Abstract
Chronic lymphocytic leukemia (CLL) is characterized by a highly variable clinical course with 2 extreme subsets: indolent, ZAP70− and mutated immunoglobulin heavy chain gene (M-CLL); and aggressive, ZAP70+ and unmutated immunoglobulin heavy chain (UM-CLL). Given the long-term suspicion of antigenic stimulation as a primum movens in the disease, the role of the B-cell receptor has been extensively studied in various experimental settings; albeit scarcely in a comparative dynamic proteomic approach. Here we use a quantitative 2-dimensional fluorescence difference gel electrophoresis technology to compare 48 proteomic profiles of the 2 CLL subsets before and after anti-IgM ligation. Differentially expressed proteins were subsequently identified by mass spectrometry. We show that unstimulated M- and UM-CLL cells display distinct proteomic profiles. Furthermore, anti-IgM stimulation induces a specific proteomic response, more pronounced in the more aggressive CLL. Statistical analyses demonstrate several significant protein variations according to stimulation conditions. Finally, we identify an intermediate form of M-CLL cells, with an indolent profile (ZAP70−) but sharing aggressive proteomic profiles alike UM-CLL cells. Collectively, this first quantitative and dynamic proteome analysis of CLL further dissects the complex molecular pathway after B-cell receptor stimulation and depicts distinct proteomic profiles, which could lead to novel molecular stratification of the disease.
Introduction
Chronic lymphocytic leukemia (CLL), the most frequent form of adult leukemia in Western countries, is characterized by a highly variable clinical course. B-CLL patients can be segregated into 1 of 2 major subsets on the basis of whether or not the immunoglobulin variable heavy chain gene (IGVH) accumulates somatic mutations.1 Indeed, patients with unmutated (UM) CLL cells have a more aggressive clinical course than those with mutated (M) CLL cells.2,3
Gene expression profiling of isolated CLL B cells had revealed that a set of genes is differentially expressed in these 2 subgroups.4,5 One of these genes encodes the ζ-associated protein of 70 kDa (ZAP70), a CD3-associated protein tyrosine kinase with a critical role in T-cell receptor (TCR) signaling and early B-cell development. ZAP70 is indeed expressed by UM-CLL, but not M-CLL, cells.5,6 This could render UM-CLL cells more sensitive to ligation of the B-cell receptor (BCR) complex.7,8 BCR signaling in normal B cells, on antigen recognition, triggers activation pathways inducing (depending on the microenvironment) proliferation, survival, differentiation, anergy, or apoptosis.9 Accordingly, a model has been proposed for CLL, suggesting that ZAP70+ UM-CLL cells would be more competent than ZAP70− M-CLL cells in responding to proliferation and/or survival signals from repeated encounters with presently unknown self-antigens or environmental antigens.
In vitro, BCR cross-linking with anti-IgM antibodies mimics antigen recognition and is followed by calcium mobilization10 and phosphorylation of a number of tyrosine-kinases (including ZAP70 when present), leading in fine to the activation of specific genes. In vitro experiments of BCR ligation in CLL cells with F(ab′)2 were shown to be followed by NF-κB and phosphatidylinositol 3-kinase/Akt kinase activation (the mutational status of CLL cells was not described in this work).11 More recent studies have indicated that UM-CLL cells are indeed more sensitive to BCR triggering than M-CLL cells and that the expression of ZAP70 is directly involved in this process.12 It should be noted, however, that the ultimate cellular response to IgM ligation is also dependent on the nature of the cross-linking agent/protocol (ie, immobilized anti-IgM ligation leads to cell proliferation/survival, whereas soluble anti-IgM cross-linking leads to cell apoptosis).13
Temporal changes in gene expression early after strong BCR cross-linking in normal B cells, M-CLL cells, and UM-CLL cells have been previously investigated by us.14 The results of this study underscored the fact that aggressive UM-CLL cells engaged a specific genetic program within hours of stimulation with increased expression of genes involved in cell-cycle regulation, proliferation, or apoptosis.
To complement our understanding of the different behavior of M- and UM-CLL cells in response to in vitro soluble anti-IgM triggering of BCR-dependent pathways, we developed a functional proteomic approach using the more recent 2-dimensional fluorescence-differential gel electrophoresis (2D-DIGE) technology,15,16 well adapted for multiple quantitative comparisons, coupled to mass spectrometry. We report here that, although the 2 CLL subsets can be distinguished at baseline by the global image of the proteome, ZAP70+ UM-CLL is particularly affected by significant changes of proteomic profile on BCR ligation. Further identification of proteins differentially expressed between the 2 groups at baseline and after stimulation led to the identification of several prime candidates potentially involved in CLL pathophysiology.
Methods
Patients and B-cell selection
B cells were purified from peripheral blood samples obtained from untreated CLL patients. All samples were drawn and used according to the institutional review board-approved protocol of each participating hospital. Cells collected from 6 patients were used for 2D-DIGE analysis: 3 had aggressive characteristics (UM IGVH and ZAP70+) and 3 indolent features (M-IGVH and ZAP70−). Protein extracts from 13 other untreated patients were used for Western blot (WB) validation of candidate proteins. Informed consent was obtained for each patient in accordance with the Declaration of Helsinki. B cells were negatively selected using the RosetteSep B-cell enrichment cocktail (StemCell Technologies) followed by density gradient centrifugation on Ficoll-Paque PLUS (GE Healthcare). Tumor cells were resuspended in RPMI 1640 medium and allowed to settle for 2 hours at 37°C/5% CO2 before stimulation.
BCR ligation and response analysis
B cells resuspended at a concentration of 107/mL were divided in two. The first half was used as control (unstimulated [US], receiving no antibody), whereas the second underwent anti-IgM stimulation (stimulated [S] cells). The latter was achieved using biotinylated goat F(ab′)2 anti–human IgM (Southern Biotechnology) at 20 μg/mL, cross-linked with 20 μg/mL avidin (Sigma-Aldrich) on ice during 5 minutes and then incubated at 37°C for 15 minutes, as adapted from Chen et al.7 The amount and duration of anti-IgM ligation had been determined previously14 to induce a brief but sustained stimulation in all cells at once. After stimulation, cells were washed, resuspended in RPMI 1640 supplemented with 10% heat-inactivated FCS (Fisher Bioblock Scientific), and incubated at 37°C/5% CO2 for up to 7 hours. Collection time points (1, 2, 4, and 7 hours) were established after a 2D-gel silver-stained pilot study (data not shown). At each collection time point, aliquots of US and S cells were washed and frozen as dry pellets for subsequent protein extractions. The degree of B-cell apoptosis after BCR ligation was evaluated by flow cytometry through FITC–annexin V binding. Concomitant labeling with propidium iodide allowed to measure cell death. Briefly, 1 × 105 S or US cells were stained with 5 μL FITC–annexin V and 10 μL propidium iodide (BD Biosciences) and analyzed on an FC 500 flow cytometer (Beckman Coulter). The percentage of cells undergoing apoptosis 6 hours after stimulation was assessed by comparing annexin V–propidium iodide staining of stimulated versus unstimulated cells. A cut-off point of 20% was set to define responders versus nonresponders.
2D-DIGE
Tumor cells isolated from the 6 patients were used for 2D-DIGE analysis. For each patient, 8 different samples were submitted to DIGE (ie, 4 US and 4 S samples corresponding to the 4 time points of cell harvesting). Migrations for 2D-DIGE were performed according to the following procedure for a total of 48 samples (24 M and 24 UM), corresponding to 24 2D-DIGE gels. It is noteworthy that such 2D-DIGE experiments can overlap up to 48 protein extracts previously mixed to form an internal standard. The use of > 48 extracts induces a risk of dilution for the less abundant proteins or shedding for major proteins. For each sample, total proteins were extracted in isoelectric focusing-specific lysis buffer containing 7M urea, 2M thiourea, 1% 3-(3-cholamidopropyl)dimethylammonio-1-propanesulfonate (CHAPS), 10% isopropanol, 10% isobutanol, 0.5% Triton X-100, 0.5% SB 3-10, 30mM Tris, 65mM dithiothreitol, 0.5% IPG buffer 3-10, and 30mM spermine. After centrifugation at 16 000g for 30 minutes at 4°C, proteins were precipitated with the Perfect-Focus Kit from G-Biosciences (Agro-Bio) and resuspended in buffer containing 7M urea, 2M thiourea, 1% CHAPS, 10% isopropanol, 10% isobutanol, 0.5% Triton X-100, 0.5% SB 3-10, and 30mM Tris. The total protein concentration of each sample was established using the Bradford Assay (Protein Assay, Bio-Rad) with BSA as standard. All protein extracts (50 μg per sample) were labeled using fluorescent Cy dyes, as per the manufacturer's instructions for minimal labeling (GE Healthcare). The internal standard (pool) used for comparisons between the 24 2D gels was a mixture of identical amounts of the 48 protein samples tested, labeled with Cy2. Cy3 and Cy5 were alternatively used to label S and US protein extracts according to the dye switch method. For each gel, the 3 labeled protein extracts, expected to comigrate, were mixed to a strip's rehydration buffer containing 7M urea, 2M thiourea, 1% CHAPS, 10% isopropanol, 10% isobutanol, 0.5% Triton X-100, 0.5% SB 3-10, 40mM dithiothreitol, and 0.5% IPG buffer 4-7 for a total volume of 460 μL. The rehydration of a 24-cm Immobiline, pH 4 to 7 DryStrip (GE Healthcare) was achieved in the dark during 16 hours. Isoelectric focusing was then performed at 20°C for a total of 85 000 Vh using the Ettan II IPGphor system (GE Healthcare). After migration, the strips were equilibrated in SDS-containing buffer (reduction and alkylation) before being loaded onto SDS polyacrylamide gels for separation according to molecular weight using an Ettan DALT 6 Electrophoresis System (GE Healthcare). After migration, DIGE gels were scanned using an Ettan DIGE Imager (GE Healthcare) according to the manufacturer's instructions. Image analysis and statistical calculations were performed using DeCyder, Version 6.5 software (GE Heathcare). All standard/sample gel images were processed using the Differential In-gel Analysis software module to detect, normalize, and quantify protein spots. Cy2 images of internal standards were used to normalize gels by calculating the standardized abundance of each spot as the ratio of both Cy3 and Cy5 signals to that of Cy2. Gel-to-gel matching of the standard maps from each gel, and statistical analyses were performed using the Biologic Variation Analysis software module. Statistical analyses were performed with the Extended Data Analysis (EDA, Version 1.0 module). First, data were filtered to retain only spots that were present in > 75% of the gels. Then, statistical tests (t test) and principal component and hierarchical cluster analyses were performed. For t test, a P value of < .05 was considered significant. The threshold chosen to appreciate variations was a 1.4-fold change (FC). Data were exported from EDA using the XML Toolbox module. Significant Analysis of Microarray (SAM) was performed on the exported data to estimate False Discovery Rate (FDR) for multiple testing. The normalized log ratio values were used to compare protein expression between groups. Statistical analyses was performed with Multiple experiment Viewer (www.tigr.org) or with Excel software using SAM.17 Differentially expressed proteins between US and S groups (at each time point with 3 biologic replicates/group for UM and or M, 2-class paired statistical analysis) were selected with SAM using a threshold of 5% for the FDR. Differentially expressed proteins between US UM and M groups (at each time point with 3 biologic replicates/group, 2-class unpaired statistical analysis) were selected with SAM using a threshold of 5% for the FDR. Time course analyses were also performed using SAM on UM paired data and M paired data to find differentially expressed proteins between unstimulated and stimulated groups, and on US unpaired data to find differentially expressed proteins between unstimulated UM and M groups.
Protein identification
Analytical gels were stained with SYPRO Ruby (Invitrogen) and used for robotized spot picking (EXQuest spot cutter, Bio-Rad). Gel plugs were washed twice with 25mM ammonium bicarbonate in 50% acetonitrile and then dehydrated in 100% acetonitrile, before being subjected to overnight in-gel trypsic digestion. Briefly, each spot was digested with a solution containing 50mM ammonium bicarbonate, 10% acetonitrile, and 20 ng/μL trypsin (G-Biosciences) on ice during 1 hour, then overnight at 37°C. The supernatants were collected and gel plugs were incubated in 0.1% trifluoroacetic acid, 50% acetonitrile in an ultrasonic bath during 10 minutes to extract residual peptides. The new supernatants were pooled to the former. Peptides were dried completely in a vacuum centrifuge and then resuspended in 0.1% trifluoroacetic acid, 50% acetonitrile, to be analyzed by mass spectrometry (MS or MS/MS). Peptides were spotted onto a matrix-assisted laser desorption ionization plate with matrix solution (50% acetonitrile, 0.1% trifluoroacetic acid, α-cyano-4-hydroxycinnamic acid at saturation) 4 times diluted in 50% acetonitrile, 0.1% trifluoroacetic acid, and analyzed by MS using an Autoflex MALDI-TOF (Bruker Daltonics). This instrument was operated in positive ion mode and externally calibrated in the peptide mass range of 700 to 3200 m/z. MS/MS analysis was performed using the Ultimate 3000 nano LC system (Dionex) coupled to an Esquire HCTultra nESI-IT-MS (Bruker Daltonics). A search for protein identity was carried out with MASCOT (www.matrixscience.com). Confident matches were defined by the MASCOT score and statistical significance (P < .05), the number of matching peptides and the percentage of total amino acid sequence covered by matching peptides.
WB analysis
Immunoblotting was carried out according to standard procedures using the following primary antibodies: PDCD4, β-actin (Sigma-Aldrich), and RAD23B (Santa Cruz Biotechnology). Cell lysates were prepared by incubating purified B-cell pellets on ice for 30 minutes in 200 μL of a lysis buffer containing 150mM NaCl, 50mM Tris, pH 8, 1% NP40, 10% glycerol, 0.5mM ethylenediaminetetraacetic acid, and protease inhibitor Complete 1X (Roche Diagnostics). The samples were then centrifuged at 4°C for 30 minutes at 16 000g. Samples containing 50 μg of protein were mixed with Laemmli buffer 1× and boiled for 5 minutes. Protein extracts were separated on 10% SDS-PAGEs and then transferred to nitrocellulose membranes (Hybond-C Extra, GE Healthcare). Nonspecific binding sites were blocked for 1 hour in 5% milk in T-TBS (50mM Tris, 150mM NaCl, 0.5% Tween-20). The membranes were then probed with primary antibodies (for PDCD4 and RAD23B) overnight at 4°C. After thorough washing in T-TBS, incubation was carried out for 1 hour in the presence of horseradish peroxidase–conjugated anti–mouse IgG or horseradish peroxidase–conjugated anti–rabbit IgG (Bio-Rad). All immunoblots were revealed by enhanced chemiluminescence using the Super Signal West Pico Substrate (Pierce Biotechnology). Data were quantified by scanning-densitometry using the Chemi-Start and Bio1D-Advanced software Version 12.10 (Vilber-Lourmat).
Results
The molecular pathogenesis of CLL remains largely enigmatic. The central role of BCR, through recognition of yet to be identified cognate and/or foreign antigen(s), is however widely acknowledged. Having recently performed a transcriptomic analysis of CLL on BCR ligation,14 we aim here to conduct a similar experiment by proteomic analysis of UM and M-CLL patients on stimulation or not of the BCR taking advantage of the highly sensitive 2D-DIGE.
Quantitative proteomic analysis at baseline and after BCR ligation
Tumor cells of 6 CLL patients selected according to their IGVH status and ZAP70 expression were used for 2D-DIGE analysis: 3 patients shared biologically indolent CLL characteristics, including M IGVH and the absence of ZAP70 expression (patients M1, M2, and M3), whereas 3 carried aggressive CLL features, including UM IGVH and ZAP70 expression (patients UM1, UM2, and UM3). Clinical and biologic characteristics of these 6 patients are shown in Table 1. B cells from 13 additional patients, segregated according to their IGVH status (supplemental Table 1; see the Supplemental Materials link at the top of the article), were used later for WB validations. Stereotyped BCR analyses were also performed for all patients according to Murray et al.18
BCR ligation was performed by a biotinylated goat anti–human IgM F(ab′)2. The protocol adapted from Chen et al7 and previously used in our gene-expression profiling was selected over other cross-linking agents, such as membrane-bound antibodies (which induce proliferation/survival) as it has the advantage of being brief but strong. Hence, aliquots of the selected CLL cells were stimulated with F(ab′)2 anti-IgM and tested for apoptosis using annexin V and propidium iodide staining by flow cytometry up to 6 hours after stimulation Flow cytometry patterns allowed to distinguish between responders and nonresponders as previously described.14 All ZAP70+ UM-CLL cells belonged to the responder group, as shown in Table 1. Two M-CLL cells were nonresponders, whereas cells from 1 M patient (#M3) were unexpectedly observed to be responder albeit being ZAP70−.
Quantitative proteomic analysis in 2D-DIGE was performed for the 48 protein extracts obtained from the 24 ZAP70− M-CLL samples, 12 US and 12 S, and the 24 ZAP70+ UM-CLL samples, corresponding to 24 2D gels as described in “2D-DIGE.” The 24 Cy2-labeled images of the internal standard (exemplified in Figure 1A) allowed to match all gel images for polypeptide spots alignment and were used for normalization. DeCyder analysis of the 72 images (24 Cy2-labeled for internal standard and 48 Cy3- or Cy5-labeled images corresponding to patients samples) led to the detection and quantification of 1873 polypeptide spots, with an average of 1328 matched spots. Of them, 928 polypeptide spots ranging between 15 and 150 kDa, with an isoelectric point between 4 and 7, present in at least 75% of the 48 analytical images, were retained for further analyses.
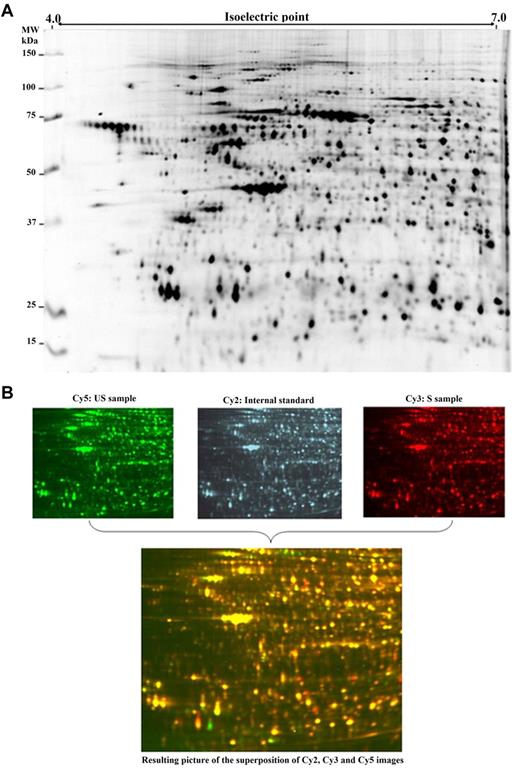
2D-DIGE gel images. (A) Representative 2D image (scanned using an Ettan DIGE Imager, GE Healthcare) of a Cy2-labeled mixture of the 48 protein extracts (50 μg of each). The total number of polypeptide spots observed and matched is 928 (only polypeptide spots present in at least 75% of analytical images were retained). (B) False-colored representation of 2D images issued from a single gel: Cy3 and Cy5 images correspond to analytical samples and Cy2 to the internal standard. Superimposition of the 3 images colored by ImageMaster 2D Platinium Version 6.0 software (GE Healthcare) allows to represent overexpressed polypeptide spots in S samples in red and overexpressed polypeptide spots in US samples in green.
2D-DIGE gel images. (A) Representative 2D image (scanned using an Ettan DIGE Imager, GE Healthcare) of a Cy2-labeled mixture of the 48 protein extracts (50 μg of each). The total number of polypeptide spots observed and matched is 928 (only polypeptide spots present in at least 75% of analytical images were retained). (B) False-colored representation of 2D images issued from a single gel: Cy3 and Cy5 images correspond to analytical samples and Cy2 to the internal standard. Superimposition of the 3 images colored by ImageMaster 2D Platinium Version 6.0 software (GE Healthcare) allows to represent overexpressed polypeptide spots in S samples in red and overexpressed polypeptide spots in US samples in green.
Figure 1B summarizes the 2D-DIGE principle of comigration of 3 protein extracts (2 analytical samples and 1 internal standard) on each gel. The merged image obtained by superimposing Cy2, Cy3, and Cy5 patterns allows to show how quantitative comparisons could be made using the Cy2-labeled internal standard. Green spots on the merged image represent proteins increased in the Cy5-labeled sample, whereas red spots identify proteins increased in the Cy3-labeled sample.
Proteomes of UM- and M-CLL cells are constitutively different
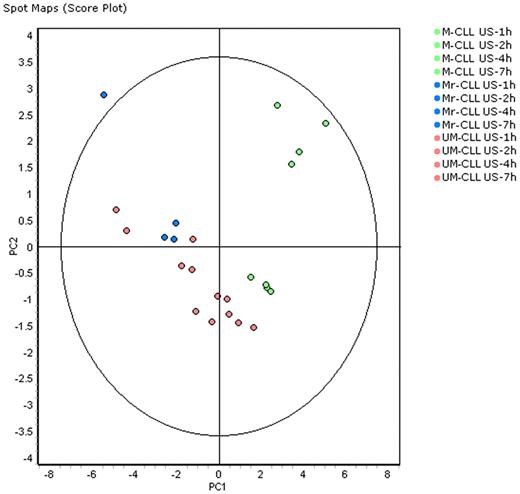
Multivariate analyses of protein expression data were first performed using the DeCyder EDA statistical analysis module. Principal component analysis (PCA) was used as an explorative tool to investigate the clustering of all US/S M- and UM-CLL samples. Each data point in the PCA plot represents an individual sample/condition and PCA performed a multivariate analysis down to the 2 most significant sources of variation (PC1 and PC2) between samples, based on global expression patterns from the subset of features present in each sample. The score plots of PC1 and PC2 are shown in Figure 2 for the 24 US samples and in Figure 3 for all US and S samples. PCA analysis demonstrated that US-CLL samples segregated into 2 groups, distinguishing baseline M-CLL and UM-CLL cells (Figure 2), except for 1 patient whose cells (although correctly identified as ZAP70− and with mutated IGVH) clustered with aggressive (ZAP70+ and with unmutated IGVH) cells. Interestingly, this patient (#M3 in Table 1) was the same for whom isolated B cells responded to BCR ligation. Samples obtained from this particular patient are represented in blue in PCA Figures 2 and 3.
PCA on samples without BCR activation distinguishes M-CLL and UM-CLL cells. PCA as a score plot of a spot map of the 24 US samples (12 M-CLL and 12 UM-CLL) color-coded according to the legend, projected onto the first 2 principal components. PC1 axis segregated most M-CLL and UM-CLL samples; #M3 patient samples colored in blue provided from M-responder-CLL (Mr) cells and segregated like UM-CLL samples. The oval englobes a 95% statistical confidence perimeter.
PCA on samples without BCR activation distinguishes M-CLL and UM-CLL cells. PCA as a score plot of a spot map of the 24 US samples (12 M-CLL and 12 UM-CLL) color-coded according to the legend, projected onto the first 2 principal components. PC1 axis segregated most M-CLL and UM-CLL samples; #M3 patient samples colored in blue provided from M-responder-CLL (Mr) cells and segregated like UM-CLL samples. The oval englobes a 95% statistical confidence perimeter.
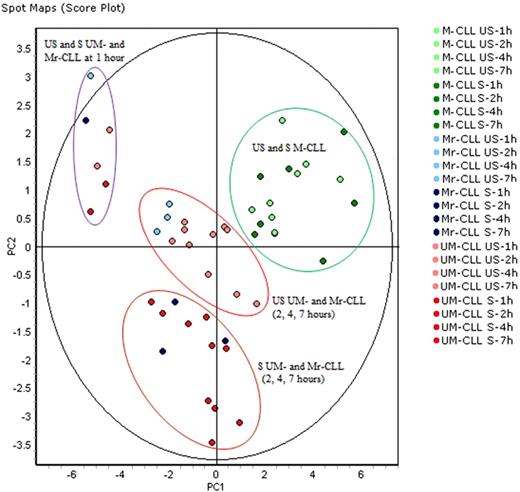
PCA of all samples differentiates US and S samples only in UM-CLL and Mr-CLL (M responder CLL) cells. PCA of all 48 samples (24 M-CLL and 24 UM-CLL) and color-coded according to the legend, projected onto the first 2 principal components. Ellipses highlight M-CLL samples (US and S samples indiscriminately), US UM-and Mr-CLL samples except at time 1 hour, S UM- and Mr-CLL samples except at 1 hour, and UM- and Mr-CLL samples at 1 hour (US and S undiscriminated). PCA analysis demonstrated that M-CLL and UM-CLL cells formed 2 distinct groups and that US and S samples at 2, 4, and 7 hours can be differentiated only in UM-CLL and Mr-CLL samples.
PCA of all samples differentiates US and S samples only in UM-CLL and Mr-CLL (M responder CLL) cells. PCA of all 48 samples (24 M-CLL and 24 UM-CLL) and color-coded according to the legend, projected onto the first 2 principal components. Ellipses highlight M-CLL samples (US and S samples indiscriminately), US UM-and Mr-CLL samples except at time 1 hour, S UM- and Mr-CLL samples except at 1 hour, and UM- and Mr-CLL samples at 1 hour (US and S undiscriminated). PCA analysis demonstrated that M-CLL and UM-CLL cells formed 2 distinct groups and that US and S samples at 2, 4, and 7 hours can be differentiated only in UM-CLL and Mr-CLL samples.
Differentially expressed polypeptide spots between baseline US M- and UM-CLL groups are presented in Table 2. Among the 26 proteins with FC > 1.4 and P < .05 selected in EDA, 6 spots corresponding to 5 unique proteins were finally retained after SAM-determined FDR application, eliminating potentially false positives. Some of the redundancies appeared to be the result of posttranslational modifications that created charge-related isoforms having different isoelectric points but nearly identical molecular masses. For example, spots #434 and #436 meet these criteria: the hematopoietic lineage cell-specific protein (HCLS1) has 1 acidic isoform (#436, isoelectric point 4.87) increased in UM-CLL cells compared with M-CLL cells, and 1 basic isoform (#434, isoelectric point 4.92) decreased in UM-CLL cells, suggesting a different phosphorylation status of HCLS1 between the 2 CLL subsets. Regarding other differentially modulated proteins, 4 were increased in UM-CLL cells: respectively, DNA-directed RNA polymerases I, II, and III subunit RPABC1 (POLR2E), cAMP-dependent protein kinase type I-α regulatory subunit (PRKAR1A), and 2 isoforms of S-formylglutathione hydrolase. One protein appeared decreased in UM-CLL cells compared with M-CLL cells, namely, the basic isoform of HCLS1. It might be noteworthy that most of the protein was present in more acidic forms in the UM-CLL subset compared with the M-CLL counterpart.
A specific proteomic response of UM-CLL cells after BCR ligation
To study the effects of BCR stimulation in both M-CLL and UM-CLL subsets, PCA analysis with all US and S samples was performed (Figure 3). This analysis highlighted that M-CLL and UM-CLL cells segregated in 4 distinct groups (with the exception of patient M3): (1) US and S M-CLL samples were all grouped in the upper right quadrant of the PCA analysis; (2) all US and S UM-CLL samples obtained at 1 hour after stimulation clustered on the left, suggesting minimal variations induced by stimulation in the first hour of the kinetics; (3) baseline US UM- and M3-CLL samples clustered together; and (4) S UM- and M3-CLL samples are clearly separated at the bottom. In short, PCA of all samples differentiated US and S samples only for UM- and M3-CLL cells. Another representation of multivariate analysis is a 2-dimensional hierarchical clustering of the 16 M-CLL samples (M3 patient samples excluded) and 24 UM-CLL samples presented in supplemental Figure 1. M-CLL samples clustered according to time, without separation between US and S samples, although UM-CLL samples clustered according to US or S status (except for 1-hour samples clustering together).
This finding was confirmed by the results of statistical calculations analyzing differentially expressed polypeptide spots between US and S samples in both M- and UM-CLL. Indeed, only 6 polypeptide spots (of 25 spots, after SAM-determined FDR application) were found increased in S M-CLL samples compared with US cells (Table 3). Despite their relative smaller number compared with S versus US UM-CLL, S versus US M-CLL did show indeed significant differences in the following polypeptides: enoyl coA hydratase ECHS1, NADH dehydrogenase (ubiquinone) iron-sulfure protein 3 (NDUFS3), ran-specific GTPase-activating protein (RANBP1), proteasome activator complex subunit 1 (PSME1), UPF0424 protein C1orf128, and catechol O-methyltransferase (COMT).
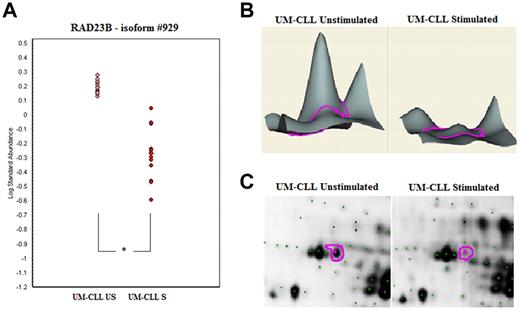
Conversely, 13 polypeptide spots were significantly increased and 12 others significantly decreased in S UM-CLL cells compared with their US counterparts (Table 4). Among the 13 proteins overexpressed after BCR stimulation, the largest variation between US and S samples was observed for cytoplasmic dynein 1 intermediate chain 2 (DYNC1I2), COMT, and HNRPK (FC of 4.1, 3.4, and 2.9, respectively). Among the 12 proteins that decreased after stimulation, the most significant were UV excision repair protein (RAD23B), lamin B2 (LMNB2), and PDCD4 (FC at −4.2, −2.8, and −2.5, respectively). The profiles of each differentially expressed protein were examined more precisely. An example of gel images and DeCyder 2D and 3D representations is shown in Figure 4 for RAD23B.
Example of 2D DIGE analysis of RAD23B protein expression. DIGE spots were analyzed with Decyder software Version 6.5 (GE Healthcare). (A) Statistical analysis of spot #929: significant decrease of RAD23B expression after BCR activation only in the UM-CLL subset. *P < .0001. (B) Decyder 3D representation of the RAD23B volume ratios difference between UM-CLL samples at baseline and after stimulation. (C) Focus of the RAD23B protein (spot #929) on US/S UM-CLL samples 2D gel images (scanned using an Ettan DIGE Imager, GE Healthcare).
Example of 2D DIGE analysis of RAD23B protein expression. DIGE spots were analyzed with Decyder software Version 6.5 (GE Healthcare). (A) Statistical analysis of spot #929: significant decrease of RAD23B expression after BCR activation only in the UM-CLL subset. *P < .0001. (B) Decyder 3D representation of the RAD23B volume ratios difference between UM-CLL samples at baseline and after stimulation. (C) Focus of the RAD23B protein (spot #929) on US/S UM-CLL samples 2D gel images (scanned using an Ettan DIGE Imager, GE Healthcare).
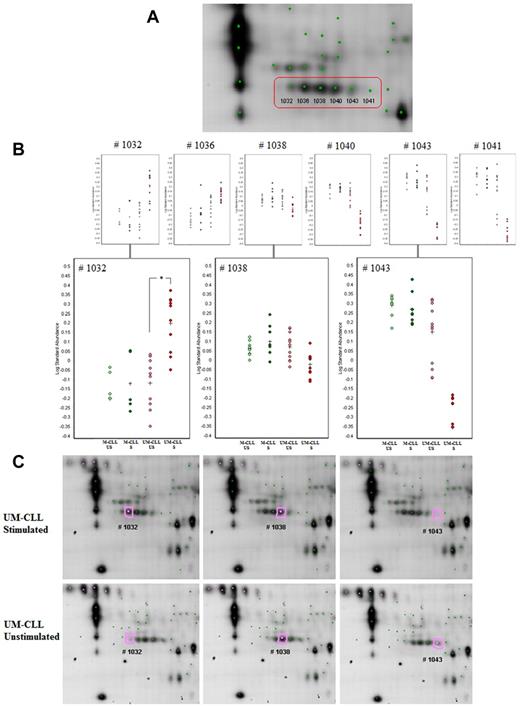
Moreover, as described in the preceding paragraph, 2D-DIGE experiments allow to distinguish different isoforms of a protein, allowing to study such posttranslational modifications as phosphorylations: the addition of 1 or several negatively charged phosphate groups to a protein decreases its isoelectric point without affecting its mass, allowing for easy detection in 2D gels (Table 5; Figure 5). Among all polypeptide spots identified in multiple forms, most of them are indeed affected by posttranslational modifications: phosphorylation of 1 amino acid residue corresponds in 2D approach to an increase of 1 or several acid isoforms associated with a decrease of 1 or several basic isoforms. We studied the variation of the different isoforms of proteins with multiple identifications and found that several of them underwent such phosphorylations after anti-IgM stimulation in UM-CLL cells (Table 5). These proteins are mostly cytoskeleton-associated proteins, such as lamin B1, lamin B2, vimentin, and plastin 2, but also proteins involved downstream the BCR pathway, such as HCLS1 or LSP1.19,20 For 1 protein (HCLS1), we observed, specifically in UM-CLL cells, a constitutive phosphorylation status (before in vitro stimulation), enhanced after anti-IgM activation. An example of posttranslational modifications of the LSP1 protein after anti-IgM stimulation in UM-CLL cells is presented in Figure 5.
Example of 2D-DIGE analysis (using DeCyder software v6.5, GE Healthcare) of multiple isoforms of LSP1. (A) Focus of the 6 identified isoforms of LSP1 on 2D gel image. (B) Statistical analysis (3 UM-CLL vs 2M-CLL patient, #M3 patient excluded) of the 6 polypeptide spots identified as LSP1 with focus on 3 isoforms #1032, #1038, and #1043: #1032 acid isoform is significantly increased after anti-IgM stimulation in UM-CLL samples (*P < .0003), #1038 isoform presents no significant modification, and #1043 basic isoform is marginally decreased after anti-IgM stimulation. Such changes (increase of acid isoform and decrease of basic isoform) in 2D electrophoresis suggest a phosphorylation mechanism. (C) Focus of LSP1 isoforms on US/S UM-CLL samples 2D gel images (scanned using an Ethan DIGE Imager, GE Healthcare).
Example of 2D-DIGE analysis (using DeCyder software v6.5, GE Healthcare) of multiple isoforms of LSP1. (A) Focus of the 6 identified isoforms of LSP1 on 2D gel image. (B) Statistical analysis (3 UM-CLL vs 2M-CLL patient, #M3 patient excluded) of the 6 polypeptide spots identified as LSP1 with focus on 3 isoforms #1032, #1038, and #1043: #1032 acid isoform is significantly increased after anti-IgM stimulation in UM-CLL samples (*P < .0003), #1038 isoform presents no significant modification, and #1043 basic isoform is marginally decreased after anti-IgM stimulation. Such changes (increase of acid isoform and decrease of basic isoform) in 2D electrophoresis suggest a phosphorylation mechanism. (C) Focus of LSP1 isoforms on US/S UM-CLL samples 2D gel images (scanned using an Ethan DIGE Imager, GE Healthcare).
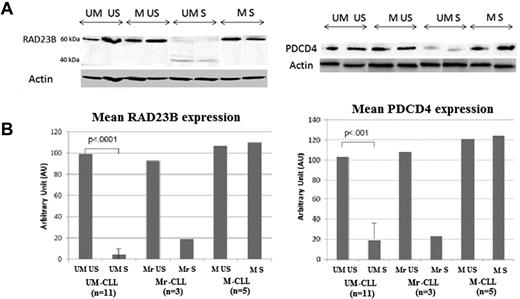
RAD23B and PDCD4 were retained for WB validations (Figure 6). WB experiments validated a significant decrease of RAD23B and PDCD4 expression after stimulation in UM-CLL cells. These WB experiments also validated the absence of any effect for BCR stimulation on M-CLL cells, except for the M3 patient and 2 of the 5 other M-CLL patients included. Interestingly, we observed in RAD23B WB experiments the appearance in S samples of a shorter band (40 kDa), suggesting a cleavage of the RAD23B 60 kDa-protein after anti-IgM stimulation, in UM-CLL cells only. Similar data for all patients are provided in supplemental Figure 2.
WB validations of PDCD4 and RAD23B profiles. (A) WB results of PDCD4 (∼ 50 kDa) and RAD23B (60 kDa) expression in US and S samples from UM- and M-CLL cells. Actin bands were used as loading controls. (B) RAD23B and PDCD4 expression levels in US and S UM-CLL (n = 11), Mr-CLL (M-responder-CLL; n = 3), and M-CLL (n = 5) samples.
WB validations of PDCD4 and RAD23B profiles. (A) WB results of PDCD4 (∼ 50 kDa) and RAD23B (60 kDa) expression in US and S samples from UM- and M-CLL cells. Actin bands were used as loading controls. (B) RAD23B and PDCD4 expression levels in US and S UM-CLL (n = 11), Mr-CLL (M-responder-CLL; n = 3), and M-CLL (n = 5) samples.
Description of an intermediate CLL subset
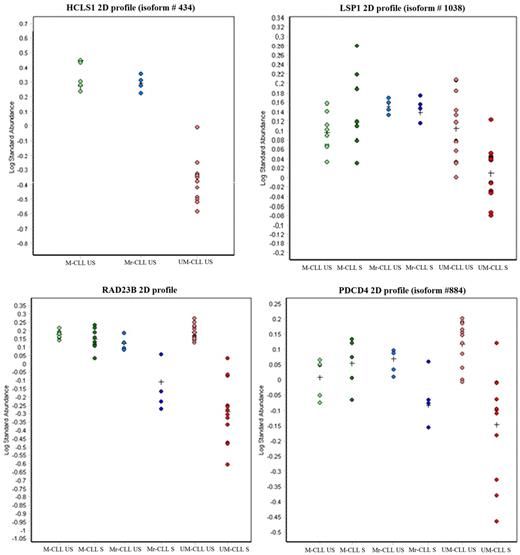
Intriguing data were indeed obtained for patient M3 (Table 1). This patient, initially included on the basis of his ZAP70− M-CLL cells, surprisingly responded to BCR ligation according to our apoptosis assay. Indeed, proteomic results showed that cells from this patient clustered with UM-CLL samples. 2D-DIGE data and WB experiments confirmed that cells from this patient expressed a hybrid proteomic profile, sharing some features of ZAP70+ UM-CLL cells (including decreased RAD23 or PDCD4) and some of ZAP70− M-CLL cells, such as increased HCLS1, whereas no modification of LSP1 after stimulation (downstream of BCR pathway; Figure 7). This specific patient's clinical features were therefore revisited and evidenced a more aggressive clinical symptomatology, including severe hyperleukocytosis having necessitated the initiation of a chemo-immunotherapy, whereas the other M-CLL patients did not need such therapy. Accordingly, the 3 UM patients had an aggressive clinical course, and all required treatment initiation within 2 years after sample collection for this study, patient UM3 actually having already died of disease progression. WB validations with cells from 13 other CLL patients reinforced this notion of intermediate form of CLL. Indeed, 2 of the 5 additional M-CLL patients whose cells were used for WB validations shared a proteomic response to BCR stimulation similar to that observed for patient M3 (PDCD4 and RAD23B WB profiles, Figure 6B). Inversely, all 8 additional UM-CLL patients had a typical aggressive profile for PDCD4 and RAD23B as did the 3 patients used for 2D experiments. These data collectively suggest that, unlike UM-CLL cases, which share a similar response profile, M-CLL cells could be dichotomized into bona fide M-CLL indolent versus aggressive profiles. It should be noted, however, that our categorization at this point is made purely on the basis of biologic information (ie, IGVH mutations, ZAP70 status, response vs nonresponse to BCR ligation, and finally the proteomic profile) and not based on the clinical phenotype of individual patients.
Proteomic profiles of #M3 patient cells (in blue). #M3 patient cells share some characteristics of ZAP70− M-CLL cells, such as HCLS1 and LSP1 profiles, but also some features of aggressive responders ZAP70+ UM-CLL cells, such as decrease of RAD23B and PDCD4 after anti-IgM activation. Points corresponded to expression levels of HCLS1, LSP1, PDCD4, or RAD23 proteins in US or S M-CLL, Mr-CLL (#M3 patient), and UM-CLL cells.
Proteomic profiles of #M3 patient cells (in blue). #M3 patient cells share some characteristics of ZAP70− M-CLL cells, such as HCLS1 and LSP1 profiles, but also some features of aggressive responders ZAP70+ UM-CLL cells, such as decrease of RAD23B and PDCD4 after anti-IgM activation. Points corresponded to expression levels of HCLS1, LSP1, PDCD4, or RAD23 proteins in US or S M-CLL, Mr-CLL (#M3 patient), and UM-CLL cells.
Gene expression profiling and proteomics comparisons
We finally compared our proteomic results with our previously published transcriptomics data on BCR-stimulated CLL cells.14 The time points selected for collecting mRNA in that study were 1 hour, 1 hour 30 minutes, 3 hours 30 minutes, and 6 hours 30 minutes; so 30 minutes shorter than time points chosen for protein collection. All the cell preparation and BCR stimulation conditions were otherwise identical. We first checked for the expression value of all the probe sets potentially coding for the various modulated proteins in M- and UM-CLL samples at baseline and after BCR stimulation (supplemental Tables 2-4). We then reanalyzed these expression data using the same methods applied in the present study. At baseline, none of the RNA coding for the modulated proteins between M- and UM-CLL samples was statistically differentially modulated (supplemental Table 2). After BCR stimulation, only 1 RNA of 6 was modulated after BCR stimulation in M-CLL cells (supplemental Table 3), whereas 11 of 25 (44%) RNA were modulated in UM-CLL (supplemental Table 4). Kinetics of the various RNA and corresponding proteins were also variable. For some RNA, kinetics were identical (either up- or down-modulated), reflecting the potential involvement of these RNA and proteins in the genetic program induced by the BCR stimulation. For others, discordant kinetics between RNA and protein could potentially reflect more complicated regulation processes as already described.21
Discussion
Since initial studies of the proteome content of CLL in the 1970s,22 progress in proteomic techniques, and especially in 2D electrophoresis, have allowed proteomic profiles comparisons of different CLL subtypes, segregated by cytogenetic abnormalities23 or IGVH mutational status with ever-growing resolution.19,24 Voss et al demonstrated a correlation between large-scale protein expression profiles and clinical data.23 The shorter survival time of CLL patients with del(11q22-q23) or del(17p13), compared with patients with del(13q14), was shown to be associated with increased levels of heat-shock protein 27 and decreased levels of thioredoxin peroxidase 2 or protein disulfide isomerase.23 Of interest, these enzymes may be associated with altered drug resistance.23 Cochran et al, using 2D electrophoresis with silver staining, showed that nucleophosmin was translated in M-CLL cells only and that levels of F-actin-capping protein subunit, 14-3–3 protein and laminin-binding protein precursor were significantly increased in M-CLL but not UM-CLL.24 In 2005, Scielzo et al evidenced, using the same technology, that M- and UM-CLL patients differed in the expression of hematopoietic lineage cell-specific protein 1 (HCLS1).19 Most of the HCLS1 protein was constitutively phosphorylated in UM-CLL patients, although this was the case for only a fraction of M-CLL patients.19 HCLS1, a protein mainly expressed in hematopoietic cells, is tyrosine phosphorylated on IgM cross-linking in B cells and binds to several cytoskeleton proteins and adapters in both normal B cells and CLL cells. Recently, its central role in the regulation of B-CLL cell trafficking and homing has been demonstrated.25
To investigate other proteins possibly modulated on BCR cross-linking, we studied temporal modifications of proteomic profiles before and at different time points after anti-IgM BCR ligation in both ZAP70− M-CLL and ZAP70+ UM-CLL subsets using the novel 2D-DIGE technology. We chose a brief and soluble BCR stimulation, using biotinylated goat F(ab′)2 anti–human IgM, for setting a punctual T0 in all stimulated cells. It should be noted that, whereas sustained BCR signaling actually promotes the survival of B CLL cells, stimulation with soluble anti-IgM leads to the induction of apoptosis.26 Because of the choice of multiple comparisons (M- and UM-CLL, US and S, with 4 time points for each patient and each condition), this 2D-DIGE study could and did include only 3 patients in each subset (3 UM and 3 M-CLL). As time course analyses using SAM were too restrictive, with very few proteins under the 5% FDR threshold, results from statistical tests at each time point were used and grouped to generate the tables of differentially expressed proteins. Nevertheless, WB analyses of protein expression levels in a larger cohort of patients did validate the initial results.
Unsupervised PCA analysis showed the ability of this technology to distinguish M- and UM-CLL subsets using the global proteomic image at baseline, whereas gene expression profiling studies demonstrated that these 2 subtypes share a common gene expression profile, with a restricted number of genes differentially expressed.4,5 Among differentially expressed proteins between US M- and UM-CLL cells, HCLS1 was demonstrated to be present mostly in its phosphorylated (acid) form in UM-CLL cells, although its basic (unphosphorylated) form was predominant in M-CLL cells. This is consistent with the results of Scielzo et al.19 Another example is the down-regulation of PDCD4 in UM-CLL cells. PDCD4 is a tumor suppressor protein inhibiting neoplastic transformation, tumor progression, and translation; loss of its expression constitutes an independent prognostic factor of poor outcome in solid tumors, especially colorectal cancer.27 Other proteins constitutively differentially expressed between M- and UM-CLL cells are mainly involved in signal transduction, immune response regulation, protein metabolism, cell growth or maintenance, and apoptosis.
After stimulation, PCA allowed to segregate US and S samples only in the UM-CLL subset, and statistical analysis confirmed that UM-CLL cells display more significant changes of proteomic profiles after BCR activation (only 6 polypeptide spot ratios were significantly different between US and S samples in M-CLL cells compared with 25 spots in UM-CLL cells). This observation is consistent with the literature suggesting that UM-CLL cells are more sensitive to anti-IgM activation than M-CLL cells.7,8 One of the proteins of interest up-regulated after anti-IgM stimulation in UM-CLL cells is HNRPK, which plays a role in pre-mRNA processing and whose involvement in the BCR signaling pathway was recently described in a functional proteomic study in the B-cell line Ramos.28 Another protein involved downstream of the BCR pathway is LSP1, which was shown modified after BCR activation only in UM-CLL samples. LSP1 is an F-actin–binding cytoskeletal protein shown as being a protein kinase C substrate in B-CLL cells.20 It has been suggested that LSP1 could be overexpressed in CLL cells compared with normal B lymphocytes since protein kinase C-dependent phosphorylation of LSP1 is more prominent in CLL cells. Our results suggest that LSP1 is phosphorylated after anti-IgM stimulation only in the UM-CLL subset. The comparison of US and S proteomic profiles allowed also to demonstrate that the amount of the tumor suppressor PDCD4 decreased after anti-IgM stimulation in UM-CLL cells (and in M-responding CLL cells). It has been recently demonstrated that PDCD4 was negatively regulated by the microRNA miR-21.29 Interestingly, Fulci et al have shown that miR-21 was dramatically overexpressed in patients with CLL,30 and a recent study suggested that miR-21 could be involved in the establishment of fludarabine resistance in CLL.31 It could therefore be hypothesized that PDCD4 could also be regulated by miR-21 in UM but not M-CLL cells, therefore distinguishing the aggressiveness of these 2 subsets. Finally, the major changes between US and S UM-CLL samples interested RAD23B, a protein that plays an important role in both DNA repair and activation and function of p53.32 2D-DIGE experiments showed that the #929 isoform of RAD23B decreased after anti-IgM stimulation in UM-CLL cells, and WB validations revealed in parallel the emergence of a shorter isoform, suggesting a cleavage of RAD23B after stimulation, as previously described in a human Burkitt lymphoma cell line.33 Indeed, Brockstedt et al33 have shown that the stimulation of BL60 cells with an anti-IgM F(ab)2 induced modifications for 12 proteins called apoptosis-associated proteins (including RAD23B but also LSP1, HNRPA1, lamin, and actin). Cleavage of some of these proteins (not RAD23B) is moreover inhibited by a selective irreversible inhibitor of caspase 3.33 It could be hypothesized that the presence of a shorter isoform of RAD23B in stimulated UM-CLL samples in our WB experiments is also related to a cleavage of the full protein during anti-IgM–induced apoptosis.
Finally, this study hints to the existence of intermediate CLL stages as suggested by the perhaps serendipitous patient M3. What could have been indeed categorized as an erroneous observation was indeed confirmed in WB experiments, which identified among additional 13 patients, 2 further M-CLL cases with “aggressive” RAD23B and PDCD4 proteomic profiles, such as UM-CLL cells. Deglesne et al had also identified a group of clinically progressive CLL patients exhibiting mutated IGVH and lack of ZAP70 expression that responded to in vitro anti-IgM stimulation (19 of 38 M-CLL).13 Accordingly, all nonresponder cases were also IGVH mutated both in this study and in theirs. Therefore, if UM-CLL cells seem to all share the same aggressiveness, the M-CLL subset appears to be more heterogeneous, with a significant number of cases displaying biologic characteristics suggestive of a possible progression to cell-activation pathways usually used by UM-CLL alone. The differentiating proteins observed here therefore deserve further exploration in this context.
In conclusion, the 2D-DIGE proteomic approach was validated here as a method to study proteome modifications after anti-IgM stimulation of CLL cells. The validity of proteomic observations in a necessarily limited series of samples was confirmed on a larger cohort of patients by WB experiments for 2 proteins: PDCD4 and RAD23B. Moreover, this technology allowed to investigate posttranslational modifications, such as phosphorylation after cell stimulation. Most importantly, this study brings out that proteomic profiles of M-CLL and UM-CLL cells, somewhat distinct at baseline segregate even more after BCR ligation, proteomic patterns changes after stimulation being selectively seen in aggressive ZAP70+ UM-CLL samples. Further studies are nevertheless needed to better identify possible targets for diagnosis/prognosis and/or therapeutic targets and explore what appear to be novel intermediate M-CLL stages.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Manuel Chapelle (Plateforme Protéomique/Spectrométrie de masse, Institut Jacques Monod, Paris, France) for allowing us to use the EXQuest spot cutter; Estelle Zink (Hématologie biologique, Strasbourg, France), Hervé Roudot, Fanny Baran-Marszak, and Florence Cymbalista (Hématologie biologique, Hôpital Avicenne, Bobigny, France) for IGVH status assessment of the CLL cells used for WB validations; Béatrice Uring-Lambert (Immunologie cellulaire, Strasbourg, France) for help in flow cytometry; and Laurent Miguet (Hématologie biologique, Strasbourg, France), Ali Dalloul (EA RHEM, Faculté de Médecine, Nancy), Iozo Delic (CEA, Fontenay aux Roses), and Christine Carapito (Laboratoire de Spectrométrie de Masse Bio-Organique, Institut Pluridisciplinaire Hubert Curien, Strasbourg, France) for critical reading of the manuscript and helpful suggestions.
This work was supported by the Ligue régionale Alsace contre le Cancer, the Association pour la Recherche contre le Cancer, Fondation pour la Recherche Médicale (FRM) and Agence Nationale pour la Recherche. A.P. was supported by a fellowship from the Fondation pour la Recherche Médicale.
Authorship
Contribution: A.P. performed experiments, analyzed data, and wrote the initial version of the manuscript; C.P. designed the 2D-DIGE study, performed experiments (especially with regards to mass spectrometry), analyzed data, and reviewed the manuscript; S.N. performed specific statistical calculations; F.D. and H.M.-B. performed the IGVH status of the CLL cells used for 2D-DIGE experiments and critically reviewed the manuscript; V.L. and R.H. included CLL patients and critically reviewed the manuscript; F.J. performed a 2D preliminary study using silver staining; M.-C.B. critically reviewed the data and the manuscript; L.V. designed and supervised the study; L.V. and S.B. analyzed data and critically reviewed and wrote parts of the manuscript; and J.G.G. made contributions in the initial design of the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Seiamak Bahram, Laboratoire d'Immunogénétique Moléculaire Humaine, Centre de Recherche d'Immunologie et d'Hématologie, 4 rue Kirschleger, 67085 Strasbourg Cedex, France; e-mail: siamak@unistra.fr.