Abstract
Hydroxyurea has been shown to be efficacious for the treatment of sickle cell anemia (SCA), primarily through the induction of fetal hemoglobin (HbF). However, the exact mechanisms by which hydroxyurea can induce HbF remain incompletely defined, although direct transcriptional effects and altered cell cycle kinetics have been proposed. In this study, we investigated potential epigenetic and alternative molecular mechanisms of hydroxyurea-mediated HbF induction by examining methylation patterns within the Gγ-globin promoter and miRNA expression within primary CD71+ erythrocytes of patients with SCA, both at baseline before beginning hydroxyurea therapy and after reaching maximum tolerated dose (MTD). Using both cross-sectional analysis and paired-sample analysis, we found that the highly methylated Gγ-globin promoter was inversely correlated to baseline HbF levels, but only slightly altered by hydroxyurea treatment. Conversely, expression of several specific miRNAs was significantly increased after hydroxyurea treatment, and expression of miR-26b and miR-151-3p were both associated with HbF levels at MTD. The significant associations identified in these studies suggest that methylation may be important for regulation of baseline HbF, but not after hydroxyurea treatment, whereas changes in miRNA expression may be associated with hydroxyurea-mediated HbF induction. This study was registered at ClinicalTrials.gov (NCT00305175).
Introduction
Hydroxyurea is a potent ribonucleotide reductase inhibitor that was initially developed as an antineoplastic agent, but was later found to be beneficial for the treatment of sickle cell anemia (SCA) primarily through its ability to induce fetal hemoglobin (HbF).1-4 Clinical trials have proven hydroxyurea to be efficacious in children, adolescents, and adults with SCA for increasing HbF.2,5-7 Hydroxyurea decreases the number and frequency of vaso-occlusive pain crises, episodes of acute chest syndrome, hospitalizations,6,8-10 and more recently has been shown to also reduce mortality.11,12 Although highly efficacious for most patients with SCA, there is considerable inter-patient variability creating a broad spectrum of HbF induction.13,14 HbF induction by cytotoxic drugs, such as hydroxyurea, has been correlated to cell cycle inhibition leading to activation of stress erythropoiesis.15-17 Other studies have suggested that HbF induction by hydroxyurea is mediated more specifically via nitric oxide-dependent transcriptional mechanisms18,19 and cyclic nucleotides.20,21 However, the precise mechanisms by which hydroxyurea can induce HbF in patients with SCA remain incompletely defined.
Expression of erythroid genes at the β-globin locus is developmentally regulated by a complex series of epigenetic and molecular processes. Proposed epigenetic mechanisms of HbF regulation in hemoglobinopathies include methylation, histone deacetylation, and chromosomal looping (reviewed in Sankaran et al,22 Bank et al,23 and Stamatoyannopoulos et al24 ). Methylation is of particular interest for understanding the reactivation of HbF because hypomethylating agents such as 5-azacytidine and decitabine were among the first compounds used to induce HbF pharmacologically, presumably by inhibition of DNA methyltransferases.25,26 Methylation of CpG dinucleotides within the γ-globin promoter has been associated with gene silencing by inhibiting the transcription of γ-globin genes in adult bone marrow.27 Compared with fetal erythroid cells, the γ-globin promoter region of adult erythroid cells is highly methylated, and this hypermethylation has been inversely correlated with HbF expression.28 Although hydroxyurea has not been identified as a hypomethylating agent, an early study suggested a concurrent decrease of methylation within the γ-globin gene in association with hydroxyurea exposure.3
Small noncoding RNA oligonucleotides (microRNA, miRNA) have emerged as ubiquitous and potent molecular regulators that modulate the expression of many protein-coding genes by inhibiting mRNA translation.29,30 Multiple miRNAs have been implicated in the regulation of cell differentiation and maturation during hematopoiesis and erythropoiesis (reviewed in Zhao et al,31 Havelange et al,32 and Laurie et al33 ). Although many studies have investigated miRNA expression during blood cell development, relatively few studies have focused on the importance of miRNA expression for HbF induction. Differential cellular miRNA expression patterns from patients with SCA compared with individuals without SCA has been reported34 ; miR-144 was found to modulate oxidative stress and was associated with the degree of anemia,35 whereas miR-15a and miR-16-1 have been linked to elevated HbF by acting via the transcription factor MYB.36
In the current study, we investigated hydroxyurea-mediated induction of HbF by analyzing epigenetic and molecular profiles in sickle erythroid cells; specifically changes in methylation patterns and miRNA expression, using circulating CD71+ erythroid progenitors isolated from SCA patients before starting hydroxyurea and after reaching the maximum tolerated dose (MTD). We examined DNA methylation of CpG islands in the Gγ-globin promoter region and analyzed changes in miRNA expression associated with HbF induction. By identifying associations of epigenetic and molecular changes with HbF after hydroxyurea treatment, we provide new insights into the mechanisms of hydroxyurea efficacy and potentially provide new targets to modify in efforts to improve treatment of SCA.
Methods
Isolation of human nucleated red blood cells (NRBC) and reticulocytes
Circulating erythroid cells were isolated from children with SCA and from healthy normal adult volunteers. Children with SCA were enrolled in the Hydroxyurea Study of Long-term Effects clinical protocol (HUSTLE, ClinicalTrials.gov NCT00305175). HUSTLE was approved by the St Jude Children's Research Hospital Institutional Review Board; all families gave written consent and when appropriate patients gave assent in accordance with the Declaration of Helsinki. As part of this protocol, 2-5 mL venous blood was collected in EDTA from each patient at baseline and after reaching treatment MTD. Blood was centrifuged at 500g for 2 minutes and the plasma supernatant containing platelets was removed. Each sample was next depleted of CD45+ cells to remove leukocytes and then enriched for CD71+ erythroid cells using an autoMACS magnetic cell sorter (Miltenyi Biotec). The average recovery of CD71+ cells were 60 × 106 and 40 × 106 cells/mL from SCA patients at baseline and at MTD, respectively. For normal controls, the average yield was 3 × 106 cells/mL, so in some cases 3 separate isolations were pooled for a total of 9 mL of blood to ensure adequate amounts of RNA could be isolated. Purity of isolated CD71+ erythroid cells was confirmed by FITC-labeled anti-CD71 and allophycocyanin (APC)-labeled anti-CD45 antibodies (BD Biosciences). These purified CD45− CD71+ populations (< 0.8% CD45+ and > 90% CD71+) contained primarily reticulocytes and nucleated erythrocytes; half of the yield was used for DNA-based methylation studies, whereas the other half was used for RNA isolation and miRNA studies.
Determination of HbF levels
At the time of sample collection, the HbF level was determined using HPLC methodology and included the acetylation peak. For consistency and because some patients had transfused blood present at baseline, %HbF was recalculated as HbF/(HbF+HbS) to more accurately measure endogenous HbF production.37 HbF values were compared by the paired t test.
Methylation quantification by pyrosequencing
To quantify DNA methylation in the Gγ-globin promoter region, genomic DNA was prepared from CD71+ erythroid cells using Puregene Blood Core Kit C (QIAGEN). Between 100-500 ng of DNA were treated with sodium bisulfite and cleaned using the EZ DNA Methylation Kit (Zymo Research) according to the manufacturer's protocol. Nested PCR was then performed using nucleotide primers designed to amplify CT-converted DNA in the promoter region of human Gγ-globin gene (HBG2; supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). PCR was initially performed using 25 touchdown cycles having an annealing temperature starting from 55°C to 48°C, followed by 40 nested cycles with standard PCR conditions of Hot Start Taq Polymerase PCR Kit (QIAGEN) with a final annealing temperature of 49°C and extension time of 1 minute. PCR products were purified using PCR Purification spin tubes (QIAGEN), and then sent to EpigenDx for quantitative pyrosequencing using 3 promoter specific primers SP1.1, SP2, and SP3. Percent methylation was determined at CpG sites located at positions −54, −51, +5, +16, and +48 within the Gγ-globin promoter. Nonerythroid peripheral blood mononuclear cells isolated from a single control individual were included in 13 separate analyses to ensure reproducibility of the method. Statistically significant differences in methylation were determined by the Wilcoxon-Mann-Whitney test.
Isolation of miRNA from CD71+ erythroid cells
miRNA was isolated from CD71+ erythroid cells by 2 different methods for use in either cross-sectional or paired-sample studies. For initial cross-sectional studies, total RNA containing miRNA was prepared from a minimum of 50 × 106 CD71+ cells freshly isolated from healthy controls, SCA patients before beginning hydroxyurea, and SCA patients at MTD. Total RNA was isolated using the miRNeasy kit according to protocol (QIAGEN) and used for subsequent microarray and real time analyses. For subsequent larger paired-sample analysis, a miRNA enriched fraction of CD71+ erythroid cells was prepared from frozen samples isolated from patients (n = 41) at baseline before beginning hydroxyurea therapy and again after reaching MTD. The miRNA enriched fraction was isolated using a modified protocol of the RNeasy kit, miRNeasy kit, and Min-elute kit (QIAGEN). In brief, mRNA was isolated from CD71+ cells using RNeasy kit and the RLT/RW1 flow-through containing small-RNAs and protein was frozen at −80°C. miRNAs were isolated from the flow-through by adding 3.5 volumes of ethanol, collecting the miRNA on the miRNeasy spin column, and then concentrating miRNA using min-Elute kit. RNA samples were used for subsequent real-time PCR assays.
Microarray analysis
Microarray analysis was used as an initial screening tool to determine differential miRNA expression in CD71+ erythroid cells using cross-sectional analysis. Relative expression of miRNA isolated from CD71+ cells of non-SCA controls, SCA patients untreated with hydroxyurea, and SCA patients treated with hydroxyurea was measured using a human miRNA microarray (G4470B; Agilent Technologies), consisting of probes for 799 miRNAs (723 human and 76 human viral) from the Sanger Version 10.1 database. Array hybridization was performed according to the manufacturer's recommended protocols. In brief, total RNA was labeled using the Agilent miRNA Labeling kit. Hybridization was carried out in an Agilent oven at 55°C for 20 hours at 20 rpm, followed by standard wash procedures. The microarray was then scanned in an Agilent scanner at 5 μm resolution, and array data were extracted using the default miRNA settings of Agilent Feature Extraction Software (Version 10.5.1.1) with the miRNA_105_Jan09 protocol. Signal intensity was normalized, scaled and log transformed. The minimum detecting miRNA expression signal was set at a threshold with greater than the 99-percentile of expression signal of the negative control probes. Analysis was performed to compare miRNA expression in CD71+ cells of non-SCA controls, SCA patients without hydroxyurea treatment, and SCA patients at hydroxyurea MTD using the Z statistic. All microarray data are available on the Gene Expression Omnibus (GEO) under accession number GSE32035.
Q-PCR of miRNA expression
miRNA expression of CD71+ reticulocytes was measured by Q-PCR in cross-sectional analysis using total RNA and paired-sample analysis using enriched miRNA samples. miRNA expression in either total RNA or enriched miRNA was measured using Taqman miRNA expression probes (supplemental Table 2; part 4427975, Applied Biosystems). For each miRNA, expression was normalized to miR-451 that was constitutively expressed and remained essentially unchanged in treatment groups across all CD71+ samples, and reported as 2ΔCT (ΔCT = CT target-CTnormalization probe). Relative fold change in miRNA expression calculated by comparative CT mathematical model was used to calculate fold change between non-sickle and sickle reticulocytes (ΔCTtargetSCA− ΔCT targetnon-SCA) for cross-sectional samples, or between baseline and MTD samples (ΔCTtargetMTD− ΔCT targetbaseline) for paired samples. Student t test of ΔCT values was used to determine significant differences in expression of specific miRNAs between non-sickle and sickle reticulocytes, and the Wilcoxon signed rank test was used to compare ΔCT values at baseline to those of MTD samples.
Correlation statistics for associations with HbF
The Pearson correlation coefficient was used to determine significant associations between age and %HbF, and between methylation results and %HbF. Significant associations between miRNA ΔCT values, and HbF levels were determined by the Spearman correlation coefficient. A mixed model was used to identify significant associations between treatment, miRNA ΔCT values and HbF levels.
Results
HbF induction
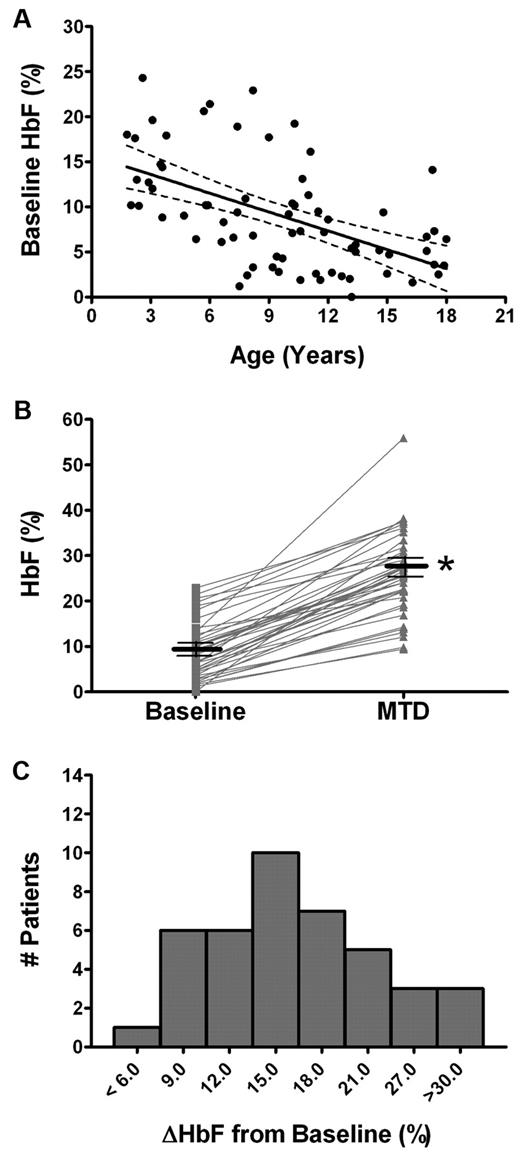
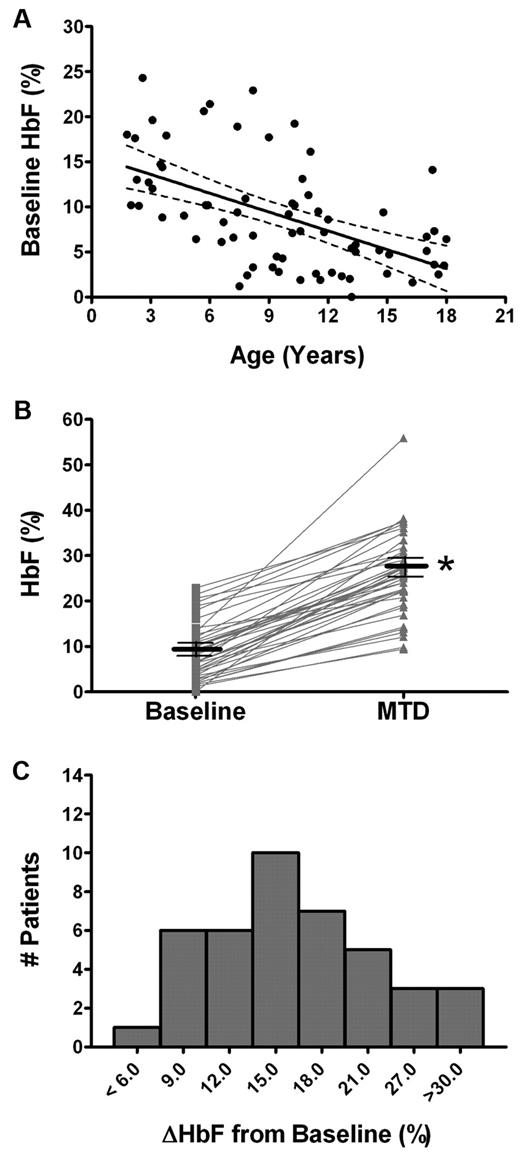
In this pediatric cohort of patients, HbF levels were analyzed from baseline before taking hydroxyurea, and after the patient reached MTD, allowing cross-sectional analysis (73 patients at baseline and 65 patients at MTD) as well as paired-sample analysis (41 patients both at baseline and MTD). Baseline HbF levels were inversely and significantly correlated to the age of the patient (r = −0.54; P < .001, Figure 1A). HbF levels significantly increased with hydroxyurea treatment in 41 paired samples. The mean HbF increased an average of 17.0% from 8.9 ± 6.1% at baseline to a mean of 25.9 ± 9.0% at MTD (Figure 1B). Although all patients displayed an increase in HbF, inter-patient variability was evident by absolute HbF changes ranging from < 6% in one child to > 30% in 3 others (Figure 1C).
Induction of HbF levels by hydroxyurea. (A) Linear regression analysis indicates an inverse correlation between age and baseline HbF levels with 95% confidence interval (dotted lines). (B) Paired-sample analysis from individual subjects at baseline (□) and at MTD (Δ) illustrates the increase in HbF with hydroxyurea treatment per patient (gray lines) and mean ± SE (black lines) for each group of samples. (C) A histogram showing the variable HbF-induction in patient samples, with the absolute change in HbF (ΔHbF) between baseline and MTD (*P < .001 baseline HbF compared with MTD by the paired t test).
Induction of HbF levels by hydroxyurea. (A) Linear regression analysis indicates an inverse correlation between age and baseline HbF levels with 95% confidence interval (dotted lines). (B) Paired-sample analysis from individual subjects at baseline (□) and at MTD (Δ) illustrates the increase in HbF with hydroxyurea treatment per patient (gray lines) and mean ± SE (black lines) for each group of samples. (C) A histogram showing the variable HbF-induction in patient samples, with the absolute change in HbF (ΔHbF) between baseline and MTD (*P < .001 baseline HbF compared with MTD by the paired t test).
Methylation changes in association with HbF induction
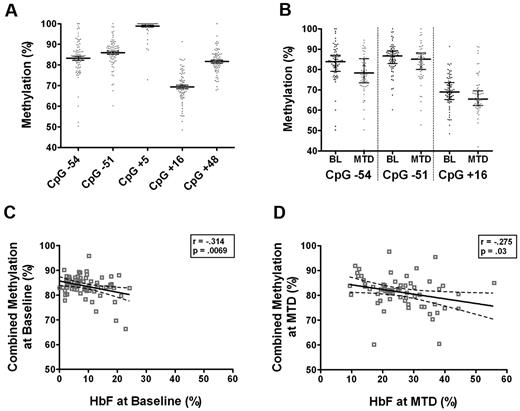
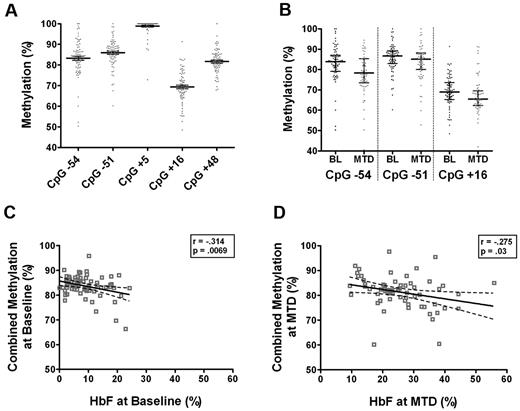
To test the hypothesis that methylation patterns in sickle reticulocytes are altered after hydroxyurea treatment, the methylation status of CpG islands within the Gγ-globin promoter region was measured before and after MTD at sites −54, −51, +5, +16, and +48. This region in sickle reticulocytes was heavily methylated at each site before hydroxyurea treatment with a median of 84% (range 69%-99%; Figure 2A). However, these CD71+ erythroid cells had a lower degree of methylation at the Gγ-globin promoter compared with peripheral blood mononuclear cells, which had a median methylation of 92.7% (range 76.2%-98%) in this region (data not shown). With hydroxyurea treatment the median methylation was only slightly though significantly decreased specifically at 3 CpG sites (−54, −51, and +16) by 4.3%, 2.5%, and 1.8% respectively (P < .035) compared with baseline values (Figure 2B). Analysis of 41 paired samples revealed a small decrease in methylation at these same sites after hydroxyurea treatment, but the changes did not reach statistical significance.
Methylation of Gγ-globin promoter in CD71+ NRBCs. (A) CpG sites −54, −51, +5, +16, and +48 of the Gγ-promoter are methylated in CD71+ NRBCs as quantitated by pyrosequencing of bisulfite treated DNA. (B) Methylation at sites −54, −51, and +16 is significantly decreased at maximum tolerated dose (MTD) of hydroxyurea compared with the baseline (BL) methylation before treatment (P < .05). (C) Data are presented as median with interquartile range. Significant inverse correlations between combined methylation of all 5 sites and HbF were detected at baseline and at MTD (D) by the Pearson correlation coefficient.
Methylation of Gγ-globin promoter in CD71+ NRBCs. (A) CpG sites −54, −51, +5, +16, and +48 of the Gγ-promoter are methylated in CD71+ NRBCs as quantitated by pyrosequencing of bisulfite treated DNA. (B) Methylation at sites −54, −51, and +16 is significantly decreased at maximum tolerated dose (MTD) of hydroxyurea compared with the baseline (BL) methylation before treatment (P < .05). (C) Data are presented as median with interquartile range. Significant inverse correlations between combined methylation of all 5 sites and HbF were detected at baseline and at MTD (D) by the Pearson correlation coefficient.
A combined methylation percentage was calculated across all 5 sites in the promoter region by averaging the measured values for each subject. Baseline methylation was significantly associated with baseline HbF levels with an inverse correlation (r = −0.314, P = .0069; Figure 2C). At MTD, a similar significant correlation was detected (r = −0.275, P = .03; Figure 2D).
miRNA expression in sickle and normal reticulocytes
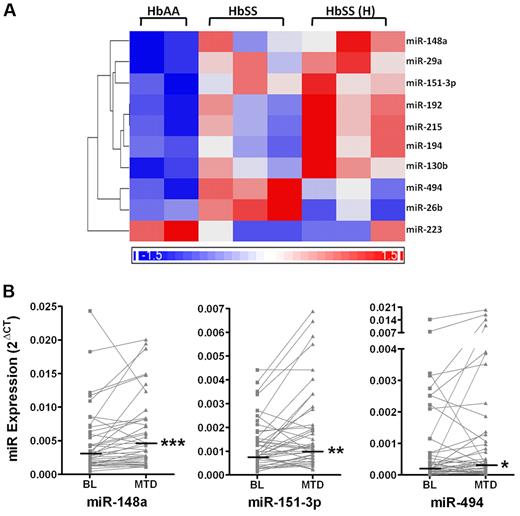
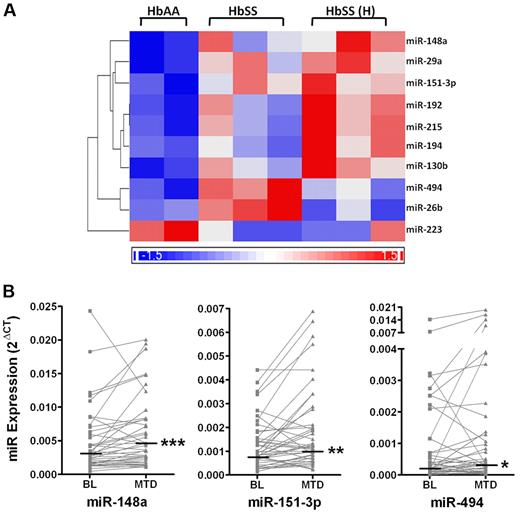
Because recent studies have identified miRNAs in mature erythrocytes that may reflect miRNA regulated processes during early erythropoiesis, we hypothesized that miRNA regulation may be involved in hydroxyurea-mediated HbF induction. First, miRNA expression in early erythroid cells was determined by microarray in a cross-sectional analysis of a small cohort of patients from the HUSTLE study. A total of 95 human miRNAs of 723 on the microarray were present at a detectable level in CD71+ reticulocytes. Unsupervised cluster analysis of the 95 expressed miRNAs showed that the normal healthy control samples clustered distinctly from sickle reticulocytes (data not shown). Analysis using the Z-statistic found that 10 miRNAs were differentially expressed among CD71+ cells from the 3 patient groups (P < .05); miR-26b, miR-29a, miR-130b, miR-148a, miR-151-3p, miR-192, miR-194, miR-215, and miR-494 were all significantly up-regulated, whereas miR-223 was down-regulated (Figure 3a). Q-PCR of larger samples sizes confirmed the trends seen by microarray, showing an increase in 9 of 10 miRNAs and a decrease of miR-223 in sickle reticulocytes compared with non-sickle reticulocytes (supplemental Table 3). Although, the miRNA profiles of non-sickle and sickle reticulocytes were very similar, specific miRNAs were identified that were distinct between the 2 populations.
Differential miRNA expression in CD71+ erythroid cells by microarray and Q-PCR analysis. (A) Heatmap illustrates relative changes in miRNA expression of 10 miRNAs that were significantly different between CD71+ cells from 2 individuals without SCA (HbAA), 3 SCA patients before hydroxyurea (HbSS), and 3 SCA patients at hydroxyurea MTD (HbSS(H)). Significant changes were identified in this cross-sectional study by Z-score statistic where P < .05. (B) Graphs illustrate the change in expression of miRNAs 148a, 151-3p, and 494 from baseline to MTD for each patient (gray symbols and lines). Median miRNA expression was significantly higher at MTD compared with baseline for each miRNA as determined by the Wilcoxon signed rank test (*P = .03; **P = .0036; ***P = .0002).
Differential miRNA expression in CD71+ erythroid cells by microarray and Q-PCR analysis. (A) Heatmap illustrates relative changes in miRNA expression of 10 miRNAs that were significantly different between CD71+ cells from 2 individuals without SCA (HbAA), 3 SCA patients before hydroxyurea (HbSS), and 3 SCA patients at hydroxyurea MTD (HbSS(H)). Significant changes were identified in this cross-sectional study by Z-score statistic where P < .05. (B) Graphs illustrate the change in expression of miRNAs 148a, 151-3p, and 494 from baseline to MTD for each patient (gray symbols and lines). Median miRNA expression was significantly higher at MTD compared with baseline for each miRNA as determined by the Wilcoxon signed rank test (*P = .03; **P = .0036; ***P = .0002).
miRNA expression is associated with hydroxyurea-mediated HbF induction
miRNA expression in erythroid cells from patients with SCA, collected both at baseline and MTD, was compared using Q-PCR in a paired-sample analysis. Although expression of 9 miRNAs increased at MTD compared with baseline, only miR-148a, miR-151-3p, and miR-494 were significantly up-regulated by hydroxyurea (P < .03; Figure 3b) with mean fold changes of 1.7, 1.6, and 2.2, respectively.
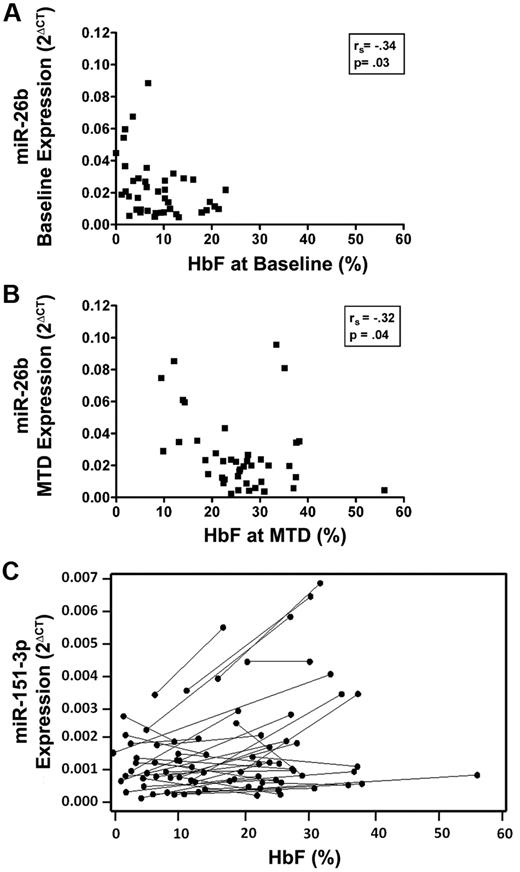
These paired samples were also used to identify significant associations between miRNA expression and HbF levels. Of the 9 miRNAs tested, only miR-26b expression was significantly associated with HbF levels. Expression of miR-26b was inversely associated with baseline HbF levels (rs = −0.34; P = .03), and a similar significant association was found between miR-26b expression and HbF at MTD (rs = −0.32; P = .040; Figure 4 A-B). Hydroxyurea treatment did not affect these associations. In the mixed model analysis aimed to identify significant associations between change in miRNA expression and change in HbF levels from baseline to MTD, miR-151-3p expression was significantly associated with HbF in response to hydroxyurea treatment (P = .047; Figure 4C). Thus, hydroxyurea treatment was associated with significant increases in miR-148a, miR-151-3p, and miR-494 expression, whereas HbF levels were significantly associated with miR-26b expression; however, only the increase in miR-151-3p expression was associated with the hydroxyurea-mediated induction of HbF.
MiRNAs 26b and 151-3p are associated with HbF. (A) Expression of miRNA 26b at baseline and at MTD was inversely associated with HbF levels at baseline and MTD. (B) respectively as identified by the Spearman correlation coefficient. (C) HbF levels and miRNA 151-3p expression in paired samples from baseline to MTD for each patient (represented by individual lines), and analysis of this data with a mixed model indicated a significant association between change in HbF and a change in miRNA 151-3p in response to hydroxyurea treatment (P < .05).
MiRNAs 26b and 151-3p are associated with HbF. (A) Expression of miRNA 26b at baseline and at MTD was inversely associated with HbF levels at baseline and MTD. (B) respectively as identified by the Spearman correlation coefficient. (C) HbF levels and miRNA 151-3p expression in paired samples from baseline to MTD for each patient (represented by individual lines), and analysis of this data with a mixed model indicated a significant association between change in HbF and a change in miRNA 151-3p in response to hydroxyurea treatment (P < .05).
Discussion
Although hydroxyurea has proven to be clinically beneficial because of its ability to induce fetal hemoglobin (HbF), the mechanisms by which HbF levels increase remain unclear. The HUSTLE clinical trial provides a unique and robust sample set to examine epigenetic and molecular changes associated with hydroxyurea treatment. Figure 1 illustrates that the HbF induction in this patient cohort compares favorably to previously published experiences treating children with hydroxyurea at MTD.4,6,14 We specifically investigated methylation and miRNA changes that occur with hydroxyurea treatment in early erythroid cells, because these 2 potential mechanisms of hydroxyurea-mediated HbF induction have not yet been carefully investigated in SCA.
Baseline methylation patterns in children with SCA (Figure 2A) revealed hypermethylation averaging 80% in the Gγ-globin promoter, similar to adult erythroid cells.28,38 Methylation in CD71+ NRBCs was inversely correlated to HbF levels at baseline and MTD (Figure 2C-D), consistent with the hypothesis that hypomethylation of the γ-globin promoter results in HbF induction. Although we used late-stage erythroblasts for analysis instead of earlier marrow precursors, the methylation levels and HbF correlations were consistent with previous findings using bone marrow. In a single adult patient, Platt et al reported 25% reduced DNA methylation within bone marrow cells at the Aγ-globin and Gγ-globin genes one week after treatment with hydroxyurea.3 In the current study, we identified significant changes in methylation, but these changes were very small with an average of < 5% decreases in methylation at key promoter sites within the Gγ-globin promoter (Figure 2B). The differences in our findings may relate to differences in the cell populations analyzed, but also differences in the hydroxyurea dosing and the time of analysis (ie, single large dose versus steady-state MTD). However, compared with known hypomethylating agents such as decitabine and 5-azacitidine that decrease methylation by 40% or more,39 and given recent data that hypomethylation alone by these agents may not be sufficient for the pharmacologic induction of HbF,40,41 it is unlikely that the observed small changes in methylation by hydroxyurea is sufficient for HbF induction. Thus methylation patterns of the β-globin locus may be a critical regulatory mechanism during normal erythroid cell development, but hypomethylation of the γ-globin promoter region is probably not an important mechanism of action for hydroxyurea-mediated induction of HbF.
In contrast to the findings with methylation status, novel and significant associations of miRNA expression with SCA, hydroxyurea, and HbF levels were identified that could indicate specific roles for miRNA in hydroxyurea-mediated HbF induction. Differential expression of 10 miRNAs between SCA and normal CD71+ reticulocytes that we identified by microarray profiling (Figure 3A) was similar to an earlier report of distinct miRNA expression patterns in erythrocytes between individuals with and without SCA.34 However, only miR-29a was common to both erythrocytes and reticulocytes in these 2 studies. Differential miRNA expression between control and SCA CD71+ erythroid cells may reflect characteristics of SCA including accelerated erythropoiesis, abnormal protein expression, and HbF production. In particular, miR-223 is of interest because down-regulation of this miRNA is required for erythroid differentiation and may inhibit important transcription factors like LMO2.42 Although targeting software predicts hundreds of potential gene targets for these miRNA species, elucidating specific targets of the differentially expressed miRNAs could provide insight into regulation of erythropoiesis and other processes that are altered in patients with SCA.
Using paired sample analysis, we found a significant up-regulation (1.6 fold or greater) for miR-148a, miR-151-3p, and miR-494 in association with hydroxyurea treatment (Figure 3B). Because a 1.5-fold induction of miR-15a and miR-16 was recently implicated in HbF expression in patients with trisomy 13,36 depending on the target transcripts of these miRNAs, a 1.6-fold modulation may be sufficient to impact hydroxyurea efficacy of HbF induction. For example, miR-148a, which has been shown to target proteins important for regulation of cell cycle and epigenetics,43-45 could, in turn, affect HbF regulation by hydroxyurea.
Two miRNAs, miR-26b and miR-151-3p, were directly associated with HbF levels (Figure 4). Expression of miR-26b was associated with HbF levels at baseline and at MTD, but the association with MTD HbF probably reflects the baseline association. In contrast, expression of miR-151-3p was not associated with HbF at baseline but was significantly up-regulated by hydroxyurea, and significantly associated with hydroxyurea-mediated HbF induction. These findings suggest that miR-26b may be associated with regulatory mechanisms of HbF, whereas miR-151-3p may be associated with pharmacologic induction of HbF by hydroxyurea. Neither miR-26b nor miR-151-3p has been described in the context of HbF expression, and their potential roles in HbF regulation or induction, and many of their target transcripts remain to be determined.
In conclusion, these studies suggest both methylation and miRNA expression in CD71+ erythroid cells are associated with HbF levels, but the mechanisms regulating endogenous HbF levels may be different from the mechanisms of hydroxyurea-mediated induction of HbF. Methylation and miR-26b expression associated with HbF levels at baseline seem to be important for endogenous regulation, but hydroxyurea treatment does not substantially modulate these processes. Altered miRNA expression associated with hydroxyurea is potentially important for the pharmacologic induction of HbF, but the possible mechanistic connections between these miRNAs and HbF need to be determined through the identification of specific transcript targets.
This prospective translational study using primary erythroid cells provides initial evidence for some epigenetic regulation of hydroxyurea-mediated HbF induction, but these findings must be evaluated with regard to the limitations of the study. By analyzing late-stage erythroblasts instead of erythroid progenitors from the bone marrow, some epigenetic or molecular changes resulting from hydroxyurea treatment may have been missed. The observed associations also do not provide direct evidence for any cause-effect linkages between methylation or miRNA expression with HbF, but instead provide initial associations that can be used as hypothesis-generating findings for future experiments. Although the exact mechanisms of HbF induction by hydroxyurea are not elucidated by our studies, the significant associations of methylation and miRNAs with HbF reinforces the need for future studies to investigate both epigenetic and alternative molecular processes as potential mechanisms for HbF expression and induction.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank St Jude Children's Hospital Clinical staff for providing patient care and hydroxyurea management, and all patients and families for study participation.
This work was supported by grant R01 HL090941 (REW) from the National Heart, Lung, and Blood Institute and by the American Lebanese Syrian Associated Charities. A.L.W. is the Lemuel Diggs Endowed Postdoctoral Fellow in Sickle Cell Disease.
National Institutes of Health
Authorship
Contribution: A.L.W. wrote the paper, designed and performed experiments, and analyzed and interpreted data; S.S. isolated RNA from samples and assisted in sample collection; T.A.H. assisted in sample collection and storage; N.M. recruited participants and served as HUSTLE study clinical coordinator; M.S. performed biostatistical analysis; Y.-D.W. performed microarray interpretation and analysis; and R.E.W. edited the paper, performed analysis, and interpreted data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Aisha L. Walker, Dept of Pharmaceutical Sciences, 262 Danny Thomas Pl, Mail Stop 313, Memphis, TN 38105; e-mail: aisha.walker@stjude.org.