Abstract
Subjects affected by Signal Transducer and Activator of Transcription 1 (STAT1) deficiency have lethal bacterial and viral infections. Complete STAT1 deficiency is inherited as an autosomal recessive disease; partial STAT1 deficiency is inherited as an autosomal recessive or autosomal dominant trait. Here, we report a patient who developed disseminated mycobacteriosis early in life and had several viral infections, including herpetic skin infection and interstitial pneumonia by cytomegalovirus with severe respiratory distress. Molecular analysis of STAT1 showed a novel homozygous mutation affecting a splice site, leading to exon 3 skipping and to synthesis of a lower molecular weight STAT1 protein. This mutation leads to marked reduction of STAT1 phosphorylation; the electromobility shift assay showed a complete defect of DNA-binding activity, which accounts for the complete impairment of peripheral blood mononuclear cell functional response to both IFN-γ and IFN-α. Moreover, analysis of natural killer cells showed a defective STAT1 phosphorylation in response to IFN-α and impaired basal cytolytic activity, suggesting that the STAT1-dependent pathway might be important for natural killer cell function. These results suggested that exon 3 skipping of STAT1 leads to abnormal signaling in response to IFN-γ and IFN-α, which is associated with susceptibility to intracellular pathogens and viruses.
Introduction
Mendelian predisposition to mycobacterial diseases is characterized by the occurrence of clinically evident infectious episodes sustained by weakly virulent mycobacteria or salmonellae in patients with no other infections. These inherited conditions are caused by germline mutations affecting genes involved in the IFN-γ and IL-12 signaling pathways.1,2 However, complete Signal Transducer and Activator of Transcription 1 (STAT1) deficiency is more severe than other deficiencies in IFN-γ receptor 1 (IFN-γR1) or IFN-γ receptor 2 (IFN-γR2) because STAT1 signaling is common to many transduction pathways and affects not only IFN-γ.3-5 Indeed, STAT1 is also required for the cellular response to many cytokines with antiviral activities, such as IFN-α/β/λ1 or cytokines with immunomodulatory functions on natural killer (NK) and B cells, such as IL-15, IL-21, and IL-27.6-8 After type I IFN stimulation, the receptor is phosphorylated by Janus kinases JAK1 and TYK2, leading to subsequent docking and phosphorylation of both STAT1 and STAT2. Active STAT1/STAT2 heterodimers are released into the cytosol and translocate to the nucleus, where they bind to type I IFN-stimulated response elements (ISREs) in the promoter of target genes.9,10 Stimulation with IFN-γ leads to activation of the Janus kinase JAK1 and JAK2, which create a docking site for 2 STAT1 molecules, which are phosphorylated and released into the cytosol as active STAT1 homodimers. The active complex translocates to the nucleus where it binds to the IFN-γ response region (GRR).9,11,12
Complete STAT1 deficiency, inherited as an autosomal recessive trait, is characterized by complete lack of STAT1 expression and abolition of STAT1-dependent responses to both IFN-γ and IFN-α. These subjects display profound susceptibility to infections with intracellular pathogens and viruses, including life-threatening infections caused by herpes simplex virus, cytomegalovirus (CMV), and Epstein-Barr virus (EBV).3,4
Partial STAT1 deficiency is inherited as an autosomal recessive or autosomal dominant disease.13-16 Patients with heterozygous mutations of STAT1, unlike patients who are homozygous, display an impaired tyrosine phosphorylation or DNA-binding activity in response to IFN-γ but retain a normal or partial response to IFN-α and are not susceptible to viral disease.13,15,17
We describe here a patient with homozygous STAT1 splicing mutation leading to skipping of exon 3 who developed generalized mycobacterial infections and severe viral disease sustained by CMV. We report that the patient's cells display a complete defect of STAT1 DNA-binding activity after stimulation with IFN-γ and IFN-α and fail to respond at even high doses of these cytokines. These biologic defects are associated with a partial impairment of NK functional activity, which might contribute to the susceptibility of the patient to infections with viral and intracellular pathogens.
Methods
Patients
The study was approved by the Institutional Review Board of Spedali Civili, Brescia, and informed consent was obtained from the parents of the children. Blood samples, with EDTA or heparin, were collected from the STAT1-deficient patient, his parents, and the healthy controls in accordance with the Declaration of Helsinki.
PBMCs and cell cultures
Peripheral blood mononuclear cells (PBMCs) were obtained from heparinized blood by density gradient centrifugation over Ficoll-Hypaque 1070 (Sigma-Aldrich). PBMCs were resuspended in RPMI 1640 medium, supplemented with 2mM glutamine, 50 μg/mL penicillin, 50 μg/mL streptomycin, and 10% heat-inactivated FCS (Euroclone) and cultured in the presence of various stimuli at the time indicated in “Results.” Phytohemagglutinin-activated T cells (PHA-T) blasts were obtained from PBMCs in presence of rIL-2 (600 U/mL) and PHA (5 μg/mL) and maintained in RPMI 1640 medium with rIL-2 (1200 U/mL). NK cells were purified by NK Cell Separation Cocktails (Rosette Sep; StemCell Technologies). The purity of the NK cells was > 96% as assessed by flow cytometry using CD56-PC5 and CD3-fluorescein isothiocyanate (Beckman Coulter; Immunotech). CD3 contamination in purified NK cells was < 1%. PBMCs (exposed or unexposed to rIL-2) and polyclonal rIL-2-activated NK cells were tested for cytolytic activity against various NK-susceptible tumor target cells, including K562 and LCL 721.221, in a 4-hour 51Cr-release assay as previously described.18,19 The effector/target ratios are indicated in the figure legends. Monocyte cells were obtained from venous blood samples; briefly, PBMCs were seeded at 5 × 105 cells/well, in RPMI 1640 medium, supplemented with 10% FCS. After 2 hours at 37°C, nonadherent cells were removed. The number of monocytes has been standardized on the basis of blood counts. B-LCLs were obtained by EBV-transformation of PBMCs from healthy control and patient and cultured in the presence of PHA 15 μg/mL (Sigma-Aldrich). STAT1-deficient U3C cells (kindly provided by Sandra Pellegrini, Pasteur Institute, Paris, France) were cultured in DMEM (Lonza Verviers) supplemented with 2mM glutamine, 50 μg/mL penicillin, 50 μg/mL streptomycin, and 10% heat-inactivated FCS.
Cytokine, ELISA assay, and flow cytometry
Cells were stimulated with the indicated doses of IFN-α, IFN-β, IFN-γ, IL-12, IL-15, IL-18 (PeproTech), IL-27 (R&D Systems), lipopolysaccharide (LPS), PHA (Sigma-Aldrich), IL-2 (Proleukin; Novartis), anti-CD3 monoclonal antibody (mAb), and anti-CD28 mAb (BD Biosciences). Cytokine concentrations in the supernatants were measured by a sandwich ELISA, using the human IFN-γ or TNF-α Screening Set (Pierce Chemical), according to the manufacturer's instructions. Each sample was tested in duplicate. Flow cytometry was performed as previously described.20 Briefly, 100 μL of whole blood or isolated cells (1.5 × 106) resuspended in 100 μL of the appropriate medium was incubated with anti-STAT1 (C-terminus) mAb (5 μL) and anti-CD3-fluorescein isothiocyanate mAb from BD Transduction Laboratories. For the phosphorylation studies, the samples were incubated at the concentrations reported in the figure legends with IFN-α or IL-15 for 20 minutes and with IL-2 or IL-15 for 12 minutes at 37°C. Intracellular staining of activated STAT protein was performed with anti-phospho-STAT1 (Y701)–phycoerythrin (PE), anti-phospho-STAT4 (Y693)–PE, anti-phospho-STAT5 (Y694)-PE, anti-CD3–fluorescein isothiocyanate, or isotype-matched mAb PE (BD Biosciences) following the manufacturer's instructions. For intracellular analysis of IFN-γ, NK cells were preincubated for 16 hours with the cytokines indicated in “Results”; cells were then stimulated with target cell line K562 and stained with 5 μg/mL PE-conjugated anti–IFN-γ for 30 minutes at 4°C. The percentage of CXCR1+ cells was evaluated on NK cells, which were gated as CD14−CD3−CD20− (lineage) and CD56+ cells. Cells were acquired using a FACSCalibur (BD Biosciences) and analyzed by the FlowJo Version 7.5 software (TreeStar).
DNA and RNA extraction, PCR sequencing, and transfection
For genetic analysis of the STAT1 gene, genomic DNA was isolated from whole blood using ABI PRISM 6100 Nucleic Acid PrepStation (Applied Biosystems) as indicated by the manufacturer. The STAT1 gene was amplified by PCR using 24 paired primers (conditions and primers available on request). PCR products were sequenced using the BigDye Terminator Kit (Applied Biosystems), and the sequences were analyzed on a 3130 Genetic Analyzer (Applied Biosystems). Total RNA was extracted from PBMCs using the RNeasy Mini Kit (QIAGEN) according to the manufacturer's instructions.
For reverse transcription (RT)-PCR experiments, 1 μg of DNase-treated total RNA was used to synthesize the first strand of cDNA by the GeneAmpRNA PCR kit (Applied Biosystems). Primers spanning from exon 1 to exon 5 were used to amplify the postulated deletion. For the RT-PCR analysis, Assays-on-Demand Products and TaqMan Master Mix from Applied Biosystems were used according to the instruction manual to analyze STAT1, STAT1Δ3, IL-6, CXCL10, IDO, TNF-[alpha], MXA, IFN-[gamma], TBX21, PIM1, c-MYC, GAPDH, EBNA1, and B2M gene expression. The eGFP STAT1 WT vector (Addgene) was provided by Alan Perantoni from the National Cancer Institute; the mutated eGFP STAT1Δ3 vector was obtained with the Phusion Site-Directed Mutagenesis kit (Finnzymes), following the manufacturer's instructions. U3C cells were transfected with eGFP STAT1 WT or eGFP STAT1Δ3 vectors or an insert-less vector (mock), using the Amaxa Nucleofector Technology (Lonza Verviers) with the SG Cell Line 4D-Nucleofector kit, according to the manufacturer's instructions.
Cytoplasmic and nuclear extracts preparation, Western blot, and electromobility shift assay
After stimulation with the indicated doses of IFN-γ, IFN-α, or IFN-β for 30 minutes, cells were lysed in cold buffer (150mM NaCl, 50mM Tris-HCl, pH 7.4, 1% nonyl phenoxypolyethoxylethanol (NP40), and protease inhibitors (Roche Diagnostics) containing 0.2 μg of aprotinin, leupeptin, and 1mM of sodium orthovanadate on ice for 15 minutes. Immunoprecipitation was performed according to instructions provided by the manufacturer. For the Western blot analysis, the cytoplasmic cell extracts were resolved on 8% polyacrylamide and subjected to immunoblots by standard procedures. Nitrocellulose membranes were first blocked for 1 hour at room temperature in Tris-buffered saline/Tween-20 containing 5% BSA and then incubated overnight at 4°C in the presence of specific primary Abs (pSTAT1, STAT1, pSTAT3, and STAT3) in the Tris-buffered saline/Tween-20 containing 0.5% BSA. Antibodies against phosphotyrosine STAT1, phosphotyrosine STAT3, and total STAT3 were purchased from Cell Signaling Technologies. Antibodies against total STAT1 (sc-346) were obtained from Santa Cruz Biotechnology, and antibodies against β-actin (A5060) were obtained from Sigma-Aldrich. Detection was carried out using horseradish peroxidase-conjugated anti–mouse or anti–rabbit IgG (GE Healthcare) and revealed using the enhanced chemio luminescance (ECL) system (GE Healthcare) according to the manufacturer's instructions.
For electromobility shift assay, after stimulation with the indicated doses of IFN-γ and/or IFN-α for 30 minutes, EBV-transformed B cells (5 × 106/condition) or STAT1wt- and STAT1Δ3-transfected U3C (2 × 106/condition) were diluted in ice-cold PBS and centrifuged twice at 300g for 5 minutes at 4°C. Nuclear extracts were prepared using a modification of the method of Dignam et al.21 Transcription factor-binding analyses were performed as described previously.22 Nuclear extracts were incubated in binding buffer in the presence of the labeled oligonucleotide STAT-binding probe from the GRR element located within the promoter of the FcγRI/CD64 gene (5-CTTTTCTGGGAAATACATCTCAAATCCTTGAAACATGCT-3) or from the ISRE element (5-GATCGGGAAAGGGAAACCGAAACTGAA-3).
Statistical analysis
All experiments were performed at least 2 times. For comparison among multiple groups, ANOVA and the Bonferroni correction test were used. The results of the statistical analysis of a single experiment are shown in the figures and reported in the figure legends, and P values < .05 were considered significant.
Results
We report a case of a 3-year-old child born to consanguineous parents of Pakistani origin living in Italy (Figure 1A). The child showed normal growth without episodes of severe infections until 10 months of age. At that time, he developed a pulmonary mycobacterial infection caused by Mycobacterium kansasii (isolated from peripheral blood). At the same time, Aspergillus fumigatus antigen was identified in the blood, but infection could not be proven. Therefore, the child was treated with antimycobacterial drugs for 9 months. After minor episodes of infection, at 17 months of age the child presented a pulmonary infection of unknown etiology that was associated with severe respiratory distress. At 23 months, the child developed an additional episode of respiratory failure resulting from interstitial pneumonia; CMV was identified in the bronchoalveolar lavage (> 100 000 copies/mL). After recovering, the child traveled to Pakistan where he again had pneumonia. At 25 months, the child presented with sepsis, disseminated intravascular coagulopathy, and seizures that were successfully treated. At 31 months of age, a relapse of pulmonary CMV infection was observed and treated with ganciclovir and anti-CMV antibodies. Because of the recurrent infections and the genetic diagnosis of STAT1 deficiency, the child was subjected to bone marrow transplantation (Table 1).
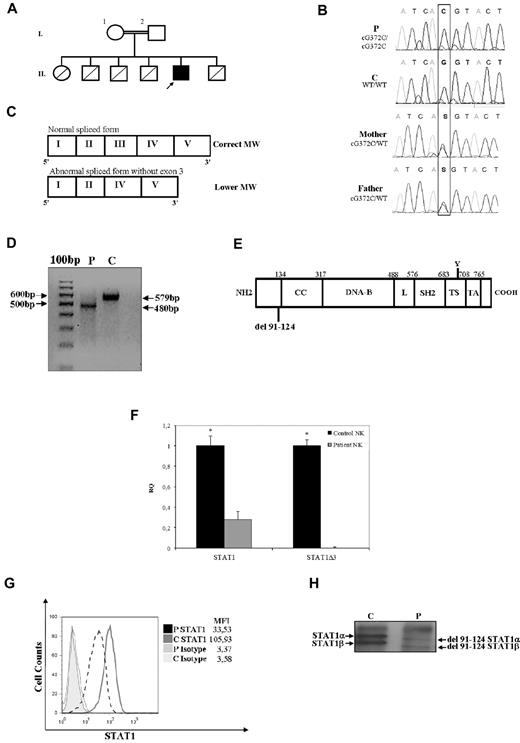
Novel STAT1 mutation associated with abnormal mRNA splicing and protein expression. (A) Family pedigree. Subjects I.1 and I.2 are first cousins. The proband is indicated with an arrow. Healthy persons are shown in white; deceased siblings are indicated by line crossing. (B) Genomic sequence analysis of exon 3 showing a homozygous nucleotide substitution at position 372 (G > C) affecting the donor splicing site. Both parents are heterozygous for the same mutation. (C) Schematic diagram of the first 5 exons of the STAT1 mRNA patient. Coding exons are indicated with Roman numerals and delineated by vertical bars. The upper band corresponds to the wild-type form. The lower band was observed for the cells of the patient and corresponds to the form of STAT1 mRNA lacking exon 3. (D) RT-PCR of STAT1 fragment spanning from exon 1 to exon 5 shows a lower size fragment in the patient. (E) Schematic representation of STAT1 protein structure, including the position of the 33 amino acid residue deletion (Δ91-124) in the N-terminal domain as a result of the G372C mutation affecting the donor splicing site of exon 3. Other domains included the coiled-coil (CC), DNA-binding (DNA-B), linker (L), Src-homology 2 (SH2), tail segment (TS), and transactivation (TA) domains. The position of Tyr701 (Y) is shown. (F) Expression of STAT1 and STAT1Δ3 mRNAs in NK cells from control and patient. Target gene expression was normalized against the housekeeping gene (GAPDH) and presented as n-fold increase over the expression in the healthy control. The experiments shown are representative of 4 independent experiments. Data are mean ± SE. *Significant difference in the response between the patient and control subject (P < .05). (G) Flow cytometry analysis of STAT1 expression in T cells (CD3+ cells), as measured by intracellular staining with anti-STAT1 mAb, was performed as described in “Cytokine, ELISA assay, and flow cytometry.” Mean fluorescence intensity is indicated. The results are representative of 4 independent experiments. (H) Immunoprecipitation and Western blot analysis of STAT1α and STAT1β were performed using an antibody against STAT1 on PBMCs (30 μg of total lysates) from the patient and a control subject. Analysis shows that the 2 bands corresponding to STAT1α and STAT1β were less abundant and at a reduced molecular weight.
Novel STAT1 mutation associated with abnormal mRNA splicing and protein expression. (A) Family pedigree. Subjects I.1 and I.2 are first cousins. The proband is indicated with an arrow. Healthy persons are shown in white; deceased siblings are indicated by line crossing. (B) Genomic sequence analysis of exon 3 showing a homozygous nucleotide substitution at position 372 (G > C) affecting the donor splicing site. Both parents are heterozygous for the same mutation. (C) Schematic diagram of the first 5 exons of the STAT1 mRNA patient. Coding exons are indicated with Roman numerals and delineated by vertical bars. The upper band corresponds to the wild-type form. The lower band was observed for the cells of the patient and corresponds to the form of STAT1 mRNA lacking exon 3. (D) RT-PCR of STAT1 fragment spanning from exon 1 to exon 5 shows a lower size fragment in the patient. (E) Schematic representation of STAT1 protein structure, including the position of the 33 amino acid residue deletion (Δ91-124) in the N-terminal domain as a result of the G372C mutation affecting the donor splicing site of exon 3. Other domains included the coiled-coil (CC), DNA-binding (DNA-B), linker (L), Src-homology 2 (SH2), tail segment (TS), and transactivation (TA) domains. The position of Tyr701 (Y) is shown. (F) Expression of STAT1 and STAT1Δ3 mRNAs in NK cells from control and patient. Target gene expression was normalized against the housekeeping gene (GAPDH) and presented as n-fold increase over the expression in the healthy control. The experiments shown are representative of 4 independent experiments. Data are mean ± SE. *Significant difference in the response between the patient and control subject (P < .05). (G) Flow cytometry analysis of STAT1 expression in T cells (CD3+ cells), as measured by intracellular staining with anti-STAT1 mAb, was performed as described in “Cytokine, ELISA assay, and flow cytometry.” Mean fluorescence intensity is indicated. The results are representative of 4 independent experiments. (H) Immunoprecipitation and Western blot analysis of STAT1α and STAT1β were performed using an antibody against STAT1 on PBMCs (30 μg of total lysates) from the patient and a control subject. Analysis shows that the 2 bands corresponding to STAT1α and STAT1β were less abundant and at a reduced molecular weight.
The susceptibility of the child to mycobacterial and viral infections suggested a defect in the IFN-γ/IFN-α pathway. Because the association of herpetic infections and the susceptibility to intracellular pathogens has been observed in patients with complete STAT1 deficiency,3,4 we sequenced the exons and the flanking intronic regions of STAT1 in the genomic DNA from the patient. Molecular analysis of the STAT1 gene in the patient showed a homozygous nucleotide substitution at position 372 affecting the donor splicing site of exon 3 (G→C; Figure 1B). His healthy parents were heterozygous for the same mutation, suggesting an autosomal recessive pattern of inheritance in this child (Figure 1B). To define the effects of the genomic mutation on mRNA synthesis, we conducted RT-PCR with primers spanning from exon 1 to exon 5 on the RNA derived from the patient and healthy control persons. Analysis of the PCR product obtained from the patient showed a smaller size fragment, suggesting skipping of exon 3 (Figure 1C-D). Direct sequencing of cDNA revealed an in-frame deletion of exon 3, which might lead to the synthesis of a protein lacking 33 amino acids (Figure 1E). To exclude any residual splicing activity of exon 3, we analyzed STAT1 mRNA expression by real-time PCR using 2 different probes: one probe was designed to recognize exon 3 sequence, whereas the second one was specific for exon 10. Real-time PCR showed no amplification of exon 3 when the specific probe was used for analysis of STAT1 mRNA in the patient, whereas a residual expression of the mRNA was observed in the cells of patient when a probe targeting the exon 10 was used (Figure 1F).
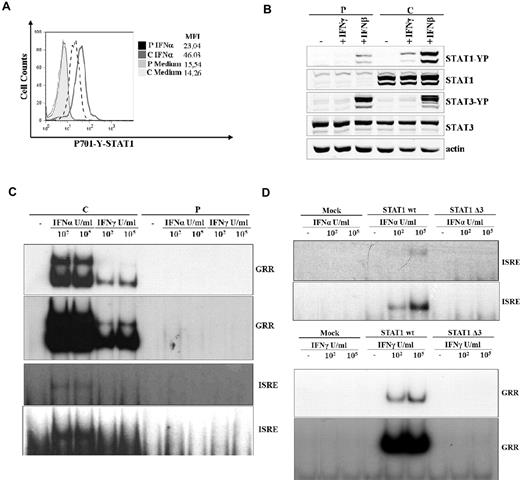
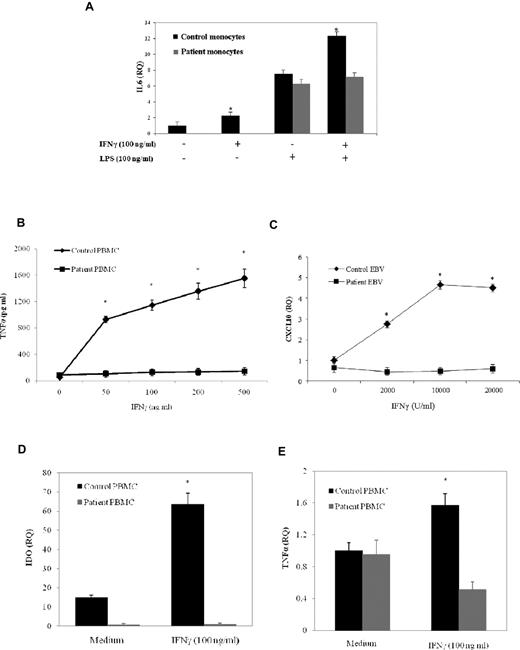
To address the effects of this mutation, we analyzed the expression level and the molecular weight of STAT1. Analysis of STAT1 expression by flow cytometry showed a reduction up to 70% of protein levels in T lymphocytes from the patient compared with those from the healthy control (Figure 1G). Immunoprecipitation and Western blot analysis of STAT1 in PBMCs from the patient showed that both the α- and β-isoforms of STAT1 were detectable. However, both isoforms displayed a lower molecular weight than the wild-type equivalent forms, and this was the expected observation of the exon 3 deletion (Figure 1H). Because STAT1 activation in response to receptor engagement is strictly associated with the phosphorylation of tyrosine-701 (Tyr-701), we assessed STAT1 activation after stimulation with IFN-α using a specific antibody directed against phosphorylated tyrosine (P-Tyr-701-STAT1). T cells from the patient displayed approximately 50% of the normal amount of P-Tyr-701-STAT1 after treatment with the cytokine, whereas control cells displayed normal phosphorylation (Figure 2A). Next, we performed a Western blot analysis of STAT1 phosphorylation using a specific anti–P-Tyr701-STAT1 antibody in EBV-transformed B cells from the patient and a control subject after stimulation with IFN-γ or IFN-β. In patient cells, we observed a dramatic reduction of STAT1 phosphorylation in response to both stimuli, which affected both isoforms (Figure 2B), whereas STAT3 phosphorylation was normal. This suggested that the STAT1 gene splicing mutation results in impaired tyrosine phosphorylation of STAT1 in response to both IFN-γ and IFN-α/β. The phosphorylation of STAT1 triggers its translocation into the nucleus and the binding to consensus promoter sequences. To address the effects of the splicing mutation on DNA-binding activity, we analyzed the electromobility shift activity of the nuclear extracts derived from EBV-transformed B cells from the patient and a control subject (Figure 2C) and from the STAT1-deficient cell line U3C after transfection with STAT1wt or STAT1Δ3 (Figure 2D). Normal STAT1 expression was detected in transfected cells by flow cytometry (data not shown). Cells were then treated with or without IFN-α or IFN-γ in the presence of the 32P-labeled oligonucleotide STAT-binding probe derived from the IFN-γ response region (GRR) or type I ISREs. In the presence of both probes, we observed that treatment with IFNs failed to induce STAT1 binding activity, even at high doses, both in EBV cells from the patient and in STAT1Δ3-transfected U3C cells, even after long exposures. Normal STAT1-binding activity was detected in the cells of the control subject and in STAT1wt-transfected U3C cells (Figure 2C-D). Monocyte cells are responsive to IFN-γ after microbial infection and are the major producer of TNF-α. To investigate a possible defect in response to IFN-γ, we evaluated the functional response of monocytes and PBMCs to IFN-γ by assessing IL-6 mRNA induction and TNF-α production, respectively. We observed that IL-6 mRNA was not induced in response to IFN-γ but still detectable after LPS stimulation in monocytes of the STAT1-deficient patient (Figure 3A). In addition, the production of TNF-α in response to IFN-γ was markedly reduced at any dose of IFN-γ in the PBMCs from the patient, whereas cells from the healthy control subject showed a dose-dependent release of TNF-α (Figure 3B). Even high doses of IFN-γ failed to induce CXCL10 in EBV-transformed B cells from the patient, suggesting a complete defect of response to IFN-γ in STAT1-deficient (Figure 3C).
G372C mutation is associated with abnormal phosphorylation and DNA binding activity. (A) Flow cytometric analysis of STAT1 phosphorylation (Y701) of T cells (CD3+) after treatment with IFN-α (40 U/μL) or medium alone for 20 minutes at 37°C. Mean fluorescence intensity is indicated. The results are representative of 4 independent experiments. (B) EBV-transformed B cells derived from the patient and a control subject were cultured for 30 minutes with IFN-γ (100 ng/mL), IFN-β (100 ng/mL), or medium alone to analyze STAT1 and STAT3 activation by Western blot. Western blot was carried out with an antibody against Tyr701-phosphorylated STAT1, Tyr705 phosphorylated STAT3, STAT1, STAT3, and actin as a reference. (C-D) DNA-binding activity was analyzed by the electromobility shift activity assay on nuclear extract and carried out in the presence of the 32P-labeled oligonucleotide STAT-binding probe derived from the IFN-γ response region (GRR) and type I ISRE. EBV-transformed B cells from patient and a healthy subject were stimulated with the indicated doses of IFN-α and IFN-γ for 30 minutes. For the GRR and ISRE probe, the top panel and the bottom panel show 2 exposures for short and long times, respectively. (C) The U3C cell line was transfected with mock, STAT1 wt, and STAT1Δ3 mutant alleles and stimulated with the indicated doses of IFN-α and IFN-γ for 30 minutes. For the GRR and ISRE probe, the top panel and the bottom panel show 2 exposures for short and long times, respectively (D).
G372C mutation is associated with abnormal phosphorylation and DNA binding activity. (A) Flow cytometric analysis of STAT1 phosphorylation (Y701) of T cells (CD3+) after treatment with IFN-α (40 U/μL) or medium alone for 20 minutes at 37°C. Mean fluorescence intensity is indicated. The results are representative of 4 independent experiments. (B) EBV-transformed B cells derived from the patient and a control subject were cultured for 30 minutes with IFN-γ (100 ng/mL), IFN-β (100 ng/mL), or medium alone to analyze STAT1 and STAT3 activation by Western blot. Western blot was carried out with an antibody against Tyr701-phosphorylated STAT1, Tyr705 phosphorylated STAT3, STAT1, STAT3, and actin as a reference. (C-D) DNA-binding activity was analyzed by the electromobility shift activity assay on nuclear extract and carried out in the presence of the 32P-labeled oligonucleotide STAT-binding probe derived from the IFN-γ response region (GRR) and type I ISRE. EBV-transformed B cells from patient and a healthy subject were stimulated with the indicated doses of IFN-α and IFN-γ for 30 minutes. For the GRR and ISRE probe, the top panel and the bottom panel show 2 exposures for short and long times, respectively. (C) The U3C cell line was transfected with mock, STAT1 wt, and STAT1Δ3 mutant alleles and stimulated with the indicated doses of IFN-α and IFN-γ for 30 minutes. For the GRR and ISRE probe, the top panel and the bottom panel show 2 exposures for short and long times, respectively (D).
Impaired IFN-γ response in a STAT1-deficient patient. (A) Monocytes of a patient and a healthy subject were stimulated in vitro for 48 hours with IFN-γ (100 ng/mL), LPS (100 ng/mL), and with a combination of IFN-γ and LPS or medium alone. Target gene expression was normalized to the housekeeping gene (GAPDH) expression and presented as n-fold of the expression in unstimulated cells from the healthy control. The experiments shown are representative of 3 independent experiments. Data are mean ± SE. *Significant difference in the response between the patient and the control subject (P < .05). (B) PBMCs (2 × 106/mL) from the patient or a healthy control were stimulated with IFN-γ (50, 100, 200, 500 ng/mL) for 48 hours. The production of TNF-α in the supernatant was determined by ELISA. (C) EBV-transformed B cells from the patient and a healthy subject were stimulated in vitro with the indicated doses of IFN-γ for 24 hours. Target gene expression was normalized against the housekeeping gene (EBNA1) and presented as n-fold increase over the expression in unstimulated cells from the healthy control. (D-E) PBMCs from the patient and a healthy subject were cultured with RPMI 1640 supplemented with 10% FBS and stimulated in vitro for 48 hours (D) or for 24 hours with IFN-γ (100 ng/mL; E). Target gene expression was normalized to the housekeeping gene (β2M) expression and presented as n-fold of the expression in unstimulated cells from the healthy control. The experiments shown are representative of 4 independent experiments. Data are mean ± SE. *Significant difference in the response between the patient and control subject (P < .05).
Impaired IFN-γ response in a STAT1-deficient patient. (A) Monocytes of a patient and a healthy subject were stimulated in vitro for 48 hours with IFN-γ (100 ng/mL), LPS (100 ng/mL), and with a combination of IFN-γ and LPS or medium alone. Target gene expression was normalized to the housekeeping gene (GAPDH) expression and presented as n-fold of the expression in unstimulated cells from the healthy control. The experiments shown are representative of 3 independent experiments. Data are mean ± SE. *Significant difference in the response between the patient and the control subject (P < .05). (B) PBMCs (2 × 106/mL) from the patient or a healthy control were stimulated with IFN-γ (50, 100, 200, 500 ng/mL) for 48 hours. The production of TNF-α in the supernatant was determined by ELISA. (C) EBV-transformed B cells from the patient and a healthy subject were stimulated in vitro with the indicated doses of IFN-γ for 24 hours. Target gene expression was normalized against the housekeeping gene (EBNA1) and presented as n-fold increase over the expression in unstimulated cells from the healthy control. (D-E) PBMCs from the patient and a healthy subject were cultured with RPMI 1640 supplemented with 10% FBS and stimulated in vitro for 48 hours (D) or for 24 hours with IFN-γ (100 ng/mL; E). Target gene expression was normalized to the housekeeping gene (β2M) expression and presented as n-fold of the expression in unstimulated cells from the healthy control. The experiments shown are representative of 4 independent experiments. Data are mean ± SE. *Significant difference in the response between the patient and control subject (P < .05).
Next, we investigated the expression of genes induced by IFN-γ.23,24 As shown in Figure 3D and E, we observed a complete impairment in the induction of IDO and TNF-α mRNAs expression on stimulation by IFN-γ (100 ng/mL) in patient PBMCs. These results suggested a severe defect of IFN-γ signaling pathway driven by STAT1 in our patient.
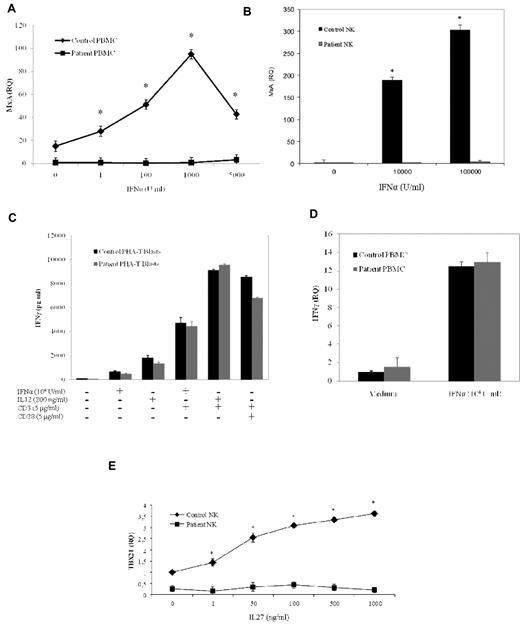
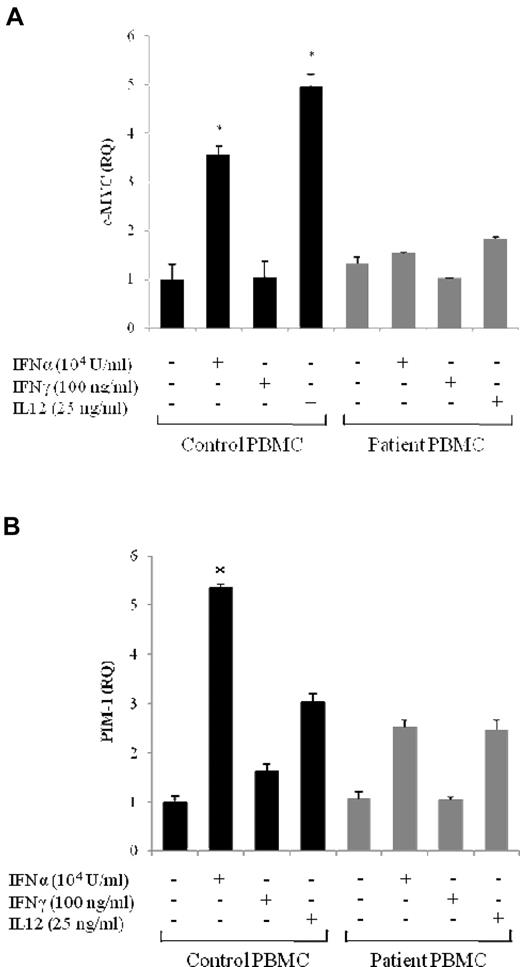
Then, we analyzed IFN-α response in STAT1-deficient cells by evaluating the induction of the antiviral factor MXA. As shown in Figure 4A, we observed a complete defect in the induction of MXA mRNA in patient PBMCs. To exclude any residual response to IFN-α, we have also assessed the up-regulation of MXA in NK cells after stimulation with high doses of the cytokine (up to 105 U/mL). At the highest dose of IFN-α, we could not detect any up-regulation of MXA mRNA in cells from the patient, whereas a strong induction was observed in NK cells from the control subject (Figure 4B). In addition, we analyzed IFN-α and IL-12 responses by analyzing the up-regulation and production of IFN-γ in PBMCs and PHA-activated T lymphocytes, respectively. We found that IFN-γ expression and production in response to both IL-12 and IFN-α were conserved in STAT1-deficient cells (Figure 4C-D). To characterize in more details the biologic effects of STAT1 mutation, we investigated the response of NK cells to IL-27 because IL-27 biologic activity requires STAT1 activation.6 Analysis of TBX21 induction in NK cells treated with increasing concentrations of IL-27 showed a complete impairment of biologic response at any dose tested, whereas cells from the healthy control subject showed a dose-dependent induction of TBX21 (Figure 4E). To determine whether STAT1-mediated gene expression was impaired in cells of the patient, we performed a real-time PCR, which evaluated the expression of genes induced either by IFN-α or IFN-γ. Analysis of c-MYC and PIM-1 mRNA levels showed reduced expression of the early genes in the patient's PBMCs stimulated with IFN-α, IFN-γ, or IL-12 compared with the gene expression in a healthy control (Figure 5).
Impaired IFN-α and IL-27 response in a STAT1-deficient patient. (A-B) PBMCs and NK cells from the patient and a healthy subject were stimulated in vitro for 24 hours with increasing amounts of IFN-α (1, 5, 100, 1000, 5000, 10 000, and 100 000 U/mL). Target gene expression was normalized against the housekeeping gene (β-2-microglobulin and GAPDH for PBMC and NK cells, respectively) and presented as n-fold increase over the expression in unstimulated cells from the healthy control. The experiments shown are representative of 4 independent experiments. Data are mean ± SE. *Significant difference in the response between the patient and control subject (P < .05). (C) PHA-T (2 × 106/mL) were stimulated with IL-12 (200 ng/mL) or IFN-α (104 U/mL) in the presence or the absence of anti-CD3 mAb (5 μg/mL) or with a combination of anti-CD3 and anti-CD28 mAb (5 μg/mL) for 24 hours in RPMI 1640 supplemented with IL-2 (1200 U/mL). Supernatants were collected after stimulation, and IFN-γ levels were evaluated by ELISA. (D) PBMCs from the patient and a healthy subject stimulated in vitro for 24 hours with IFN-α (104 U/mL). Target gene expression was normalized to the housekeeping gene (β2M) expression and presented as n-fold of the expression in unstimulated cells from the healthy control. The experiments shown are representative of 4 independent experiments. Data are mean ± SE. *Significant difference in the response between the patient and control subject (P < .05). (E) NK cells from the patient and a healthy subject were cultured without IL-2 for 16 hours and then stimulated in vitro with the indicated doses of IL-27 for 24 hours. Target gene expression was normalized against the housekeeping gene (GAPDH) and presented as n-fold increase over the expression in unstimulated cells from the healthy control subject. The experiments shown are representative of 4 independent experiments. Data are mean ± SE. *Significant difference in the response between the patient and control subject (P < .05).
Impaired IFN-α and IL-27 response in a STAT1-deficient patient. (A-B) PBMCs and NK cells from the patient and a healthy subject were stimulated in vitro for 24 hours with increasing amounts of IFN-α (1, 5, 100, 1000, 5000, 10 000, and 100 000 U/mL). Target gene expression was normalized against the housekeeping gene (β-2-microglobulin and GAPDH for PBMC and NK cells, respectively) and presented as n-fold increase over the expression in unstimulated cells from the healthy control. The experiments shown are representative of 4 independent experiments. Data are mean ± SE. *Significant difference in the response between the patient and control subject (P < .05). (C) PHA-T (2 × 106/mL) were stimulated with IL-12 (200 ng/mL) or IFN-α (104 U/mL) in the presence or the absence of anti-CD3 mAb (5 μg/mL) or with a combination of anti-CD3 and anti-CD28 mAb (5 μg/mL) for 24 hours in RPMI 1640 supplemented with IL-2 (1200 U/mL). Supernatants were collected after stimulation, and IFN-γ levels were evaluated by ELISA. (D) PBMCs from the patient and a healthy subject stimulated in vitro for 24 hours with IFN-α (104 U/mL). Target gene expression was normalized to the housekeeping gene (β2M) expression and presented as n-fold of the expression in unstimulated cells from the healthy control. The experiments shown are representative of 4 independent experiments. Data are mean ± SE. *Significant difference in the response between the patient and control subject (P < .05). (E) NK cells from the patient and a healthy subject were cultured without IL-2 for 16 hours and then stimulated in vitro with the indicated doses of IL-27 for 24 hours. Target gene expression was normalized against the housekeeping gene (GAPDH) and presented as n-fold increase over the expression in unstimulated cells from the healthy control subject. The experiments shown are representative of 4 independent experiments. Data are mean ± SE. *Significant difference in the response between the patient and control subject (P < .05).
Impaired both IFN-γ and IFN-α responses in a STAT1-deficient patient. (A-B) PBMCs from the patient and a healthy subject were stimulated in vitro for 4 hours with IFN-α (10 000 U/mL), IFN-γ (100 ng/mL), IL-12 (25 ng/mL), or medium alone to determine c-Myc (A) and PIM-1 (B) expression. The experiments shown are representative of 4 independent experiments. Target gene expression was normalized to the housekeeping gene (β-2-microglobulin) and presented as n-fold increase of the expression over unstimulated cells from the healthy control. Data are mean ± SE. *Significant difference in the response between the patient and control subject (P < .05).
Impaired both IFN-γ and IFN-α responses in a STAT1-deficient patient. (A-B) PBMCs from the patient and a healthy subject were stimulated in vitro for 4 hours with IFN-α (10 000 U/mL), IFN-γ (100 ng/mL), IL-12 (25 ng/mL), or medium alone to determine c-Myc (A) and PIM-1 (B) expression. The experiments shown are representative of 4 independent experiments. Target gene expression was normalized to the housekeeping gene (β-2-microglobulin) and presented as n-fold increase of the expression over unstimulated cells from the healthy control. Data are mean ± SE. *Significant difference in the response between the patient and control subject (P < .05).
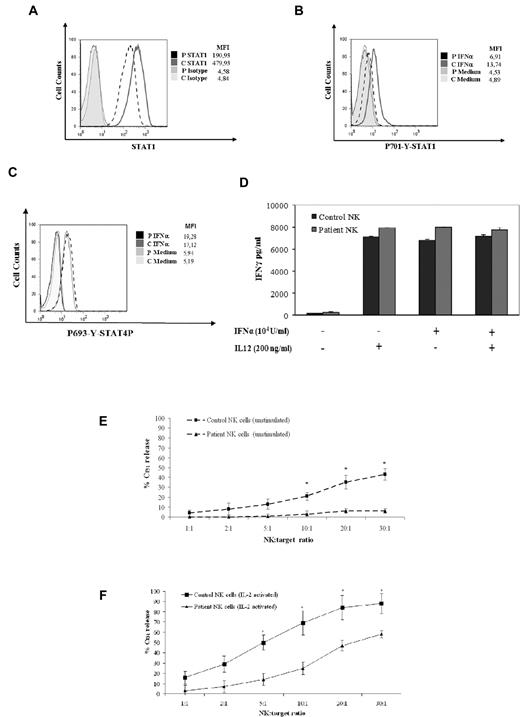
NK cells have an important role in the activation of monocytes/macrophages.7 Indeed, there is evidence that treatment of NK cells with IFN-α results in the activation of the antiviral machinery and in the ability to respond to other cytokines that regulate NK-cell activation, such as IL-15.8,25 Therefore, we investigated the expression and activation of STAT1 in NK cells from the STAT1-deficient patient. Analysis of STAT1 expression in NK cells by intracellular staining showed that the protein level was reduced up to 60% (Figure 6A). Moreover, IFN-α stimulation of NK cells resulted in severely impaired STAT1 phosphorylation (Figure 6B).
Defective NK-cell activation and cytolytic activity in NK cells from a STAT1-deficient patient. (A) Cytometric analysis of STAT1 expression in IL-2–activated NK cells using intracellular staining with anti-STAT1 mAb was performed as described in “Cytokine, ELISA assay, and flow cytometry.” Mean fluorescence intensity is indicated. The results are representative of 3 independent experiments. (B) We performed a flow cytometric analysis of STAT1 phosphorylation (Y701) of IL-2–activated NK cells after treatment with IFN-α (40 U/μL) or medium alone for 20 minutes at 37°C using intracellular staining with an anti-phospho-STAT1-PE mAb. (C) Cytometric analysis of STAT4 phosphorylation in IL-2–activated NK cells using intracellular staining with anti-phospho-STAT4-PE mAb was performed as described in “Cytokine, ELISA assay, and flow cytometry.” Mean fluorescence intensity is indicated. The results are representative of 3 independent experiments. (D) IL-2–activated NK cells were stimulated with IL-12 (200 ng/mL) or IFN-α (104 U/mL) for 24 hours. Supernatants were collected after stimulation, and IFN-γ levels were evaluated by ELISA. (E-F) Defective cytolytic activity of the STAT1-deficient patient against NK-susceptible target cells. Freshly isolated NK cells derived from the patient (▴) or from a healthy donor (■) were tested against the K562 target cells either before (E) or after overnight incubation with rIL-2 (F). The results shown here are representative of 3 independent experiments. Data are mean ± SE. *Significant difference in the response between the patient and the control subject (P < .05).
Defective NK-cell activation and cytolytic activity in NK cells from a STAT1-deficient patient. (A) Cytometric analysis of STAT1 expression in IL-2–activated NK cells using intracellular staining with anti-STAT1 mAb was performed as described in “Cytokine, ELISA assay, and flow cytometry.” Mean fluorescence intensity is indicated. The results are representative of 3 independent experiments. (B) We performed a flow cytometric analysis of STAT1 phosphorylation (Y701) of IL-2–activated NK cells after treatment with IFN-α (40 U/μL) or medium alone for 20 minutes at 37°C using intracellular staining with an anti-phospho-STAT1-PE mAb. (C) Cytometric analysis of STAT4 phosphorylation in IL-2–activated NK cells using intracellular staining with anti-phospho-STAT4-PE mAb was performed as described in “Cytokine, ELISA assay, and flow cytometry.” Mean fluorescence intensity is indicated. The results are representative of 3 independent experiments. (D) IL-2–activated NK cells were stimulated with IL-12 (200 ng/mL) or IFN-α (104 U/mL) for 24 hours. Supernatants were collected after stimulation, and IFN-γ levels were evaluated by ELISA. (E-F) Defective cytolytic activity of the STAT1-deficient patient against NK-susceptible target cells. Freshly isolated NK cells derived from the patient (▴) or from a healthy donor (■) were tested against the K562 target cells either before (E) or after overnight incubation with rIL-2 (F). The results shown here are representative of 3 independent experiments. Data are mean ± SE. *Significant difference in the response between the patient and the control subject (P < .05).
Next, we have evaluated the role of STAT4 for IFN-γ induction in NK cells of the patient with STAT1 mutation. To this aim, flow cytometric analysis of intracellular expression of P-STAT4 after stimulation with IFN-α was performed. Both the STAT1-deficient patient and a healthy control showed normal response to IFN-α (Figure 6C). To examine NK biologic response to cytokines that activate the STAT4 pathway,26 we studied the production of IFN-γ in response to IFN-α and IL-12. We demonstrated that production of IFN-γ in response to both IL-12 and IFN-α was maintained in STAT1-deficient cells (Figure 6D). Because NK cytotoxicity is an important response against viruses, we studied the cytolytic activity of freshly isolated NK cells before and after overnight culture with IL-2. The cytolytic response against the NK-susceptible erythroleukemia cell line K562 and the LCL 721.221 B-EBV–infected cell line was evaluated as percentage of Cr51 release. The cytolytic activity of NK cells, both unstimulated and activated with IL-2, was reduced in the patient compared with healthy control (Figure 6E-F; data not shown). To analyze the maturation of NK cells, we have evaluated the number of CD56dim and CD56 bright subsets by analysis of CD56 and CXCR1 staining. We observed that both NK CD56dim/CXCR1+ and CD56bright/CXCR1− were present in blood of STAT1-deficient patient (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Moreover, analysis of CD56dim CD16+ NK cells subset showed normal expression of KIRs and normal intracellular staining of the lytic effector molecules perforin and granzyme B (data not shown). These observations suggest that NK cells of the STAT1-deficient patient display the phenotypical features of terminally differentiated NK cells.
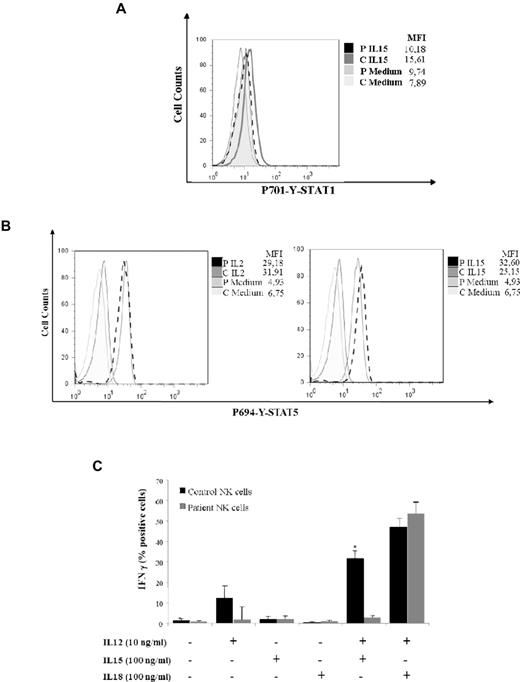
To investigate the consequences of the impaired STAT1 phosphorylation of NK cells in response to IFN-α, we analyzed the effects of IL-15–mediated STAT1 phosphorylation in IFN-α–treated NK cells. Under these experimental conditions, NK cells derived from 4 normal subjects showed a moderate but consistent phosphorylation of STAT1, whereas cells from the STAT1-deficient patient remained unresponsive to IL-15 stimulation (Figure 7A). To exclude NK cell defects because of abnormal γ-chain cytokine family signaling, we evaluated the STAT5 phosphorylation after treatment with IL-2 and IL-15. Both STAT1-deficient patient and healthy control showed comparable levels of P-STAT5 (Figure 7B). Therefore, we investigated the production of IFN-γ by NK cells cultured in the presence of activating cytokines, such as IL-12, IL-18, and/or IL-15, together with the target cell line K562. We found that the combined treatment of NK cells with IL-15 and IL-12 in the presence of K562 target cells resulted in significant production of IFN-γ in the NK cells from the normal subject, whereas the NK cells of the STAT1 patient secreted lower levels of the cytokine (Figure 7C). This observation suggests that IL-15 activation of STAT1 might be important for activation of IFN-γ production and up-regulation of NK cytotoxic activity.
Defective NK-cell activation and cytolytic activity in response to IL-15 in cells from a STAT1-deficient patient. (A) Activated NK cells derived from a healthy subject or from the STAT1-deficient patient were deprived from IL-2 for 16 hours and then preincubated with IFN-α (104 U/mL) for 48 hours and stimulated with IL-15 (100 ng/mL) or medium alone for 20 minutes. STAT1 phosphorylation of IL-15–stimulated NK cells was assessed by intracellular staining with anti-phospho-STAT1-PE mAb. Mean fluorescence intensity is indicated. The results are representative of 3 independent experiments. (B) Activated NK cells from patient and healthy control were IL-2 deprived for 16 hours and then stimulated with IL-2 (100 ng/mL) and IL-15 (100 ng/mL) or medium alone for 20 minutes. STAT5 phosphorylation was assessed by intracellular staining with anti-phospho-STAT5-PE mAb. Mean fluorescence intensity is indicated. The results are representative of 3 independent experiments. (C) Activated NK cells derived from a healthy subject or from the STAT1-deficient patient were stimulated with IL-12 (10 ng/mL), IL-15 (100 ng/mL), IL-18 (100 ng/mL), and with a combination of IL-12 and IL-15, or IL-12 and IL-18 or medium alone in the presence of the target cell line K562. The intracellular expression of IFN-γ by NK cells of the healthy subject (black bar) or of the patient (gray bar) are shown as the percentage of IFN-γ–positive cells. Data are mean ± SE. *Significant difference in the response between the patient and control the subject (P < .05).
Defective NK-cell activation and cytolytic activity in response to IL-15 in cells from a STAT1-deficient patient. (A) Activated NK cells derived from a healthy subject or from the STAT1-deficient patient were deprived from IL-2 for 16 hours and then preincubated with IFN-α (104 U/mL) for 48 hours and stimulated with IL-15 (100 ng/mL) or medium alone for 20 minutes. STAT1 phosphorylation of IL-15–stimulated NK cells was assessed by intracellular staining with anti-phospho-STAT1-PE mAb. Mean fluorescence intensity is indicated. The results are representative of 3 independent experiments. (B) Activated NK cells from patient and healthy control were IL-2 deprived for 16 hours and then stimulated with IL-2 (100 ng/mL) and IL-15 (100 ng/mL) or medium alone for 20 minutes. STAT5 phosphorylation was assessed by intracellular staining with anti-phospho-STAT5-PE mAb. Mean fluorescence intensity is indicated. The results are representative of 3 independent experiments. (C) Activated NK cells derived from a healthy subject or from the STAT1-deficient patient were stimulated with IL-12 (10 ng/mL), IL-15 (100 ng/mL), IL-18 (100 ng/mL), and with a combination of IL-12 and IL-15, or IL-12 and IL-18 or medium alone in the presence of the target cell line K562. The intracellular expression of IFN-γ by NK cells of the healthy subject (black bar) or of the patient (gray bar) are shown as the percentage of IFN-γ–positive cells. Data are mean ± SE. *Significant difference in the response between the patient and control the subject (P < .05).
Discussion
Although the innate immune response against viruses is usually initiated by the release of type I IFN, other cell types, such as NK cells, and cytokine pathways are required for the complete clearance of viruses. In particular, by integrating signals from inhibitory and activating receptors, NK cells are able to recognize cells already infected by viruses.27,28 Virus-derived proteins can activate receptors (eg, NKG2D), although these signals can be obscured by receptors transducing negative signals.29 This delicate balance of multiple activating and inhibitory receptors of NK cells is regulated by cytokines, such as IL-15, IL-12, and type I IFN, which are able to enhance the NK cytotoxic activity and the production of IFN-γ.7,8
Our studies suggest that STAT1 deficiency had an impact on the functional activation of NK cells. Because this cell type was recognized in the role of the immune response against intracellular pathogens and viral infections, this observation might be important for understanding the mechanisms of susceptibility in STAT1-deficient patients to mycobacterial and viral infections. We observed that unstimulated NK cells displayed a cytotoxicity defect against the cell line target K562. It is probable that the defect of NK-mediated cytolysis was related to the immunomodulatory role of STAT1 and to its balanced equilibrium with STAT4 expression. Although STAT4 is expressed at higher levels in NK cells and is required for IFN-γ production in response to type I IFN, STAT1 is barely detectable in NK cells, although its expression is dramatically induced during early viral infections.30,31 NK cells have an unusual sensitivity for type I IFN activation of STAT4 because its high basal expression, but STAT1 levels had been demonstrated to limit access to STAT4. In the case of reduction of STAT1 levels, as observed in the patient reported, STAT4 activation cannot be regulated by recruitment of STAT1 to type I IFN receptor.30,31 In addition, NK cytotoxicity is regulated by other cytokines that require STAT1 for their signaling pathway. In particular, IL-15 promotes STAT1 phosphorylation, induces STAT1 DNA binding to the interferon promoter GAS in NK cells, and improves the cytotoxicity of NK cells via up-regulating the expression of the NKG2D receptor and the effector molecules perforin and TRAIL.7,8 Moreover, we observed that IFN-γ production by NK cells activated with IL-15 and IL-12 in the presence of target cells is impaired in STAT1-deficient cells, suggesting that the IL-15 response involves STAT1 signaling.26
In addition, our results highlighted the broad genetic and clinical heterogeneity of STAT1 deficiency in humans. The mutation identified in this patient led to the synthesis of the STAT1α and STAT1β isoforms, which both lacked 33 amino acid residues and were expressed at lower levels in the cells of the patient. Although this mutation is classified as a hypomorphic mutation because it did not abrogate STAT1 phosphorylation in response to both IFN-γ and IFN-α, it resulted in a severe defect of DNA binding complexes as assessed by the electromobility shift assay. In keeping with this observation, biologic studies of the cellular response to IFN-γ demonstrated that the cells of the patient were refractory even to high doses of the cytokine, which was evaluated on the basis of TNF-α production, whereas activated PBMCs displayed a normal response to IL-12 and IFN-α in terms of IFN-γ production. Moreover, IFN-α failed to induce, in PBMCs from the patient with STAT1 deficiency, the expression of MXA, a protein that is associated with an antiviral response.
It is interesting that the previously reported forms of complete STAT1 deficiency were characterized by mutations that prevented STAT1 protein synthesis, thereby leading to an absolute defect of cellular response to both IFN-γ and IFN-α.3,4 The patient we described carried a splicing mutation, which preserved protein synthesis and, to partial extent, the tyrosine phosphorylation of STAT1 in response to both IFN-α and IFN-γ, but prevented the cellular responses to the same cytokines. These immunologic findings are also consistent with the clinical manifestations of STAT1 deficiency in this patient, which were characterized by disseminated mycobacterial infections and multiple CMV infection episodes. In contrast to other patients with complete STAT1 deficiency,3,4 reduced STAT1 phosphorylation in response to IFN-γ or IFN-α was detected, but DNA binding activity and functional response to the cytokines were abolished. Patients with the autosomal recessive partial form of STAT1 deficiency presented severe but curable intracellular bacterial and viral diseases, and these patients showed reduced but not abolished responses to both IFN-γ and IFN-α and weaker STAT1 DNA-binding activity.14,16 The divergence between the severe defects in the biologic response to IFN-γ and IFN-α and the preserved expression and tyrosine phosphorylation of STAT1 might be related to the effects of the novel splicing mutation. Analysis of the N-terminal domain has shown that this region is characterized by a well-defined hydrophobic core that is highly conserved across the STAT proteins.32,33 Indeed, there is evidence that the N-terminal region of STAT1 plays a crucial role in the stabilization of STAT1 dimer-dimer interactions34 and is required for interaction between STAT1 and the transcriptional coactivator protein CBP.35 Therefore, mutations affecting the N-terminal domain might have an impact on several functions of the protein. Study of STAT1 truncated mutants of the protein lacking the first 131 amino acid residues has shown that the mutant did not show cooperative binding for STAT1-STAT1 dimer interaction36 and that the N-terminal region is involved in deactivation of the phosphorylated protein by reorientation of STAT1 dimers to an antiparallel form that facilitates easy access to phosphatase.37,38
STAT1 deficiency constitutes a rare immunodeficiency that affects the innate immune response against viruses and intracellular pathogens, and this condition displays patterns of both recessive and dominant inheritance and a whole spectrum of biologic responses to IFN-γ or IFN-α. The heterogeneity of the clinical and genetic features of STAT1 deficiency require a complete assessment of IFN-γ, IL-12, and IFN-α responses for every patient who presents with severe infection by intracellular pathogens and/or viruses.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by PRIN 2009, Fondazione Cariplo (NOBEL grant), European Union (grant FP7 HLH-cure; project 201461; R.B.), and Fondazione Nocivelli (S. Giliani). D.V. is the recipient of a Brescia Rotary Fellowship; N.T. is a recipient of a Fondazione Italiana per la Ricerca sul Cancro fellowship.
Authorship
Contribution: D.V. performed genetic analysis, protein expression, and phosphorylation studies and wrote the manuscript; L.T. carried out the functional studies and wrote the manuscript; G.T. and S.P. performed NK studies; N.T., S. Gasperini, and F.B. performed phosphorylation and DNA-binding activity studies; A.P., F.P., and L.D.N. were in charge of the patients' follow-up; S. Giliani led the genetic, protein expression, and phosphorylation studies; and R.B. supervised the functional studies and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Raffaele Badolato, Istituto di Medicina Molecolare Angelo Nocivelli, Università di Brescia, c/o Spedali Civili, 25123 Brescia, Italy; e-mail: badolato@med.unibs.it.
References
Author notes
S. Giliani and R.B. contributed equally to this study and share senior authorship.