Abstract
Abstract 2653
Non-Hodgkin's lymphoma (NHL) is a heterogeneous disease. The current International Prognostic Index (IPI) is useful, but not perfectly predictive and there is a need for a more precise prognostic index. The host immune response to cancer has been shown to predict outcome in multi-variate analyses in many types of cancer. We hypothesized that its genetics may contribute to prognostication. We focused on an evaluation of inherited functional polymorphisms in key immune genes that have previously been shown by others to influence outcome in various cancers. The objective was to determine whether an index of SNPs that alter function of critical immune system genes would enhance the predictive value of the IPI in patients with aggressive NHL.
PATIENTS AND METHODS: Our study cohort consisted of 192 patients with NHL enrolled at diagnosis between 1990 and 1996. Patients provided informed consent and demographic, clinical and outcome data as well as tumor and blood samples were collected and stored on all patients. Patients were treated as per the standard of care at the time. 124 were of the aggressive type while 68 were low grade. We focused on the aggressive cohort in this analysis. DNA was extracted from blood, bone marrow and biopsy tissue. We evaluated the following genes; IL1A rs1800587, CXCR2 rs1126580, IL4R rs2107356, TNF rs1800629, IL4R175V rs454078, IL8 rs4073, FCGR3A rs396991, P2RX7 rs3751143, IL12B rs3212227. Genotyping was performed using allele-specific PCR primers tagged with GC tails to enhance melting point discrimination. PCR reactions were set up with the aid of a robot and run on a real-time PCR instrument. Kaplan-Meyer curves were generated for the overall survival of each genotype. Univariate analysis was performed to compare genotypes with IPI score, IPI factors and disease bulk.
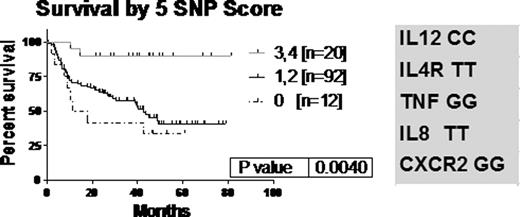
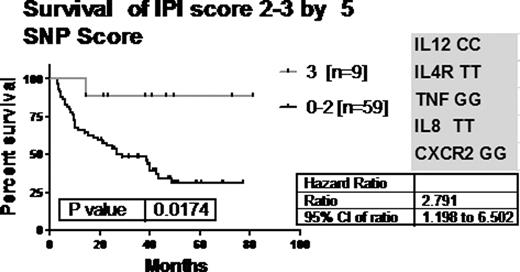
We evaluated relationships to existing standard prognostic factors. We found a significant relationship between IL12B CC carriers and IPI score 0–1 (p=0.0012). Patients with IL12B CC had less extranodal disease, lower stage and bulk. We modeled the data to identify combinations of SNPs with the most predictive power. The most predictive genotype combination included the genes for IL8, IL12B, IL4R, CXCR2 and TNF. A SNP score, determined by the number of favourable genotypes, showed significance in survival, p=0.004 [fig.1]. The group of 68 patients with an IPI score of 2–3 was analyzed to determine whether the SNP score could distinguish outcome within this IPI group [fig 2]. Patients with 3 of these favourable alleles had significantly improved survival [HR= 2.791]. Scoring with an index made up of three genes-IL8, CXCR2 and TNF alone -was also significant, p=0.02 [not shown].
We evaluated 10 SNPs with functional consequences on the immune response in a population of previously untreated aggressive lymphoma patients undergoing standard management. We found that a combination of SNPs statistically predicted outcome of these 124 patients with aggressive lymphoma. Although Habermann et al [Blood 2008 112(7); 2694–2702] had created a SNP score based on IL1A, IL8RB (CXCR2), IL4R and TNF, we have found that outcome is best predicted in our cohort with IL8, IL12, CXCR2, IL4R and TNF. A useful model could also be obtained using 3 SNPS (IL8, CXCR2 and TNF). Our results are important in that we have shown that this combination of SNPs can further enhance prognostification beyond IPI. Specifically statistically significant overall survival differences in IPI 2,3 patients was found based upon the SNP score using IL8, IL12, CXCR2, IL4R and TNF and the three-SNP combination. Incorporating SNP scores into IPI may be a useful strategy to further identify patients with increased likelihood of improved or adverse outcomes.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.