Abstract
Abstract 4883
Ras gene mutations especially N-Ras gene are found in up to 30% of AML cases. The mutation of Ras gene that most commonly occurs by one base substitution in codon 12, 13 or 61 and leads to a constant activity of the Ras protein which can induce uncontrolled cell proliferation and escape from apoptosis. While some groups insist Ras mutations are related with a better outcome, others suppose mutated Ras genes are associated with short survival. The purpose of this study was to detect the frequency and distinct features of N-ras mutation in adult Korean patients with de novo AML and to compare the performance of a pyrosequencing analysis to a direct sequencing method for detecting N-ras mutations.
This study selected newly diagnosed 70 patients with AML that were treated at the Gachon University Gil Hospital from May 2004 to February 2011. The sample collection from each patient occurred different times, and follow-up collections were conducted among 6 patients who had mutation. We analyzed 78 bone marrow samples of 70 de novo AML patients for detecting N-ras codon 12, 13, and 61 mutations using pyrosequencing method, and all data was confirmed by direct sequencing.
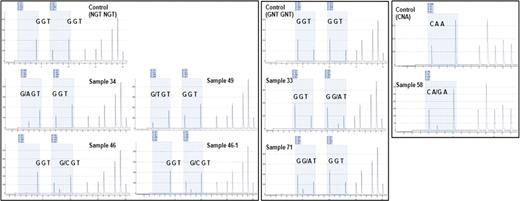
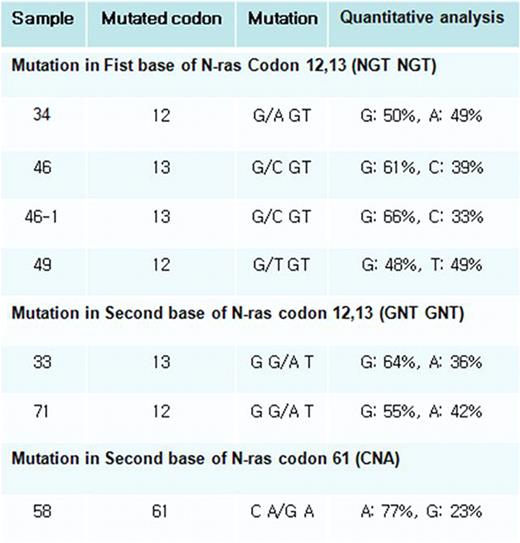
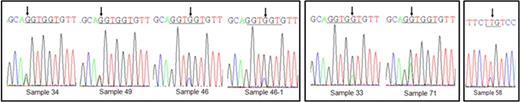
N-ras mutations were detected in 7 samples of 6 of the 70 patients examined (Figure 1 and Table 1). Mutations at codon 12, 13 and 61 were found in 3 patients, 2 patients and 1 patient, respectively. Mutations in this study were point mutation and they induced amino acid substitution. All base substitutions in codon 12 subsequently led to the change of wild type glycine (G12S, G12C, G12D). Mutations at codon 13 also induced amino acid change of wild type glycine (G13D, G13R). Mutation in codon 61 caused substitution of glutamine (Q61R). There is no difference in methods of mutation detection and all mutations disappeared after chemotherapy. And 4 of 6 patients were expired, 1 of 6 was associated with treatment failure, and only one patient was diagnosed to complete remission (Table 1). We can not establish statistically significant differences between the N-ras positive and negative groups in age, sex, WHO classification, cytogenetic abnormality or complete blood count.
Incidence of Ras mutations and patient characteristics.
| Sample . | Age . | Sex . | Mutation . | WHO classification . | Karyotype . | Mutation in follow up sample . | FLT mutation . | Clinical course . | Survival period after diagnosis . |
|---|---|---|---|---|---|---|---|---|---|
| N-ras codon12 (incidence: 4.3%) | |||||||||
| 34 | 59 | M | G12S | AML with MRC | 46,XY[20] | Not detected | Not detected | F/U loss | |
| 49 | 52 | M | G12C | AML, NOS (Acute myelomonocytic leukemia) | 46,XY[20] | Not done | Not detected | Expire | 3 days |
| 71 | 81 | F | G12D | AML, NOS (Acute monocytic leukemia) | 46,XX[20] | Not done | D835Y | Expire | 1 month |
| N-ras codon13 (incidence: 2.9%) | |||||||||
| 33 | 54 | F | G13D | AML, NOS (Acute myeloblastic leukemia, without maturation) | 46,XX,der(16) t(1;16)(q21; q12.1)[14]/46,XX[6] | Not detected | Not detected | Expire | 3 months |
| 46 | 67 | M | G13R | AML, NOS (Acute myeloid leukemia with maturation and basophilia) | 46,XY,del(9) (q21q22) [25]/46, XY[5] | G13R | Not detected | Alive, complete remission | 23 months |
| N-ras codon61 (incidence: 1.4%) | |||||||||
| 58 | 47 | F | Q61R | AML, NOS (Acute myeloid leukemia with maturation) | 46,XX[20] | Not detected | D835Y | Alive, induction failure, allo- BMT | 14 months |
| Sample . | Age . | Sex . | Mutation . | WHO classification . | Karyotype . | Mutation in follow up sample . | FLT mutation . | Clinical course . | Survival period after diagnosis . |
|---|---|---|---|---|---|---|---|---|---|
| N-ras codon12 (incidence: 4.3%) | |||||||||
| 34 | 59 | M | G12S | AML with MRC | 46,XY[20] | Not detected | Not detected | F/U loss | |
| 49 | 52 | M | G12C | AML, NOS (Acute myelomonocytic leukemia) | 46,XY[20] | Not done | Not detected | Expire | 3 days |
| 71 | 81 | F | G12D | AML, NOS (Acute monocytic leukemia) | 46,XX[20] | Not done | D835Y | Expire | 1 month |
| N-ras codon13 (incidence: 2.9%) | |||||||||
| 33 | 54 | F | G13D | AML, NOS (Acute myeloblastic leukemia, without maturation) | 46,XX,der(16) t(1;16)(q21; q12.1)[14]/46,XX[6] | Not detected | Not detected | Expire | 3 months |
| 46 | 67 | M | G13R | AML, NOS (Acute myeloid leukemia with maturation and basophilia) | 46,XY,del(9) (q21q22) [25]/46, XY[5] | G13R | Not detected | Alive, complete remission | 23 months |
| N-ras codon61 (incidence: 1.4%) | |||||||||
| 58 | 47 | F | Q61R | AML, NOS (Acute myeloid leukemia with maturation) | 46,XX[20] | Not detected | D835Y | Alive, induction failure, allo- BMT | 14 months |
Mutation of Ras gene occurred low incidence (8.6%) in the Korean AML patients compared with Western population (Europe: 11–44%, America: 12–25%, Australia: 12–21%) and other Asian population (Japan: 14%, China: 18%, Thailand: 13%). There was no preference for specific mutation in Korean AML patients. Four of six patients with N-ras mutation were expired, these finding showed a trend to worse survival although we did not observe statistical significance. Comparison of both methods revealed identical results. Pyrosequencing is more rapid and simple technique than direct sequencing and uniquely provide quantitative information of detected mutations (Figure 1.B).
A.
B.
C.
The detection of N-ras mutation by pyrosequencing and direct sequencing methods.
The detection of N-ras mutation by pyrosequencing and direct sequencing methods.
A. Pyrogram from pyrosequencing method. Control means wild type sample and others are mutation detected samples. First box means mutation in first base of N-ras codon 12, 13 and second box presents mutation in second base of N-ras codon 12, 13. Third reveals mutated sample in N-ras codon 61. B. Quantitative date of mutation is gained from pyrosequencing method. C. The result of pyrosequencing is confirmed by directed sequencing method.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.