Abstract
Multiple myeloma (MM) develops from a premalignant plasma cell proliferative disorder, and with time can progress to a more aggressive disease in extramedullary locations. The gradually clinical evolution is supported by clonal expansion of cells that acquire genetic lesions over years. This model of cancer evolution based on ongoing genomic instability mechanism may apply to development of most MM cases. However, in a small fraction of newly diagnosed MM who relapse quickly and finally die within 2 years, the gradual model appears to be untenable. Analysis of high resolution copy number profiles obtained using single nucleotide polymorphism array data from 764 newly diagnosed MM identified large numbers of genomic rearrangements with the hallmarks of chromothripsis in 1.3% of samples. Moreover, this catastrophic event confers a poor outcome. Because chromothripsis appears to occur in a single crisis, our results suggest that high-risk MM patients use this novel way of cancer evolution.
Introduction
Multiple myeloma (MM) is a hematopoietic cancer emblematic of gradual evolution model. MM is almost always preceded by a benign premalignant plasma cell disorder, monoclonal gammopathy of undetermined significance; then progression of intramedullary MM is associated with severe clinical features and, in a fraction of patients, the tumors acquire the ability to proliferate in extramedullary sites, such as blood. In this case, it is called plasma cell leukemia, a more aggressive disease.1-3 These hallmark features of cancer evolution are coupled with complex spectra of diverse genetic alterations apparent at diagnosis and acquisition of additional changes with progression of the disease, indicating a striking genomic instability. Understanding the mechanisms underlying genomic instability in MM cells is critical to delineate pathogenesis, overcome drug resistance, and improve patients' outcome.4 A new mechanism of genomic instability called chromothripsis defined by tens to hundreds of chromosomal rearrangements involving localized genomic regions in cancer cells has been recently described.5 A large survey of single nucleotide polymorphism (SNP) array data suggests that chromothripsis occurs in 2% to 3% of primary tumors. The authors provide evidence arguing that the massive genomic alterations are generated in 1 or occasionally 2 events supporting the idea that chromothripsis probably occurs in cancers that develop after a “punctuated equilibrium” model of evolution. To assess whether this phenomenon occurs in cancer with progressive acquisition of genomic alterations that finally lead to an aggressive malignant phenotype, we analyzed high-resolution copy number profiles of 764 newly diagnosed MM using SNP arrays.
Methods
Patients and DNA sample preparation
Bone marrow from 764 patients with MM was obtained during standard diagnostic procedures in the Intergroupe Francophone du Myélome centers and shipped overnight to the Hematology Department at University Hospital in Nantes. Informed consent was obtained from all patients. Plasma cell purification was performed as previously described.6 Purified plasma cells were frozen at −80°C in lysis buffer. DNA was extracted using the DNA AllPrep DNA/RNA MiniKit (QIAGEN) in accordance with the manufacturer's instructions. DNA quality and quantity were assessed using the NanoDrop Spectrophotometer (NanoDrop Technologies). Approval for this study was obtained from the University Hopsital of Nantes.
Genomic analysis
All DNA samples were hybridized to Affymetrix Genome-Wide Human SNP Array 6.0 according to the manufacturer's instructions (Affymetrix). Affymetrix CEL files were analyzed using Affymetrix Genotyping Console software, Version 4.0 for initial quality control, followed by use of the Affymetrix Birdseed algorithm, Version 2.0 to generate SNP genotype calls. Genotyping using the Birdseed algorithm was performed using at least 44 arrays in each analysis. All samples passed the Affymetrix recommended contrast quality control and SNP call rates threshold. Copy number, allele ratio, and allele specific copy number data analysis were performed on CEL files and CHP files using Partek Genomic Suite software, Version 6.5, build 6.10.0212 (Partek Inc; http://www.partek.com). To locate segments with copy number changes, we used genomic segmentation algorithm of Partek Genomic Suite. DNA gains and losses arising from B-cell antigen receptor gene rearrangements at 2p11.2 (IGK@), 14q32.33 (IGH@), and 22q11.22 (IGL@) were excluded from the analysis.
Accession codes
Minimum information about a microarray experiment-compliant data have been deposited at Gene Expression Omnibus with accession number GSE27560.
Results and discussion
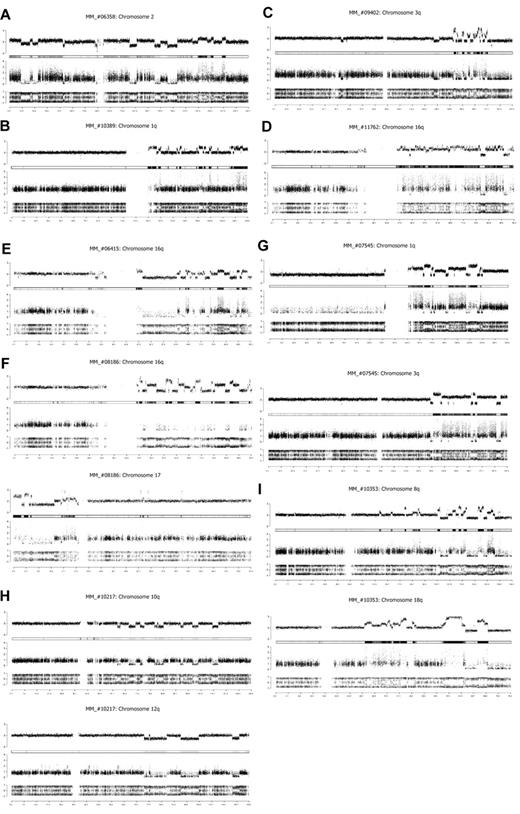
Copy number profiles derived from SNP arrays data indicated a complex genomic rearrangement with the hallmarks of chromothripsis in 1.3% of primary MM (10 of 764). The genomic alteration patterns identified in these newly diagnosed MM patients recapitulate all characteristics observed by Stephens et al.5 Genomic chaos can affect an entire chromosome (Figure 1A), a chromosome arm (Figure 1B), or focalized region of a chromosome (Figure 1C). We identified more than 50 rearrangements involving chromosome 16q with a copy number profile rapidly alternating between 2 states of copy number 3 and copy number 4 in the MM_#11762 (Figure 1D). Chromosome 16q remodeling was observed in 2 other cases; however, genomic alteration patterns are completely different among the patients. The copy number changes alternate between 3 states: one copy, 2 and 3 in the MM_#06415 case (Figure 1E), and predominantly between one copy, 2 and 6 in the MM#_08186 case (Figure 1F). Furthermore, in 4 patients, we identified similar copy number profiles on 2 chromosomes (Figure 1F-I), suggesting that cocoordinated rearrangements occur in MM patients, although the number of chromosomes involved is limited.
Chromothripsis patterns in newly diagnosed MM patients. Assessment of DNA copy number changes by using genotyping SNP6 microarrays. (Top row) The total copy number (log scale), with the heat map below. (Middle row) The contribution of each allele to the copy number (minimum allele is in black and maximum allele in gray). (Bottom row) Allelic ratio for each SNP (homozygous SNPs cluster at ratios ∼ −1 and 1 and heterozygous SNPs ∼ 0). Genomic location (Mb) is indicated on the bottom. (A) MM_#06358 patient, copy number profile of chromosome 2. (B) MM_#10389 patient, copy number profile of chromosome 1q. (C) MM_#09402 patient, copy number profile of chromosome 3q. (D) MM_#11762 patient, copy number profile of chromosome 16q. (E) MM_#06415 patient, copy number profile of chromosome 16q. (F) MM_#08186 patient, copy number profile of chromosomes 16q and 17. (G) MM_#07545 patient, copy number profile of chromosomes 1q and 3q. (H) MM_#10217 patient, copy number profile of chromosomes 10p and 12q. (I) MM_#10353 patient, copy number profile of chromosomes 8q and 18q.
Chromothripsis patterns in newly diagnosed MM patients. Assessment of DNA copy number changes by using genotyping SNP6 microarrays. (Top row) The total copy number (log scale), with the heat map below. (Middle row) The contribution of each allele to the copy number (minimum allele is in black and maximum allele in gray). (Bottom row) Allelic ratio for each SNP (homozygous SNPs cluster at ratios ∼ −1 and 1 and heterozygous SNPs ∼ 0). Genomic location (Mb) is indicated on the bottom. (A) MM_#06358 patient, copy number profile of chromosome 2. (B) MM_#10389 patient, copy number profile of chromosome 1q. (C) MM_#09402 patient, copy number profile of chromosome 3q. (D) MM_#11762 patient, copy number profile of chromosome 16q. (E) MM_#06415 patient, copy number profile of chromosome 16q. (F) MM_#08186 patient, copy number profile of chromosomes 16q and 17. (G) MM_#07545 patient, copy number profile of chromosomes 1q and 3q. (H) MM_#10217 patient, copy number profile of chromosomes 10p and 12q. (I) MM_#10353 patient, copy number profile of chromosomes 8q and 18q.
Our results obtained in a large cohort of newly diagnosed MM patients indicate that chromothripsis occurs in a hematopoietic cancer known to gradually evolve, although its frequency is lower than those of all cancers. Given the size of the cohort investigated, we assume that we have a representative picture of this phenomenon in MM. The hallmarks of chromothripsis in MM are as follows: (1) a small number of chromosomes affected by copy number rearrangements, including chromosomes 1q, 2, 3, 8q, 10, and 16q; of these, only 1q and 16q are known to be frequently altered in MM7-9 ; (2) a large spectrum of copy number that oscillates between copy number of 1 and 2, 2 and 4, 3 and 4, and 1 and 6 depending on the MM patient; (3) frequent (4 of 10) interchromosomal rearrangements that affect a limited number of chromosomes, as judged by similar alteration patterns; and (4) the average number of DNA breaks found in the 10 cases is < 30.
Clinical follow-up of the newly diagnosed MM with chromothripsis pattern revealed that half (5 of 10) experienced rapid relapse within 10 months after diagnosis; 4 of them died, and 3 deaths occurred within 12 months (Table 1), suggesting that this phenomenon confers a poor outcome in MM. It is highly probable that the massive rearrangements resulting from chromothripsis provoke dysregulation of a large number of genes leading to high clonal aggressiveness that reduces or bypasses monoclonal gammopathy of undetermined significance stage as exemplified by the 34-year-old patient with plasma cell leukemia who died 6 months after diagnosis. Although these patients are categorized as newly diagnosed patients, they clearly represent different biologic entities than in patients who live for longer than 10 years.
Chromothripsis occurs in MM at diagnosis and has clinical implications. Although it is infrequent compared with bone tumors, it delineates a new high-risk entity in MM. Such a pattern of DNA rearrangement is easily detectable using SNP arrays and could be done in routine laboratory to identify patients with aggressive disease.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Elise Douillard, Magali Devic, Emilie Maurenton, and Nathalie Roi for excellent technical expertise.
This work was supported by Intergroupe Francophone du Myélome, the French National Research Agency (grant R08079NS) (S.M.), the French Institute National du Cancer (grant R09076NN) (H.A.L.), and National Institutes of Health (grant PO1 CA155258–01) (S.M., H.A.-L., N.C.M.).
National Institutes of Health
Authorship
Contribution: F.M., H.A.-L., N.C.M., and S.M. designed the study and analyzed the data; H.A.-L. provided study materials or patients and collected clinical follow-up data; and S.M. wrote the report.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stéphane Minvielle, Inserm U892, Centre de Recherche en Cancérologie Nantes-Angers, Institut de Recherche Thérapeutique de l'Université de Nantes (IRT-UN), 8 quai Moncousu-BP 70721, 44007 Nantes Cedesc 1, France; e-mail: stephane.minvielle@chu-nantes.fr.