increased transcription of chemokine genes in LPS-treated macrophages, whereas a decrease in NPM1 leads to a huge amount of chemokine production. Interestingly, the decreased expression of NPM1 in mouse cells also affects the cytokine response to LPS. Thus, NPM1 may regulate chemokine gene transcription in a dose-dependent manner in LPS-activated macrophages, thereby modulating macrophage polarization. The human NPM1 gene on the 5q35 locus is frequently affected by chromosome translocation, mutation, and deletion in hematopoietic disorders.36-38 The common feature in these cases is that the total amount of NPM1 is decreased because of the reduction of the NPM1 gene dosage to heterozygosity, and npm1+/− mice develop a hematologic syndrome with features of human myelodysplastic syndrome in which the deregulated production of cytokines and altered polarization of macrophages could possibly play a role. Complete inactivation of the npm1 gene in the mouse germline leads to a series of developmental defects and embryonic lethality at midgestation because of severe anemia that results from defects in primitive hematopoiesis.39
LPS stimulation of macrophages has demonstrated a rapid and extended effect on gene expression,40,41 including the sustained activation of several inflammatory genes mediated by a transcriptional network in which NF-κB played an important role.42 More than 20 years ago, NPM1 was demonstrated to bind to secondary structures of single-stranded and double-stranded DNA.43 More recently, NPM1 was shown to be an NF-κB coactivator for the induction of the human SOD2 gene13 and to recruit histone deacetylases on genes whose repression is associated with retinoic acid–induced differentiation, thus behaving like a transcriptional corepressor.14,16 NPM1 is also a histone chaperone involved in chromatin organization and transcription control.44 Here, we have shown that NPM1 is localized on the MCP1 promoter in LPS-activated macrophages, and we provide a demonstration that NPM1 may function as a negative regulator in this context. NPM1 and NF-κB are part of the same transcriptional regulatory complex on the MCP1 promoter on LPS stimulation that also enforces NPM1 interaction with the histone deacetylase HDAC1 on the MCP1 promoter.
NPM1 is only one of the caspase and cathepsin B targets in M-CSF–treated monocytes. We have shown previously that key proteins can be protected by chaperones from caspase-mediated cleavage when these enzymes are activated along a differentiation process.45,46 Here, we show how the limited proteolysis of a cellular protein actively participates in the tightly controlled differentiation process and is rapidly inhibited on activation of the differentiated cells to exert other functions. Future studies will also determine whether NPM1 is a protease target in other cell types in which proteases are requested for the differentiation program, for example, in erythroid cells undergoing erythropoietin-induced maturation.25,47,48 More generally, these results underline the potential contribution of proteases in the fine-tuning of a differentiation process.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to André Bouchot for microscopy analyses, Jennifer Fraszczak for providing TLR ligands, and Pier-Paolo Pandolfi for kindly providing the npm+/− mice.
L.G. obtained a doctoral fellowship from the Ligue Nationale Contre le Cancer (LNCC). Our group (E.S.) is supported by grants from the LNCC (Label), ANR, INCA, Centpoursanglavie, and Fondation de France. The UMR1009 equipment was supported by the Association pour la Recherche sur le Cancer. T.V.B. obtained a postdoctoral fellowship with the FWO-Vlaanderen, and P.V. is holder of a Methusalem grant from the Flemish government (BOF09/01M00709). Research in P.V.'s group is supported by Apo-Sys, FP7-200767, and Euregional PACT II, IAP6/18, FWO G.0875.11, FWO G.0973.11, UGent MRP GROUP-ID, and VIB.
Authorship
Contribution: L.G., N.B., G.J., A.J., R.C., and N.D. performed in vitro experiments; L.G., T.G., C.P., E.D., and T.V.B. performed in vivo experiments; B.M. and P.V. provided animal models; and E.S. directed the work and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Eric Solary, Inserm UMR 1009, Institut Gustave Roussy, 114 Rue Edouard Vaillant, 94804 Villejuif Cedex, France; e-mail: eric.solary@igr.fr.