Abstract
Induction of antibody-mediated immunity against hematologic malignancies requires CD4+ T-cell help, but weak tumor antigens generally fail to induce adequate T-cell responses, or to overcome tolerance. Conjugate vaccines can harness alternative help to activate responses, but memory B cells may then be exposed to leaking tumor-derived antigen without CD4+ T-cell support. We showed previously using lymphoma-derived idiotypic antigen that exposure to “helpless” antigen silences the majority of memory IgG+ B cells. Transfer experiments now indicate that silencing is permanent. In marked contrast to IgG, most coexisting IgM+ memory B cells exposed to “helpless” antigen survive. Confirmation in a hapten (NP) model allowed measurement of affinity, revealing this, rather than isotype, as the determinant of survival. IgM+ B cells had Ig variable region gene usage similar to IgG but with fewer somatic mutations. Survival of memory B cells appears variably controlled by affinity for antigen, allowing a minority of low affinity IgG+, but most IgM+, memory B cells to escape deletion in the absence of T-cell help. The latter remain, but the majority fail to undergo isotype switch. These findings could apply to other tumor antigens and are relevant for vaccination strategies aimed to induce long-term antibody.
Introduction
Tumor antigens are generally weakly immunogenic, and T-cell responses can be subject to tolerogenic pressure.1 To avoid this, vaccines have been designed as conjugates of tumor antigens with immunogenic molecules, such as KLH.2 Conjugation captures the large undamaged CD4+ T-cell repertoire, providing T-cell help for the antitumor immune responses.2,3 Idiotypic (Id) Ig-KLH vaccines aimed to suppress B-cell tumors via an anti-Id response are one example of the promise of this approach.4-6 We have used a similar strategy to deliver Id fusion proteins via DNA vaccines.7,8 The Id is composed of tumor-derived single chain Fv and is linked to the fragment C (FrC) of tetanus toxin. DNA vaccines encoding Id-FrC fusion proteins induce high levels of anti-Id antibody and protective immunity against B-cell lymphoma.9
Continuous suppression of lymphoma cell growth probably requires maintenance of antibody levels. Although long-lived plasma cells can secrete antibody for extended periods,10 memory B cells are also required. Survival factors for memory B cells are unclear, but antigen appears unnecessary.11,12 For antibody responses induced by conjugate vaccines, the question of the effect of confronting memory B cells with free unconjugated “helpless” antigen, potentially derived from residual or emergent tumor, arises. In the case of Id antigen, or for carbohydrate antigens, this led to dramatic loss of memory B cells.13 We found previously that the major fraction of the IgG component of the memory B-cell response induced by a DNA Id-FrC fusion vaccine was completely and specifically silenced by Id protein alone.14 In contrast, Id protein coupled to FrC protein rescued and boosted the anti-Id IgG response, indicating the critical importance of CD4+ T-cell help for maintenance of the memory B-cell response.
We have now shown that silencing of the anti-Id IgG response by unconjugated antigen is permanent, but the surprising observation was that the IgM response appeared relatively unscathed. To assess the wider applicability of these findings and to investigate the effects of antigen on memory B cells at different maturational stages, we have used a model antigen, the well-defined hapten NP (4-hydroxy-3-nitrophenyl) acetyl-carrier model. Clearly, NP itself is unlikely to induce CD4+ T-cell responses whereas the carrier protein will, mimicking the situation with Id antigen. In addition, there is a relatively high frequency of NP-specific B cells within the natural B-cell repertoire in wild-type C57/BL6 mice, and these can be tracked using antigen.15,16 We show that, in the absence of T-cell help, silencing of a proportion of IgG+ memory B cells occurs in this model and that it is permanent. In contrast, coexisting IgM+ memory B cells show only transient silencing but appear unable to regenerate a high-affinity IgG response. The NP model allowed the further conclusion that the key distinguishing feature appears to be affinity for antigen, with rheostatic control of the fate of memory B cells which, in the absence of T-cell help, allows only low-affinity, mainly IgM+, B cells to survive. It also confirms the generality of the findings, which probably apply to all weak immunogens, especially tumor antigens.
Methods
Proteins and immunization
In the Id model, C57BL/KaLwRij mice (8-12 weeks old) were given priming injections of 25 μg of p.Id-FrC DNA vaccine into each rear anterior tibialis muscle where indicated injections of 50 μg Id Ig were administered subcutaneously 6 weeks later. Id Ig was purified from sera of 5T33 tumor-bearing mice using a G-protein column. Where indicated, a booster injection was given subcutaneously with 100 μg Id-FrC fusion protein. The Id-FrC fusion protein was generated in 293f cells (Invitrogen) and purified using an anti-FrC antibody affinity column.
In the NP model, C57BL/6 mice (8-12 weeks old) were primed by intraperitoneal injection with 100 μg of NP16-KLH (Biosearch Technologies) in alum; where indicated, mice were also boosted intravenously with 20 μg of NP16-KLH 6 weeks later or at times indicated. In suppression experiments after immunization with NP16-KLH, mice received 200 μg of “helpless” antigen NP conjugated to mouse serum albumin (NP-MSA) or MSA (Invitrogen) as a control subcutaneously. NP-MSA was prepared as described.17
All animal experimentation was conducted within local Ethical Committee and United Kingdom Coordinating Committee for Cancer Research (London, United Kingdom) guidelines under Home Office License.
Adoptive transfer
In the Id model, donor mice were immunized with DNA vaccine and then injected with 50 μg Id Ig 6 to 7 weeks later. After a further 3 weeks, B cells were purified using a B-cell separation kit (Miltenyi Biotec) following the manufacturer's instruction. T cells were purified from FrC-primed mice using a T-cell separation kit (Miltenyi Biotec). B and T cells were injected itravenously into RAG2KO mice. Individual mice received a total of 1 × 107 B cells and 4 × 106 T cells. The recipients were boosted with 100 μg of Id-FrC protein intravenously one week later and bled after a further one week. Anti-Id and FrC antibodies were measured by ELISA.
In the NP model, for establishing B-cell memory, donor mice were primed and boosted with NP16-KLH as described.18 Three weeks after boost, they received either 200 μg NP-MSA or the same amount of MSA alone. After a further 2 weeks, 5 × 106 B cells purified as for the Id model were transferred into C57Bl/6 recipient mice primed with 100 μg KLH in alum. For NP, it was not necessary to use RAGKO recipients because, in contrast to Id, the higher frequency of NP-specific memory B cells did not require the homeostatic expansion of injected cells for detection. One week later, the recipients were boosted with 20 μg NP16-KLH intravenously and bled after a further one week. Antibody responses were measured by ELISA.
Detection of antibody and antigen-specific B cells
Anti-Id antibody levels were measured by ELISA as described.14 For NP, ELISA plates were coated with 5 μg/mL of NP16-bovine serum albumin (BSA, Biosearch Technologies), and then the same protocol used for Id antigen was followed. For IgM, antimouse IgM antibody (Sigma-Aldrich) was used at 1/1000 dilution. Antigen-specific B cells secreting IgM and IgG were measured by ELISPOT. Briefly, 96-well plates (Millipore, MAIPS4510) were coated with either the Fab fragment of 5T33Id IgG (1 μg/mL) or FrC (3 μg/mL) or NP16-BSA (5 μg/m) overnight in sterile phosphate-buffered saline. To validate the specificity of NP responses, control plates were coated with 5 μg/mL BSA with subsequent sample incubation. Plates were blocked using RPMI 1640 supplemented with 10% fetal calf serum. Splenocytes were plated at varying densities and incubated overnight in RPMI 1640 supplemented with 10% fetal calf serum, 2-mercaptoethanol, penicillin, and streptomycin. Horseradish peroxidase-labeled antimouse IgG (The Binding Site) or anti-IgM antibody (Sigma-Aldrich) was used for detection with a subsequent incubation with 3-amino-9-ethylcarbazole (Sigma-Aldrich) for 10 minutes. Spots were enumerated using an ELISPOT reader (AID). The values were plotted as the number of antibody-secreting cells (ASCs) per million lymphocytes after subtraction of nonspecific background values.
Detection of NP-specific memory B cells
Mice were primed with NP16-KLH in alum followed by a booster injection 7 weeks later. For detection of memory B cells, splenic lymphocytes were harvested 3 weeks after the last injection. The cells (5 × 106) were initially preincubated with CD16/32 blocking antibody (clone 93, eBioscience) at 1 μg per 106 cells. This was followed by staining with anti-IgD fluorescein isothiocyanate (clone 11–26c), anti-CD19 peridinin chlorophyll protein-Cy5.5 (clone eBio1D3) (both eBiosciences), anti-IgG1 allophycocyanin (clone X56, BD Biosciences), biotinylated anti-IgM (clone R6–60.2, BD) antibodies, and incubation with 1–2 μg NP25-phycoerythrin (PE; Biosearch) for 40 minutes on ice. This was followed by incubation with streptavidin-PE-Cy7 (BD Biosciences). Fluorescence-activated cell sorter (FACS) analysis was performed using a FACSCanto with FACSDiva software (BD Biosciences).
Antibody affinity
The affinity of NP-specific IgG and IgM was expressed using a formula: affinity = U/mL to NP4-BSA/U/mL to NP16-BSA. Binding to each molecule (coated at 5 μg/mL to ELISA plates) was determined by ELISA and expressed in units relative to a known standard. For IgM affinity, serum samples were mildly treated with 2-mercaptoethanol (Sigma-Aldrich) followed by blocking of released SH groups with iodoacetamide (Sigma-Aldrich) as described.19,20 Sample dilutions were then incubated with plates for 2 hours at room temperature. This was followed by secondary antibodies as described for detection of antibody.
VH gene analysis
For VH-gene analysis, mice were primed and boosted with NP16-KLH as above and then given a second booster injection of NP16-KLH 3 weeks later. Spleens were harvested 3 days later and stained with NP25-PE and anti-CD138 allophycocyanin-labeled antibody (clone 281–2, BD Biosciences). NP-specific plasmablasts were sorted as a NP surface positive, CD138+ population from 3 mouse spleens. Bulk sorting was performed using a FACSAria, and cells were used for RNA purification. RNA was purified from a total of 105 cells using the Rneasy kit (QIAGEN). cDNA was produced using the Superscript II first-strand synthesis kit (Invitrogen) with random hexamers. The cDNA was used as template for V-gene amplification by Hot star Taq polymerase (Promega) using a framework one forward J588a primer or primer mix A, B, and C (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) and a μ- or γ-constant region reverse primer. Polymerase chain reaction used the following conditions: 94°C for 10 minutes, then 30 cycles of 94°C for 30 seconds, 55°C for 30 seconds, 72°C for 1 minute, followed by 7 minutes at 72°C. Purified polymerase chain reaction products were ligated into pGEM-T vector (Promega) followed by transformation into competent Escherichia coli (XL-1 blue strain). DNA sequencing was carried out on 48 IgG and 36 IgM colonies. Sequencing reactions were run on an ABI3130xl genetic analyzer, and the data were analyzed using MacVector software (Acceltys Version 7.2.3).
Statistical analysis
Statistical analysis was performed using the Mann-Whitney test.
Results
Contrasting effects of “helpless” antigen on the IgM and IgG responses against Id Ig
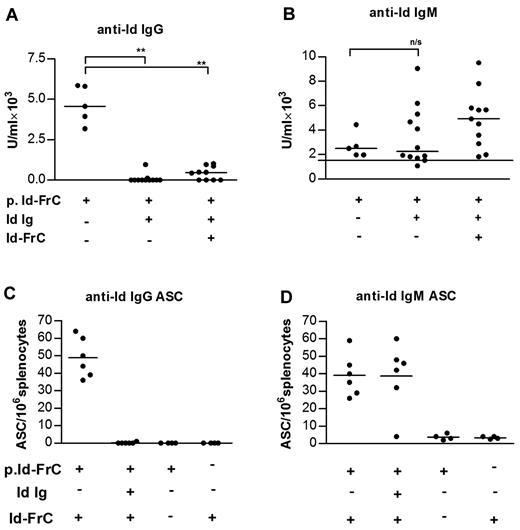
A DNA vaccine encoding Id single-chain Fv fused to FrC (p.Id-FrC) was used to establish a conventional IgG anti-Id antibody response, detectable at 7 weeks after priming as serum antibody (Figure 1A). At this stage, the majority of germinal centers (GCs) have contracted and B-cell memory is established.15 ASCs were detectable in the spleens after challenge with Id-FrC protein (Figure 1C). After an injection of unconjugated Id antigen 6 weeks after priming, both antibody (Figure 1A) and ASCs (Figure 1C) were reduced to undetectable levels. When suppressed mice were challenged with conjugated Id (Id-FrC) 3 days after giving Id antigen, they also failed to produce significant IgG antibody (Figure 1A).
Unconjugated idiotypic Ig induces suppression of anti-Id IgG but not IgM antibody and memory B-cell responses. (A-B) Mice were primed with p.Id-FrC fusion DNA vaccine; and 6 weeks later, they either were injected with Id Ig or received no injection. Three days later, the injected animals were divided into 2 groups, and one was boosted with Id-FrC fusion protein (n = 11) and the other group remained unboosted (n = 12). Anti-Id IgG and IgM antibodies were measured one week later by ELISA. (C-D) For ASC, mice were primed and then either received Id Ig (n = 6) or not (n = 6); and 3 days later, both groups were boosted with 100 μg of Id-FrC protein. ASCs were measured 48 hours after the booster injection by ELISPOT. The unboosted control (n = 4) and a separate group, which received a booster injection only, were also included (n = 4). These are combined data of 2 independent experiments. The horizontal bars represent median values. The horizontal line in panel B represents a background level in naive mice (average of 3). **Statistical significance (P < .01). n/s indicates not significant.
Unconjugated idiotypic Ig induces suppression of anti-Id IgG but not IgM antibody and memory B-cell responses. (A-B) Mice were primed with p.Id-FrC fusion DNA vaccine; and 6 weeks later, they either were injected with Id Ig or received no injection. Three days later, the injected animals were divided into 2 groups, and one was boosted with Id-FrC fusion protein (n = 11) and the other group remained unboosted (n = 12). Anti-Id IgG and IgM antibodies were measured one week later by ELISA. (C-D) For ASC, mice were primed and then either received Id Ig (n = 6) or not (n = 6); and 3 days later, both groups were boosted with 100 μg of Id-FrC protein. ASCs were measured 48 hours after the booster injection by ELISPOT. The unboosted control (n = 4) and a separate group, which received a booster injection only, were also included (n = 4). These are combined data of 2 independent experiments. The horizontal bars represent median values. The horizontal line in panel B represents a background level in naive mice (average of 3). **Statistical significance (P < .01). n/s indicates not significant.
Data on the IgG response confirm previous findings14 but can be contrasted with the lack of effect of the same suppressive procedure on the IgM response. Serum anti-Id IgM was clearly detectable 7 weeks after priming (Figure 1B). Splenic IgM+ ASCs (Figure 1D) were also detectable after challenge with Id-FrC protein (Figure 1D). However, injection of unconjugated Id failed to reduce either antibody or ASCs (Figure 1B,D). Indeed, surviving IgM+ B cells were able to boost serum IgM levels after an injection of conjugated Id antigen (Figure 1B), showing that functional memory cells remained. The fact that most of the IgG response was lost indicates that few of the IgM+ B cells were capable of switching, at least by 1 week after injection of conjugate.
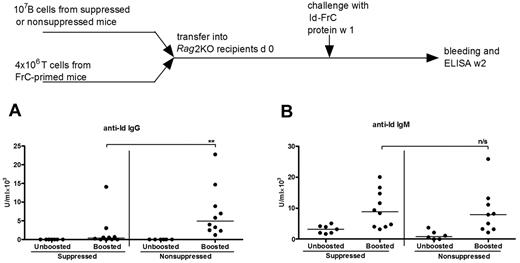
To assess the fate of B cells of each isotype after confrontation with “helpless” antigen, purified B cells from suppressed or control mice were transferred into RAG2KO recipients together with carrier-primed (FrC) CD4+ T cells. One week later, the ability of transferred B cells to respond to Id-FrC protein conjugate was measured and compared with an unboosted control. No Id-specific IgG was detected in unboosted controls in either group (Figure 2A). On booster injection, IgG anti-Id antibody was clearly produced by nonsuppressed B cells (Figure 2A), with 10 of 10 responders producing antibody with a median level of 4917 U/mL. However, suppressed B cells (Figure 2A) produced significantly less IgG anti-Id antibody, with 4 of 10 nonresponders, 4 low responders (< 700 U/mL) and 2 responders with levels comparable to the nonsuppressed group. The median level of antibody in the suppressed group was 292 U/mL, approximately 17-fold less than in the nonsuppressed group (P < .01).
Id-specific IgG+ B cells are lost irreversibly, but IgM+ B cells are unaffected by “helpless” Id antigen. Purified B cells (107) from either suppressed or nonsuppressed donor mice were mixed with 4 × 106 FrC-primed purified CD4+ T cells and injected into Rag2KO recipients. One week after transfer, mice in each recipient cohort were divided into 2 groups and one was challenged with 100 μg Id-FrC fusion protein (suppressed, n = 10; nonsuppressed, n = 9). The remaining groups were unchallenged controls. Mice were bled a week later, and sera were analyzed for anti-Id IgG (A) and IgM (B) by ELISA. The horizontal bars represent median values. These are combined data of 2 independent experiments. **Statistical significance (P < .01). n/s indicates not significant.
Id-specific IgG+ B cells are lost irreversibly, but IgM+ B cells are unaffected by “helpless” Id antigen. Purified B cells (107) from either suppressed or nonsuppressed donor mice were mixed with 4 × 106 FrC-primed purified CD4+ T cells and injected into Rag2KO recipients. One week after transfer, mice in each recipient cohort were divided into 2 groups and one was challenged with 100 μg Id-FrC fusion protein (suppressed, n = 10; nonsuppressed, n = 9). The remaining groups were unchallenged controls. Mice were bled a week later, and sera were analyzed for anti-Id IgG (A) and IgM (B) by ELISA. The horizontal bars represent median values. These are combined data of 2 independent experiments. **Statistical significance (P < .01). n/s indicates not significant.
The IgM anti-Id response detected in parallel was also low in unboosted mice in both groups (Figure 2B). After boosting with Id-FrC conjugate, levels in nonsuppressed mice were enhanced, with 9 of 9 responders and a median level of 8775 U/mL (Figure 2B). However, in contrast to the IgG response, suppressed B cells were also able to produce an anti-Id IgM after boosting, with 11 of 11 responders and a median level of 9825 U/mL (Figure 2B). For IgM, the levels in the nonsuppressed or suppressed groups were not significantly different (P > .05). These data show that the loss of the IgG+ B cells is irreversible but that the transferred IgM+ B cells had been largely unaffected by exposure to “helpless” Id antigen.
Induction of IgG and IgM memory antibody responses against NP
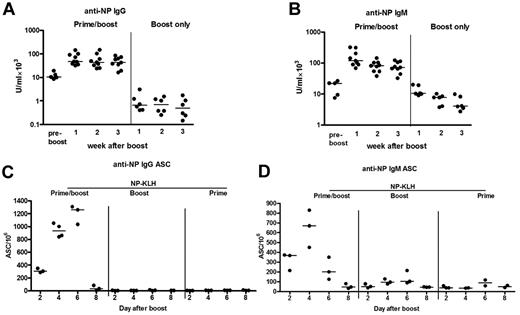
A different antigen (NP) was then investigated, both to assess the generality of the findings and to probe the nature of the antibody responses. The first question was whether NP conjugated to KLH could induce a detectable secondary IgM response, in parallel with an IgG response. Priming with NP-KLH, followed after 6 weeks by intravenous boosting with the same antigen, induced anti-NP serum antibody of both IgG (Figure 3A) and IgM (Figure 3B) isotypes. Both responses increased by week 1 (Figure 3A-B) and remained high for the 3-week period of analysis. A control injection of NP-KLH into unprimed mice produced a low level primary response of IgG and IgM antibodies over the same period (Figure 3A-B). The results were paralleled by splenic ASCs with the IgG response peaking at day 6, whereas the IgM response peaked at day 4 and fell rapidly by day 8 (Figure 3C-D), again with insignificant levels induced by a booster injection only. These findings indicate that IgM+ B cells, detectable in the spleen, form part of the memory response.
NP-specific memory antibody responses are of both IgG and IgM isotypes. Six weeks after priming, mice were boosted with NP-KLH and the levels of IgG (A) and IgM (B) antibody in sera (n = 9) were compared with unboosted controls (n = 5) and controls, which received a booster injection only (n = 6) as measured by ELISA at time points indicated. (C-D) IgG+ and IgM+ antibody-secreting cells in the spleens (n = 3) were measured by ELISPOT on the days indicated. Controls that received a booster injection only (n = 3) as well as an unboosted control group were included. Horizontal bars represent median values. These data are representative of 2 independent experiments.
NP-specific memory antibody responses are of both IgG and IgM isotypes. Six weeks after priming, mice were boosted with NP-KLH and the levels of IgG (A) and IgM (B) antibody in sera (n = 9) were compared with unboosted controls (n = 5) and controls, which received a booster injection only (n = 6) as measured by ELISA at time points indicated. (C-D) IgG+ and IgM+ antibody-secreting cells in the spleens (n = 3) were measured by ELISPOT on the days indicated. Controls that received a booster injection only (n = 3) as well as an unboosted control group were included. Horizontal bars represent median values. These data are representative of 2 independent experiments.
Effect of “helpless” antigen on NP-specific memory IgG and IgM B cells
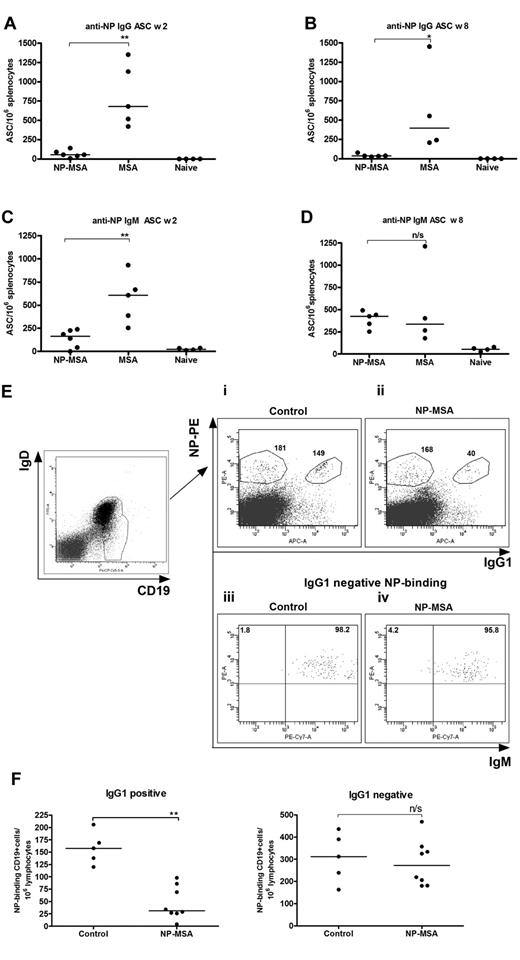
Splenic memory B cells are clearly able to respond to a booster injection of NP-KLH, and the next question was the effect on these B cells of removing the KLH component of the conjugate, thereby exposing them to “helpless” antigen. To address this, we injected NP coupled to the autologous protein MSA (NP-MSA) at week 6. This was followed by a conventional booster injection of NP-KLH 2 or 8 weeks later, with a subsequent measurement of ASC levels at day 4.
Direct effects in immune mice.
The effect of NP-MSA on the IgG response mirrored that found previously for Id14 (ie, almost complete loss of the ASCs). This had occurred by 2 weeks after injection (Figure 4A) and was still evident at 8 weeks (Figure 4B). Control MSA had no effect. In contrast, the IgM responses, although reduced at 2 weeks (Figure 4C), recovered by 8 weeks (Figure 4D) to levels similar to mice injected with MSA alone. It appeared therefore that, although IgG+ B cells were silenced by soluble “helpless” NP-MSA antigen, IgM+ B cells were only reduced transiently, with recovery to nonsuppressed levels. Because the injection of NP-KLH alone into naive mice induced no significant IgM responses at this time point (Figure 4C-D), recovery appears to be via reversal of the transient suppression of the IgM+ memory B cells. The failure to detect IgG+ B cells again suggests that the splenic IgM+ B cells do not undergo significant switch to IgG after the boosting injection with NP-KLH.
Permanent suppression of NP-specific IgG+ memory cells and transient suppression of IgM memory B cells caused by “helpless” antigen NP-MSA. Mice were primed with NP-KLH in alum; and 6 weeks later, mice were injected with NP-MSA (n = 6) or unhaptenated MSA (n = 5, week 2; n = 4, week 8). Two or 8 weeks later, mice were boosted with 20 μg of NP-KLH and ELISPOT was performed after 4 more days. Initially (week 2), suppression of both IgG+ (A) and IgM+ (C) B cells was observed. By week 8, whereas IgG+ memory cells remain suppressed (B), IgM memory recovered (D). Horizontal bars represent median values. **Statistical significance (P < .01). *Statistical significance (P < .03). n/s indicates not significant. These data are representative of 2 independent experiments. (E-F) Loss of NP-specific IgG+ memory cells after exposure to “helpless” antigen. Mice were primed and boosted with NP-KLH and then injected either with 200 μg of NP-MSA or 200 μg MSA (control) 3 weeks later. After a further 6 days, splenocytes were taken and analyzed by FACS. B cells were gated on CD19 and the IgD+ and IgD− populations and then analyzed further for antigen binding and for expression of IgG1. (E) Representative FACS profile showing loss of NP-specific IgG1+ but not IgG− B cells in mice injected with NP-MSA versus those injected with MSA (i-ii). The numbers of NP-specific B cells (gated) per 106 lymphocytes (i-ii) are shown. IgG− NP binding cells were positive for surface IgM in both controls and mice injected with NP-MSA (iii-iv). The quadrant statistics of IgG1− gates are shown (iii-iv). (F) Enumeration of NP-specific IgG+ and IgG− memory cells in the 2 groups. Horizontal bars represent median values. **Statistical significance (P < .01). These are combined data of 2 independent experiments.
Permanent suppression of NP-specific IgG+ memory cells and transient suppression of IgM memory B cells caused by “helpless” antigen NP-MSA. Mice were primed with NP-KLH in alum; and 6 weeks later, mice were injected with NP-MSA (n = 6) or unhaptenated MSA (n = 5, week 2; n = 4, week 8). Two or 8 weeks later, mice were boosted with 20 μg of NP-KLH and ELISPOT was performed after 4 more days. Initially (week 2), suppression of both IgG+ (A) and IgM+ (C) B cells was observed. By week 8, whereas IgG+ memory cells remain suppressed (B), IgM memory recovered (D). Horizontal bars represent median values. **Statistical significance (P < .01). *Statistical significance (P < .03). n/s indicates not significant. These data are representative of 2 independent experiments. (E-F) Loss of NP-specific IgG+ memory cells after exposure to “helpless” antigen. Mice were primed and boosted with NP-KLH and then injected either with 200 μg of NP-MSA or 200 μg MSA (control) 3 weeks later. After a further 6 days, splenocytes were taken and analyzed by FACS. B cells were gated on CD19 and the IgD+ and IgD− populations and then analyzed further for antigen binding and for expression of IgG1. (E) Representative FACS profile showing loss of NP-specific IgG1+ but not IgG− B cells in mice injected with NP-MSA versus those injected with MSA (i-ii). The numbers of NP-specific B cells (gated) per 106 lymphocytes (i-ii) are shown. IgG− NP binding cells were positive for surface IgM in both controls and mice injected with NP-MSA (iii-iv). The quadrant statistics of IgG1− gates are shown (iii-iv). (F) Enumeration of NP-specific IgG+ and IgG− memory cells in the 2 groups. Horizontal bars represent median values. **Statistical significance (P < .01). These are combined data of 2 independent experiments.
To further confirm the finding that IgG+ NP-specific B cells were permanently removed after NP-MSA injection, we used FACS analysis for direct detection of NP-specific B cells. Mice were primed and boosted with NP-KLH and given NP-MSA or MSA only (controls) 3 weeks after the boost and analyzed after a further 6 days. To identify memory B cells, we gated on CD1921 and then analyzed the IgD+ (for IgM+ cells)22 and IgD− (for IgG+) populations21 for antigen binding and for expression of the predominant IgG subclass, IgG1 (Figure 4E). A representative FACS profile shows that IgG1+ NP-specific memory B cells (149 per 106 lymphocytes) could be detected in the control mouse (Figure 4Ei). The test mouse, injected with NP-MSA (Figure 4Eii), had 40 IgG1+ NP-specific B cells, indicating an approximately 4-fold decrease. In contrast, the IgG1−, NP-binding population did not decrease significantly with 168 cells per 106 in the test mouse (Figure 4Eii) and 181 in the control (Figure 4Ei). Further analysis revealed that the IgG1−, NP-binding population expressed IgM in both control and test mice (Figure 4Eiii-iv).
Figure 4F shows data compiled from groups of mice treated as in Figure 4E. The data confirm that the levels of the IgG1+, NP-binding memory B cells in the test group that received NP-MSA (n = 8, median of 30) were significantly lower (P < .01) than that in the control group (n = 5, median of 160). The IgG− (IgM) NP-binding cell numbers in contrast were comparable, with a median of 271 in mice that received NP-MSA versus 311 in the controls (P > .05). These data indicate that injection of NP-MSA has caused loss of the majority of the IgG+ NP-specific memory B cells from the spleen while IgM+ cells were relatively unaffected.
The IgM+ B-cell population will necessarily include naive B cells, and it was possible that some could recognize the NP-PE antigen.22,23 To determine the level of these in the repertoire, we measured antigen-binding IgM+ B cells in the spleens of naive mice. A minor population (25-40 cells/106 lymphocytes) was detected, which could account for only less than 20% of the total antigen-binding B cells. It is clear, therefore, that the majority of antigen-specific IgM+ B cells, which resist exposure to NP-MSA, are memory B cells.
Effects revealed by adoptive transfer.
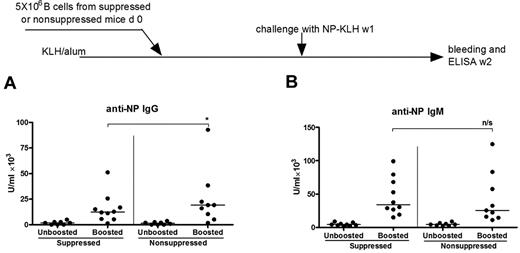
To assess the long-term effects of exposure to “helpless” antigen on B cells expressing IgG or IgM, adoptive transfer was used. B cells were isolated from mice primed and boosted with NP-KLH and then injected with NP-MSA (suppressed) or no protein (nonsuppressed). Cells were then transferred into recipients primed with KLH/alum alone (Figure 5). Each recipient group was divided into 2, with one group receiving a booster injection of NP-KLH and another being an unboosted control. One week later, serum NP-specific IgG and IgM were measured. In both unboosted groups, the levels of IgG and IgM antibodies remained similarly low between suppressed and nonsuppressed groups (Figure 5). In boosted groups, IgG responses were reduced from a median of 19 250 U/mL in the nonsuppressed group to 12 250 U/mL in the suppressed group (P < .05), although, in contrast to the Id model, a proportion of B cells able to produce IgG antibody remained significant (Figure 5A). For IgM, there was no detectable effect of the prior injection of donors with NP-MSA, with similar levels in both suppressed (34 000 U/mL) and nonsuppressed mice (25 300 U/mL). The conclusion is that, in the NP model, the effect of “helpless” antigen exposure in the donors has been to partially deplete the IgG response, but not to affect the IgM response. Because affinity for antigen may differ in the 2 isotypic populations, the next question was the role of affinity in determining survival or loss of the memory B cells.
The effect of “helpless” antigen on NP-specific IgG+ and IgM+ memory B cells as revealed by adoptive transfer. A total of 5 × 106 purified B cells from either suppressed or nonsuppressed donor mice were transferred into recipients primed with KLH in alum. One week later, the recipient mice were challenged with 20 μg NP-KLH (suppressed, n = 11; nonsuppressed, n = 10), and serum samples were analyzed for IgG (A) and IgM (B) by ELISA after further one week. *Statistical significance (P < .05). n/s indicates not significant. These are combined data of 2 independent experiments.
The effect of “helpless” antigen on NP-specific IgG+ and IgM+ memory B cells as revealed by adoptive transfer. A total of 5 × 106 purified B cells from either suppressed or nonsuppressed donor mice were transferred into recipients primed with KLH in alum. One week later, the recipient mice were challenged with 20 μg NP-KLH (suppressed, n = 11; nonsuppressed, n = 10), and serum samples were analyzed for IgG (A) and IgM (B) by ELISA after further one week. *Statistical significance (P < .05). n/s indicates not significant. These are combined data of 2 independent experiments.
Loss of memory B cells depends on affinity for antigen
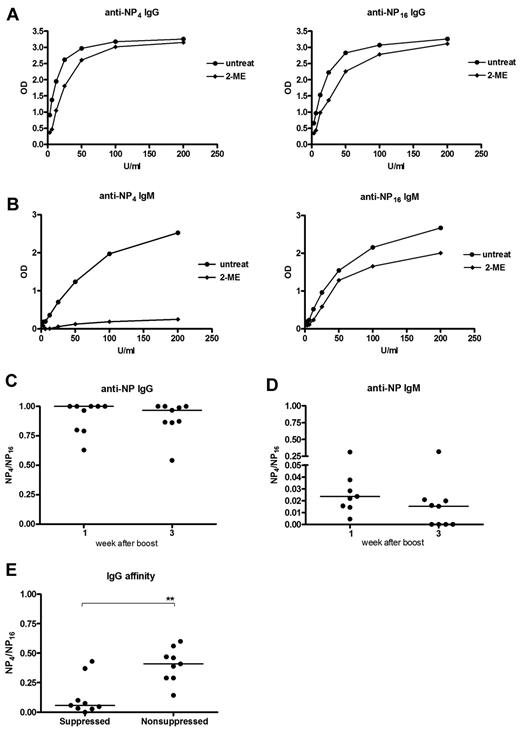
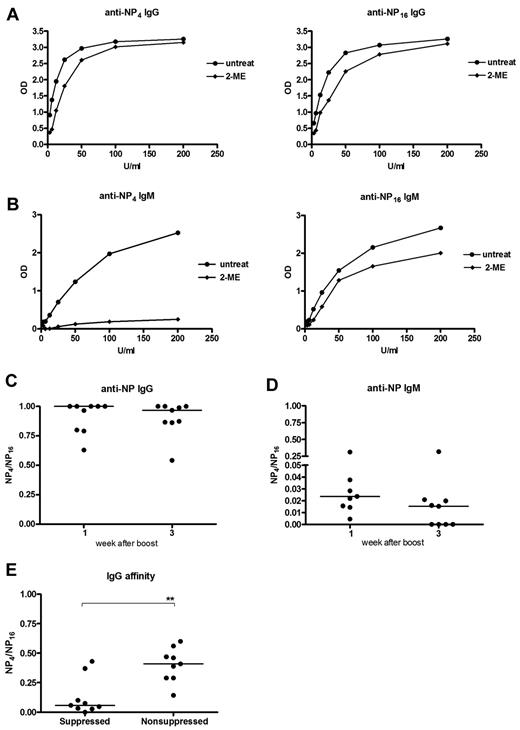
The NP model allows analysis of the binding affinity of the induced antibodies.18,24 Highly haptenated antigen (NP16) binds to both low- and high-affinity Ig, whereas less haptenated antigen (NP4) binds only to high-affinity Ig. For an equivalent assessment of IgG and IgM, it was necessary to reduce the pentameric IgM to monomer (“Antibody affinity”). Pooled serum IgG taken 3 weeks after a booster injection of NP-KLH, bound similarly to either NP4 or NP16 (Figure 6A) with or without reduction, indicating that most of the IgG was of high affinity and reduction did not affect binding to NP. In contrast, for IgM, this destroyed almost all the NP4 binding, with no significant effect on NP16 binding (Figure 6B). Binding affinities can be expressed as a ratio of binding of NP4 to NP16 of approximately 1 for IgG and approximately 0.02 for IgM at week 1 (Figure 6C-D), decreasing only slightly at week 3 (Figure 6C-D). These data from individual mice confirm the overall findings on pooled sera (Figure 6A-B) in showing a large difference in affinity between the 2 isotypes.
Characterization of affinity of memory IgG and IgM antibodies. NP-specific antibody affinity was measured by binding to either highly haptenated antigen (NP16; total Ig affinity) or less haptenated antigen (NP4; high-affinity Ig). Sera were mildly treated by 2-mercaptoethanol (2-ME) to reduce pentameric IgM. Mice were primed and boosted with NP-KLH, and affinity was measured with or without 2-ME in pooled sera samples 3 weeks after boost (A-B) or individual samples taken at 1 or 3 weeks after boost (C-D). (A-B) Representative of 2 independent experiments. (C-D) Combined data of 2 independent experiments. (E) Affinity of IgG in suppressed and nonsuppressed groups in samples from the adoptive transfer experiments shown in Figure 5. The samples were taken one week after challenge with NP-KLH after an adoptive transfer one week before challenge. Horizontal bars represent median values. **Statistical significance (P < .01). These are combined data of 2 independent experiments.
Characterization of affinity of memory IgG and IgM antibodies. NP-specific antibody affinity was measured by binding to either highly haptenated antigen (NP16; total Ig affinity) or less haptenated antigen (NP4; high-affinity Ig). Sera were mildly treated by 2-mercaptoethanol (2-ME) to reduce pentameric IgM. Mice were primed and boosted with NP-KLH, and affinity was measured with or without 2-ME in pooled sera samples 3 weeks after boost (A-B) or individual samples taken at 1 or 3 weeks after boost (C-D). (A-B) Representative of 2 independent experiments. (C-D) Combined data of 2 independent experiments. (E) Affinity of IgG in suppressed and nonsuppressed groups in samples from the adoptive transfer experiments shown in Figure 5. The samples were taken one week after challenge with NP-KLH after an adoptive transfer one week before challenge. Horizontal bars represent median values. **Statistical significance (P < .01). These are combined data of 2 independent experiments.
The next question concerned the affinity of the small population of IgG+ memory B cells, which had escaped silencing by “helpless” antigen. For this, we analyzed the IgG antibody induced after adoptive transfer of memory B cells exposed to “helpless” antigen (Figure 5A). The first observation unrelated to suppression was of an overall reduction of antibody affinity in the recipients (median, 0.4; Figure 6E) compared with the donors (median 1.0; Figure 6C). This is probably because only memory B cells are transferred, whereas the serum of the donors contains IgG from bone marrow-derived plasma cells, potentially of higher affinity.18
The ratios of binding to NP4 and NP16 showed that the surviving IgG+ B cells were capable of producing only low-affinity antibody (median 0.05), approximately 10-fold lower that nonsuppressed B cells (Figure 6E). This ratio is in the same range as the levels measured in the IgM response in the donors (Figure 6D). The findings indicate that suppression has dramatically affected the affinity of the IgG response, reducing it to the range of the largely unaffected IgM response. The fact that this fraction of the IgG response can escape suppression points to affinity rather than isotype as a determinant of survival. It was not possible to probe for IgM affinity as the binding to NP4 after reduction was too low for analysis in both groups.
Analysis of somatic mutations in anti-NP VH sequences
To gain further insight into the nature of the IgG and IgM memory responses, we performed VH sequence analysis. For the NP-specific IgG antibody response, there is a characteristic VH gene usage with certain somatic mutations known to be associated with increased affinity.25-27 To analyze VH genes, IgG+ and IgM+ memory B cells in immune mice were differentiated by booster injection; and after 3 days, splenic plasmablasts were FACS sorted using anti-CD138 and NP-PE to identify a double-positive population (“VH gene analysis”). Sequencing of 36 IgG-derived VH-Cγ amplificates from 4 polymerase chain reaction reactions showed all to be mutated (mean 4.4%; Table 1). Among these were 22 separate clones, with additional intraclonal variants. Analysis of the clones confirmed the known preferential usage of the V1-72 gene (V186.2; 13 of 22),25 with additional usage of V14-3 and a few other individual VH genes (Table 1). The 29 IgM-derived VH-Cμ amplificates contained 17 separate clones, most being mutated but with lower levels (mean 0.8%). The IgM-derived clones showed a similar profile of the preferential usage of V1-72 (6 of 17), with a proportion using V14-3 (Table 1).
Further analysis of the V1–72-derived clones indicated that the D gene, D1–1 (DFL16.1) was the most common in both isotypes, with DQ52 found in a minority (Table 2). Three J genes (J1, J2, and J4) were identified. The signature mutation associated with increased affinity for NP lies at position 33 in CDR1 (W to L).26 This was found in 9 of 13 IgG-derived clones (69%) and in 4 of 6 (67%) IgM-derived clones. A second alteration, considered to arise during VDJ recombination and also associated with increased affinity, lies at position 99 (Y to G).28 This was detected in 4 of 13 IgG-derived clones but was not found in IgM, possibly because of the smaller cohort (Table 2). As reported previously, the 2 changes were always found in separate clones. Overall, the level of mutation differs between the isotypes, but the pattern of mutations was similar.
To probe the relationship between the IgG+ and IgM+ B-cell populations, we sought clonal identity by analyzing CDR3 sequences. We detected one V1-72-DFL16.1-J1 sequence where common truncations and N-additions at the D-J junction indicate a shared clonal origin of IgG- and IgM-derived sequences (Figure 7). Clonal relationships were consolidated by common mutational changes in other parts of the VDJ sequences, including at position 33 W to L (Figure 7). A second case involved a V1-72-DQ52-J1 sequence, with very close similarity between intraclonal variants of an IgM-derived sequence and the corresponding IgG-derived sequences. Similarity was revealed by N-insertions at the V-D boundary, together with shared base changes at other positions (Figure 7). These examples reveal coexistence of IgG and IgM isotypic variants of a common parental B cell.
Coexistence of clonally related IgG and IgM isotypic variants among NP-specific memory B cells. V-genes from memory B-cell pool were sequenced as described in “VH gene analysis.” Sequence comparison reveals shared CDR3 among 2 V-D-J rearrangements in IgG and IgM clones as indicated. Shared clonal origin was further confirmed by similar mutation patterns, including L to W at position 33.
Coexistence of clonally related IgG and IgM isotypic variants among NP-specific memory B cells. V-genes from memory B-cell pool were sequenced as described in “VH gene analysis.” Sequence comparison reveals shared CDR3 among 2 V-D-J rearrangements in IgG and IgM clones as indicated. Shared clonal origin was further confirmed by similar mutation patterns, including L to W at position 33.
Discussion
Targeting cell surface molecules by passive antibody has clear value in the clinic, and the approach of active vaccination to induce and maintain antibody levels is attractive. Induction of antibody against weakly immunogenic tumor antigens, such as Id or Muc1, usually requires activation of T-cell help via conjugation to strong immunogens.2,29,30 This appears also to be the case for prophylactic vaccines against polysaccharides of pathogens, such as Streptococcus pneumoniae31,32 which may not elicit adequate T-cell help. Various vaccination strategies can therefore induce high levels of antibody, and plasma cells may persist for some time. However, it is probable that memory B cells are required for maintenance of antibody production. In our study, we have not investigated the longevity of surface Ig− plasma cells but have focused instead on the memory B-cell population. Our concern was that for cancer, and possibly for some infections, release of unconjugated “helpless” antigen could occur naturally and may affect memory B cells.
We found previously using Id antigen that the entire anti-Id memory IgG B-cell response induced by a DNA Id-FrC fusion vaccine is completely and specifically silenced by unconjugated Id Ig.14 To extend the concept to other antigens, we have reproduced these findings in the NP model. Adoptive transfer in both systems revealed that a significant proportion of antigen-exposed IgG+ memory B cells is lost. In our setting, GCs would be expected to have contracted.15 However, the effect on memory B cells appears similar to that observed in GCs.33,34
In contrast to IgG+ B cells, transferred IgM+ B cells survive, but the key to survival is not in the isotype expressed, but in the affinity for antigen. In the NP system, it was possible to compare affinities of IgG with monomeric IgM, thereby reflecting the status of cell surface IgM. It is clear that the IgM anti-NP response is of low affinity, whereas the majority of the IgG response is of high affinity. The small population of IgG+ memory B cells, which survives exposure to “helpless” antigen, was of low affinity, indicating this, rather than the isotype, as the determinant. Analysis of somatic mutations supported the link between mutational level and affinity. However, the signature W to L mutation in CDR1, which increases affinity by 10- to 100-fold,26 was observed in IgM+ clones, indicating that this change does not take affinity to the threshold of vulnerability.
Until recently, the IgG isotype has largely served as a memory marker in mice.15,35,36 However, this is not the case for human B cells,37-39 and evidence is mounting for the existence of T-dependent IgM+ memory B cells in mice, possibly with a lower level of somatic mutation.40 The origin and behavior of these cells are of interest for immunotherapeutic strategies because IgM antibody could attack tumor cells. One possibility is that they derive from the marginal zone41 with some being capable of entering follicles and undergoing somatic mutation.42 When formed, these IgM+ memory B cells appear to be scattered in follicles.43 This differs from their IgG+ counterparts, which are clustered in close proximity to contracted GCs.43
Recently, a new mouse model of memory B-cell labeling revealed long-term IgM+ B cells induced by a particulate T-dependent antigen. These displayed both a GC and non-GC phenotype and coexisted with IgG+ B cells.44 The IgM+ population responded to antigenic challenge, producing IgM+ and IgG+ centroblasts and some IgM-secreting plasmacytes. A soluble antigen (NP-CGG), although differing in the longevity of induced GC structures, also induced IgM+ memory B cells. Analysis of the intronic sequences of the IgM VH genes revealed that 22% to 51% were mutated at variable levels. The findings argue for a post-GC origin and do suggest that at least a proportion of the formed IgM+ memory B cells retain the ability to undergo isotype switch.
Our findings are in line with this model in that the IgM+ memory cells, which survive “helpless” antigen exposure, appear capable of secreting IgM. It is an open question whether IgM antibody of low affinity but high avidity can be useful in attacking pathogens or cancer cells, and it would be useful to investigate this in patients. Switch events can certainly occur but may generate IgG+ B cells of low affinity, which also survive, but, as in the model, secretion of IgG is not detectable.
Support for the ability of at least a proportion of memory IgM+ B cells to contribute to the isotype-switched memory pool has been recently reported.22 The capacity to switch appeared to be influenced by the affinity of coexisting antibody, with high affinity being inhibitory.22 This could be a factor in our system; but, even if IgM+ cells were able to switch after antibody has declined, a practical question is whether any of these cells can undergo further affinity maturation or whether naive B cells have to be activated by further vaccination.
The new insights indicate that vaccination with conjugated tumor antigens is probably effective only in patients in remission. If successful, there should be no problem for maintenance of memory. However, if tumor emerges, unconjugated antigen could attack high-affinity memory B cells. A simple strategy to bypass this would be to maintain T-cell help by repeated injections of conjugated antigen, possibly with long-term expression via adjuvant depots or via DNA delivery. The benefits of repeated vaccinations have been seen already in one study where patients became anti-Id antibody positive with prolonged vaccination.45
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Delin Zhu for design of sequencing primers and Ms Isla Wheatley for generating some sequencing data.
This work was supported by Leukemia and Lymphoma Research, United Kingdom.
Authorship
Contribution: N.S. designed and performed experiments and wrote the manuscript; M.S. and A.S. performed experiments; G.B. analyzed data; and F.K.S. designed experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Freda K. Stevenson, Genetic Vaccine Group, Cancer Sciences Division, School of Medicine, University of Southampton, Southampton, SO16 6 YD, United Kingdom; e-mail: fs@soton.ac.uk; and Natalia Savelyeva, Genetic Vaccine Group, Cancer Sciences Division, School of Medicine, University of Southampton, Southampton, SO16 6 YD, United Kingdom; e-mail: ns1@soton.ac.uk.