Abstract
CD160 is a human natural killer (NK)-cell–activating receptor that is also expressed on T-cell subsets. In the present study, we examined 811 consecutive cases of B-cell lymphoproliferative disorders (B-LPDs), and demonstrated CD160 expression in 98% (590 of 600) of chronic lymphocytic leukemia (CLL) cases, 100% (32 of 32) of hairy cell leukemia (HCL) cases, 15% (5 of 34) of mantle cell lymphoma (MCL) in the leukemic phase, and 16% (23 of 145) of other B-LPD cases. CD160 transcript and protein were absent in the normal B-cell hierarchy, from stem cells, B-cell precursors, maturing B cells in the germinal center, and circulating B cells, including CD5+CD19+ B1 cells in umbilical cord. CD160 positivity was significantly higher in CLL and HCL in terms of percentage (65.9% and 67.8%, respectively, P < .0001) and median fluorescence intensity (552 and 857, respectively, P < .0001) compared with all other B-LPD cases. Lymph node CLL samples were also CD160+. Using the disease-specific expression of CD5, CD23, and CD160, a score of 3 characterized CLL (diagnostic odds ratio, 1430); a score of 0 excluded CLL, MCL, and HCL; and the CD23/CD5 ratio differentiated CLL from leukemic CD23+ MCL. In the B-cell lineage, CD160 is a tumor-specific antigen known to mediate cellular activation signals in CLL, and is a novel target for therapeutic manipulation and monitoring of minimal residual disease.
Introduction
CD160 is an Ig-like activating natural killer (NK) cell receptor expressed on the majority of circulating NK cells and on a subset of circulating cytotoxic T cells, but not on B cells or EBV-transformed B-cell lines.1,2 In contrast to the majority of NK cell receptor genes located on chromosomes 12 and 19,3 the CD160 gene is located on chromosome 1q42.3.4 CD160 is expressed by most peripheral blood TCRγδ lymphocytes, a minor subset of circulating CD8bright+ TCRαβ cells, and all small intestinal intraepithelial T lymphocytes, phenotyped as CD3+TCRα/β+CD56−.5 A minor population of CD4+ T cells also express CD160.6 CD160 mRNA expression was shown to be highly restricted to NK cells and not detected in myeloid and B-cell lines by Northern blot analysis.5 Outside of the immune system, CD160 is expressed on endothelial cells of neoangiogenic microvessels at the periphery of tumors.7
NK cells play a key role in innate immunity, having potent cytolytic activity against virally infected and tumor cells.8 NK-cell activity is regulated by inhibitory and activatory receptors expressed at the cell surface and their interaction with associated ligands.9 CD160 binds to MHC class Ia and Ib with low affinity10 and triggers cytotoxic function in peripheral blood NK cells, as well as cytokine production, including IFN-γ, TNF-α, and IL-6.11,12 Only a limited number of human activating NK-cell receptors have been demonstrated to induce cytokine production and release in addition to cytotoxicity.13 The PI3K signaling molecule is required for CD160-mediated cytokine release, with involvement of the signaling molecules Syk and ERK upstream and downstream of PI3K, respectively.14
Recent work has demonstrated CD160 expression in malignant human B cells.15 CD160 expressed on the surface of B-cell chronic lymphocytic leukemia (CLL) mimicked CD160 functions in normal NK and T cells: cellular activation, up-regulation of BCL-2 and BCL-XL, and improved in vitro cell survival and cytokine production, specifically IL-6 and IL-8. PI3K/Akt signaling was required for CD160-mediated functions in CLL cells.15 Similar “aberrant” expression of a signaling molecule, CD3-receptor-associated protein tyrosine kinase or ζ-associated protein-70 (ZAP-70), was reported in CLL.16,17 Like CD160, ZAP-70 was initially described exclusively in T cells and NK cells,18 but was subsequently detected in mature and immature human B-lymphoid malignancies,19-21 as well as normal murine and human B cells.22,23
In the present study, we investigated normal and malignant human B cells for expression of CD160. This extensive study established that the NK cell receptor antigen CD160 shows restricted expression in the B-cell lineage to malignant compared with normal B cells. Moreover, the varying expression of CD160 can be exploited diagnostically, as shown in test and validation sets consisting of > 970 cases of B-cell lymphoproliferative disorders (B-LPDs).
Methods
Patients and samples
This study involved a test cohort of 811 consecutive patient samples referred for investigation of B-LPD between 2002 and 2008, for which a complete analysis was performed. Standard diagnostic criteria were used to establish the diagnosis of CLL (n = 600), mantle cell lymphoma (MCL, n = 34), hairy cell leukemia (HCL, n = 32), and other B-LPDs (n = 145) incorporating acute lymphoblastic leukemia (ALL) and other mature B-cell malignancies.24,25 Correlation with BM aspirate and trephine, lymph nodes, and spleen by flow cytometry, histology, and karyotyping was performed when required.
The validation cohort included 163 consecutive cases of mature B-LPD (CLL, n = 113; MCL, n = 8; HCL, n = 3; and other B-LPDs, n = 39). The median fluorescence intensities (MFIs) for malignant populations given in this study were generated from the validation cohort.
Nondiagnostic analyses were approved by the National Research Ethics Service, East London, and the City HA Local Research Ethics Committee, and written informed consent was obtained in accordance with the Declaration of Helsinki. After informed consent, control blood samples were taken from normal healthy donors (n = 30), donor BM aspirates (n = 6), and cord blood sampling (n = 5). When possible, tissue perfusion samples, including spleen (n = 5), lymph-node biopsy (n = 5), and tonsil biopsy (n = 4) were obtained from both healthy donors and patients .
Immunophenotypic analysis using CD160FCA
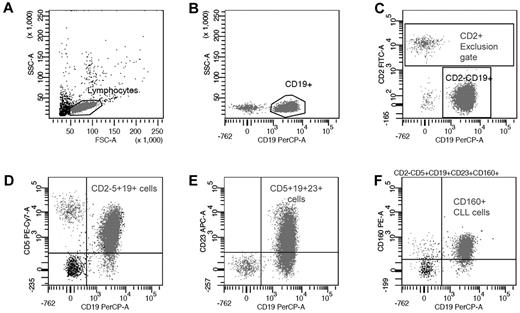
The 5-color CD160 Flow Cytometric Assay (CD160FCA) incorporated CD2-FITC (clone S5.2), CD5-APC (clone L17F12), CD19-PerCP (clone 4G7), and CD23-APC (clone EBVCS-5; all from BD Biosciences); and CD160-PE (clone BY55 IgM isotype; Immunotech). A combination of internal negative controls and an isotype IgM were used (Immunotech). Whole blood samples were analyzed within 24 hours. Leukocytes (1 × 106) were labeled with the appropriate pretitered antibody for 15 minutes at room temperature in darkness. Erythrocyte lysis was performed using Pharm Lyse (Becton Dickinson), a buffered ammonium chloride–based lysing solution, according to the manufacturer's recommendations, then washed in BD Cell Wash (BD Biosciences). A minimum of 10 000 lymphocyte-gated events was acquired for each patient on a FACSCanto (BD Biosciences), with data being acquired and analyzed by BD FACSDiva Version 6.1.3 clinical software for enhanced acquisition analysis. Positivity was defined as > 20% of leukemic cells expressing a given surface antigen. A sequential gating strategy for the specific identification of CD160 on CD5+CD19+ B cells using multicolor flow cytometry was established (Figure 1). The BD FACSDiva software calculated the MFI of the CD2−CD5+CD19+CD23+CD160+ population.
Sequential gating strategy for CD160 detection on malignant CD19+ B cells. (A) Total B cells were identified using forward and side scatter to gate the lymphoid region and exclude any apoptotic cells and debris. (B) CD19+ B cells were compared with side scatter to exclude any nonspecific binding. (C) CD19+ B cells were further isolated by gating the CD2+ events and generating a “NOT” (exclusion) gate. (D) The malignant B cells were separated from the normal residual B cells using a CD2−CD5+CD19+ gate. (E) CD23 expression was calculated from the malignant population and used to generate the mini-CLL score and CD23r. (F) In the context of CLL, CD160 positivity defined on the CD2−CD5+CD19+CD23+ population was then calculated from this “pure” malignant B-cell population.
Sequential gating strategy for CD160 detection on malignant CD19+ B cells. (A) Total B cells were identified using forward and side scatter to gate the lymphoid region and exclude any apoptotic cells and debris. (B) CD19+ B cells were compared with side scatter to exclude any nonspecific binding. (C) CD19+ B cells were further isolated by gating the CD2+ events and generating a “NOT” (exclusion) gate. (D) The malignant B cells were separated from the normal residual B cells using a CD2−CD5+CD19+ gate. (E) CD23 expression was calculated from the malignant population and used to generate the mini-CLL score and CD23r. (F) In the context of CLL, CD160 positivity defined on the CD2−CD5+CD19+CD23+ population was then calculated from this “pure” malignant B-cell population.
CD160FCA-derived scores
A mini-CLL score (0-3) was calculated using the markers CD5, CD23, and CD160, with one point for each marker. CD23r, a ratio of CD23 to CD5 expression, was calculated from the percentage of CD19+ B cells expressing CD23 compared with the CD19+CD5+ percentage. Combining these parameters as (mini-CLL score) × (CD23r) allowed the immunophenotypic data to be expressed as a single numerical value, the “diagnostic discriminant.”
RNA extraction, reverse transcription, cDNA amplification (RT-PCR), and sequencing for CD160
PBMCs from EDTA-anticoagulated venous blood obtained from healthy donors and patients were isolated by density gradient centrifugation (Lymphoprep; Amersham Bioscience). Fresh CD19+ B cells were isolated using a MACS and a CD19+ cell-isolation kit according to the manufacturer's recommendations (Miltenyi Biotec). CD19+ cell purity was shown to be > 97%.
Total RNA was isolated using TRIzol reagent according to the manufacturer's instructions (Invitrogen). For each reverse transcription, 5μg of RNA was used. Reverse transcription was performed using 500 ng of an oligo-dT primer (Invitrogen) and the SuperScript First Strand Synthesis kit (Invitrogen) in a total volume of 20 μL. Specific primers for the amplification of CD160 (GenBank accession no. NM_007053) cDNA were designed on the basis of published sequences. The CD160 primers were as follows: BY01 (5′-TGCAGGATGCTGTTGGAACCC-3′, forward) and BY3UN (5′-CCTGTGCCCTGTTGCATTCTTC-3′, reverse). β-actin cDNA amplification was performed in parallel as an internal control. The synthesis of specific cDNA fragments was achieved using 1 μL of the reverse-transcribed product according to a standard procedure (Invitrogen) in a total volume of 20 μL. Each sample was subjected to denaturation (94°C for 30 seconds), annealing (60°C for 30 seconds), and extension (72°C for 90 seconds) steps for 35 cycles. The amplified products were separated on a 1% agarose gel. For the CD160 cDNA sequencing, the open reading frame was amplified by PCR with BY01 and BY3UN primers and Taq High-Fidelity (Invitrogen). The PCR product was purified (QIAEX II; QIAGEN) and analyzed with the following primer sequences: forward primers BY01 (5′-TGCAGGATGCTGTTGGAACCC-3′) and BY03 (3′-TCAGCCTGAACTGAACTGAGAGTGCCTTC-5′); reverse primers BY02 (5′-CAGCTGAGACTTAAAAGGGATC-3′), BY04 (3′-CACCAACACCATCTATCCCAG-5′), and BY3UN (5′-CCTGTGCCCTGTTGCATTCTTC-3′).
Tissue microarray and IHC
Biopsy samples were obtained from patients who had given informed consent, including 88 cases of CLL, 97 MCL samples prepared on tissue microarrays (TMAs), 2 sample sections of HCL, and a control group of tissue from paraffin-embedded tissue samples. TMAs were prepared from paraffin-embedded histological blocks. The TMAs and the sections were dewaxed twice in xylene for 5 minutes and then washed in 100% ethanol, followed by 2 initial peroxidase-blocking steps for 5 minutes each. Tissues were then heated in a pressure cooker containing a Tris-citrate–based buffer (Vector Laboratories) for approximately 10 minutes, washed in running tap water, and transferred back into the buffer. The slides underwent a blocking step using horse serum for 20 minutes and were then air-dried. The primary CD160 antibody, clone CL1-R2 (IgG1) was produced in-house, applied at a concentration of 10 μg/mL, and incubated overnight at 4°C. Negative control samples were prepared by omitting the primary CD160 antibody. IHC staining was performed using the Super Sensitive Polymer-HRP Detection System (Biogenex). Slides were subjected to a Super Enhancer Reagent (Biogenex) for 20 minutes, followed by a poly-HRP reagent conjugated to an anti–mouse antibody for 30 minutes at room temperature. The antibody-enzyme complex was then incubated with 3,3′-diaminobenzidine, immersed and counterstained with Gills II hematoxylin, dipped in acid/alcohol, washed in running water, dehydrated using ethanol followed by xylene, and finally mounted with a xylene-based DPX mount (Sigma-Aldrich).
Statistical analysis
Standard approaches were used to calculate diagnostic indices of sensitivity and specificity. Statistical analysis was performed using Stats Direct Version 2.7.6 software and Prism 5 software for Macintosh (GraphPad). The diagnostic odds ratio was used as a single measure of efficacy of a diagnostic test; this ratio is unaffected by prevalence and effectively compares the odds of positivity in disease (sensitivity) relative to the odds of positivity in patients without the disease (1 − specificity). The ratio can range from 0 to infinity, with larger numbers indicating a better test performance. Indices are quoted as the statistic (95% confidence interval [CI]). When reporting the MFI, indices are quoted as the mean (95% CI of the mean).
Results
Expression of CD160 on normal B cells
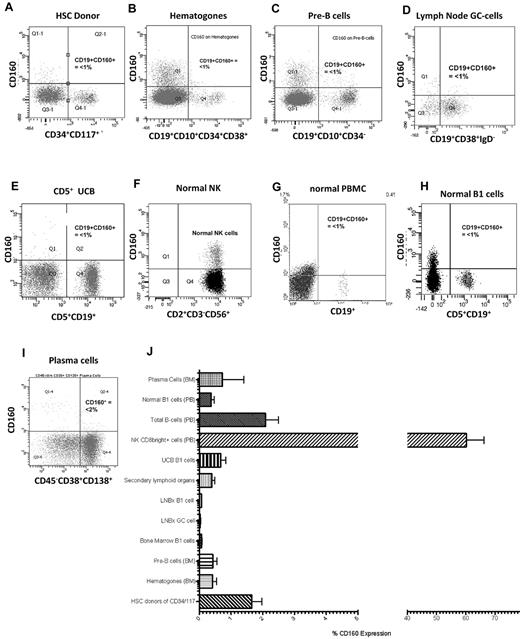
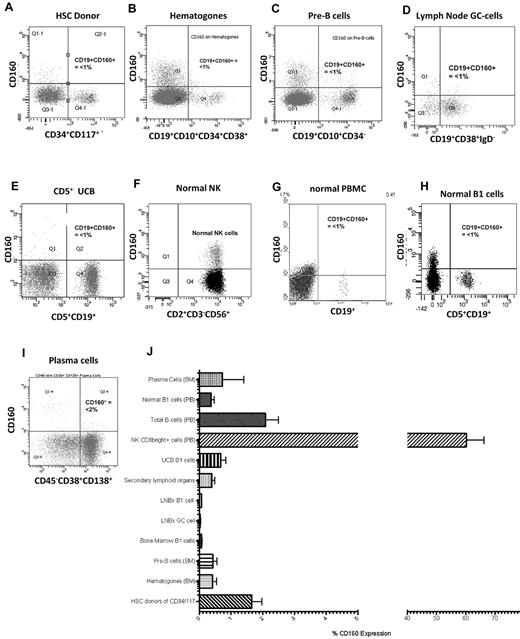
In normal peripheral blood, CD160 protein is expressed on the cell membrane of 15%-20% of CD2+ lymphocytes, and the CD160 transcript is highly restricted to NK and T cells1,2,5 (Figure 2F). To investigate the expression of CD160 protein on normal B cells, the B-cell hierarchy was studied from immature HSCs to the mature terminally differentiated plasma cell. Hematopoietic stem cells from harvest donors (n = 5) were identified using a combination of CD34+/CD117+/CD38−/dim/CD133+. The mean percentage expression of CD160 in this population was 1.7% (range, 0.77%-2.54%) (Figure 2A and J). Similarly, immature BM B cells, which are reactive benign B-cell precursors such as hematogones (n = 6) (CD19+/CD10+/CD34+/CD38+), and pre-B cells lacking surface immunoglobulin (n = 6; CD19+/CD10+/CD34−/sIg−) were negative for CD160 expression: means, 0.78% (range, −0.05%-1.61%) and 0.43% (range, 0.09%-0.76%), respectively (Figure 2B-C and J).
CD160 expression in normal lymphocytes is restricted to NK and T cells. (A) CD160 expression on normal donor HSCs gated on the CD34+CD117+ cells (n = 5). (B) CD160 expression on immature reactive benign B-cell precursors, termed hematogones, that are typically CD19+CD10+CD34+CD38+ (n = 5). (C) CD160 expression on pre-B cells from BM samples that lack surface immunoglobulin but are CD19+CD10+CD34− (n = 6). (D) CD19+CD38+IgD− GC cells from both tonsils and lymph nodes (n = 6). (E) UCB showing CD160 and CD19 expression on CD5+ cells (n = 5). (F) Normal peripheral blood NK cells demonstrating CD160 positivity (n = 30). (G) Normal peripheral blood mononuclear B cells (n = 30). (H) Normal B1 cells from a healthy donor (n = 30). (I) CD45−/dimCD38+CD138+ terminally differentiated plasma cells from BM samples (n = 5). (J) Percentage CD160 expressed on individual cell types. PB indicates peripheral blood; LNBx, lymph node biopsy. Secondary lymphoid organs are splenic and tonsillar material.
CD160 expression in normal lymphocytes is restricted to NK and T cells. (A) CD160 expression on normal donor HSCs gated on the CD34+CD117+ cells (n = 5). (B) CD160 expression on immature reactive benign B-cell precursors, termed hematogones, that are typically CD19+CD10+CD34+CD38+ (n = 5). (C) CD160 expression on pre-B cells from BM samples that lack surface immunoglobulin but are CD19+CD10+CD34− (n = 6). (D) CD19+CD38+IgD− GC cells from both tonsils and lymph nodes (n = 6). (E) UCB showing CD160 and CD19 expression on CD5+ cells (n = 5). (F) Normal peripheral blood NK cells demonstrating CD160 positivity (n = 30). (G) Normal peripheral blood mononuclear B cells (n = 30). (H) Normal B1 cells from a healthy donor (n = 30). (I) CD45−/dimCD38+CD138+ terminally differentiated plasma cells from BM samples (n = 5). (J) Percentage CD160 expressed on individual cell types. PB indicates peripheral blood; LNBx, lymph node biopsy. Secondary lymphoid organs are splenic and tonsillar material.
Mature polyclonal B cells in secondary lymphoid organs (lymph nodes, n = 5; spleen, n = 5; tonsils, n = 4) were isolated by repeated perfusion, and germinal center (GC) B cells were identified by their CD19+/CD38+/IgD− expression. The mean expression of CD160 on lymph node GC cells was 0.79% (range, 0.29%-1.28%; Figure 2D and J); normal polyclonal B cells isolated from spleen and tonsil biopsies also lacked CD160, with a mean expression of 0.39% (range, 0.20%-0.60%; Figure 2J).
Umbilical cord blood (UCB) is rich in naive B cells (CD19+CD5+ B1 cells). UCB B1 cells did not express CD160 (mean, 0.68%; range, 0.24%-1.11%; Figure 2E and J). Progressing through the B-cell hierarchy, CD160 was expressed in 2.08% (range, 1.21%-2.95%) of peripheral blood total B cells (Figure 2G) and in 0.37% (range, 0.16%-0.58%) of B1 cells (Figure 2H). As a percentage of the total leukocyte population, CD160 expression on total B cells was 0.11% (range, 0.04%-0.18%). Finally, CD45−/dim/CD19−/CD38+/CD138+ terminally differentiated plasma cells (n = 5) from normal BM failed to express CD160 (mean, 0.73%; range, −2.14%-3.61%; Figure 2I). There were no significant differences within the B-cell hierarchy with regard to percentage CD160 expression (P > .05; Figure 2J).
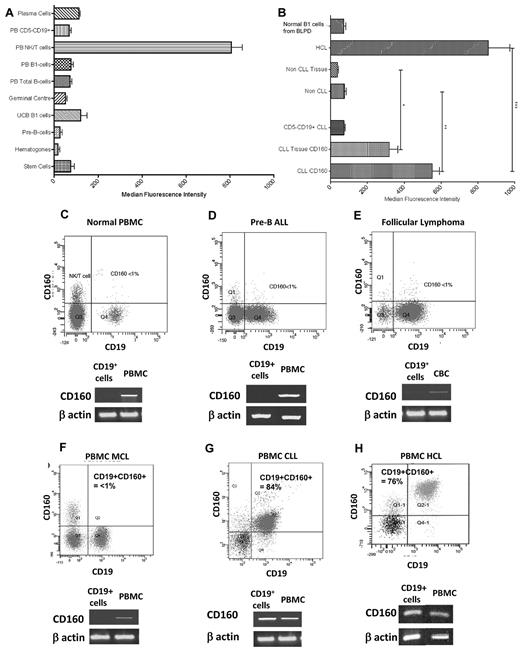
Circulating NK and T cells acted as a positive control, representing 11.05% of all CD2+ events (CI: 7.90-14.20). A total of 60.21% of NK cells expressed the CD160 antigen with an MFI of 806.2 (CI: 701.8-910.5; Figure 3A). The MFI of CD160 on immature B cells—HSCs (75.3, CI: 35.36-115.3), hematogones (17.2, CI: −4.74-39.07), and pre-B cells (26.5, CI: −0.46-53.46)—isolated from normal donors was significantly less than normal NK and T cells (P < .0001; Figure 3A). Similarly, mature B-cell populations in both tissue and peripheral blood had low CD160 MFIs: GC B cells, 38.8 (CI: 23.06-54.54); naive UCB B cells, 121.3 (CI: 29.2-213.3); and circulating B cells—B1 cells, 76.53 (CI: 53.7-99.4) and CD5− B cells, 69.26 (CI: 52.2-86.4). Normal BM plasma cells also had a low CD160 MFI of 112.3 (CI: 84.9-139.8, n = 5; Figure 3A).
CD160 protein and transcript expression is restricted to CLL and HCL. (A) MFI of CD160 in normal B cells. Stem cells were isolated from normal HSC donors. Hematogones and pre-B cells were analyzed from BM samples. (B) MFI of malignant B cells demonstrating significantly increased expression in CLL and HCL. “Non-CLL” represents all B-LPDs excluding CLL and HCL cases. All tissue fields include lymph nodes, spleen, BM, and tonsillar material. Non-CLL tissue includes pre-B-ALL BM samples. *P < .0001; **P < .0001; ***P = .0114. (C-H) Representative flow cytometric images of normal PBMCs (C), pre-B ALL (D), follicular lymphoma (E), MCL (F), CLL (G), and HCL (H). Below each flow cytometric plot is the corresponding cDNA amplification using specific primers for CD160 after reverse transcription of total RNA extracted from highly purified CD19+ B cells (isolated using a magnetic-activated cell sorter; purity > 97%). β-actin cDNA synthesis was used as an internal control.
CD160 protein and transcript expression is restricted to CLL and HCL. (A) MFI of CD160 in normal B cells. Stem cells were isolated from normal HSC donors. Hematogones and pre-B cells were analyzed from BM samples. (B) MFI of malignant B cells demonstrating significantly increased expression in CLL and HCL. “Non-CLL” represents all B-LPDs excluding CLL and HCL cases. All tissue fields include lymph nodes, spleen, BM, and tonsillar material. Non-CLL tissue includes pre-B-ALL BM samples. *P < .0001; **P < .0001; ***P = .0114. (C-H) Representative flow cytometric images of normal PBMCs (C), pre-B ALL (D), follicular lymphoma (E), MCL (F), CLL (G), and HCL (H). Below each flow cytometric plot is the corresponding cDNA amplification using specific primers for CD160 after reverse transcription of total RNA extracted from highly purified CD19+ B cells (isolated using a magnetic-activated cell sorter; purity > 97%). β-actin cDNA synthesis was used as an internal control.
Expression of CD160 in malignant B cells
Malignant B cells at different stages of maturation were analyzed for CD160 protein and mRNA. Immature progenitor cells of pre-B-ALL were negative for CD160 expression (n = 5; Figure 3D). A test cohort of 811 cases of mature B-LPD with same-day analysis of CD160 expression was investigated during the study period. Within the test cohort, 600 cases were diagnosed as CLL using standard criteria, of which 590 (98.3%) expressed CD160, with a mean CD160 positivity of 65.9%. All 32 of 32 cases of HCL were positive for the CD160 antigen (Figure 3H), with a mean percentage positivity of 67.8%, but a higher MFI than CLL (857.2 vs 552.5, P = .01). Malignant B cells of CLL and HCL had a characteristic weak to weak-moderate staining for CD160, with a single displaced peak showing a Gaussian distribution of staining intensity. Of the 179 other B-LPD cases, 28 (15.6%) were positive for the CD160 antigen. These B-LPDs included 34 cases of MCL in the leukemic phase, with 29 (85.3%) negative and 5 (14.7%) positive for CD160. Representative cases of CD160− immature pre-B-ALL (Figure 3D), follicular lymphoma (Figure 3E), and MCL (Figure 3F) are shown, with CD160 representing < 1% of the malignant population.
To confirm the flow cytometric protein-expression data, total RNA was extracted from highly purified CD19+ B cells. As expected, the CD160 transcript was detected in all control mononuclear cell fractions (representing normal NK and T cells), but was absent from purified normal B cells (Figure 3C), pre-B-ALL (Figure 3D), follicular lymphoma (Figure 3E), and MCL (Figure 3F) samples. To control for contaminating normal NK and T cells in the purified CD19+ fraction, a CD2 PCR control was used, which was negative in all cases (data not shown). CD160 transcript was only detected in purified B cells of CLL (Figure 3G) and HCL (Figure 3H). Sequence analysis of the CD160 transcript in CLL cells corresponded to the CD160 coding sequence published previously (GenBank no. NM_007053). These findings indicate that CD160 protein and RNA expression are absent in normal human B cells and the majority of mature B-cell malignancies, but are expressed in CLL and HCL cells.
With respect to the intensity of CD160 expression, the non-CLL and non-HCL cases were grouped together for statistical comparison as “non-CLL” (Figure 3B), including cases of myeloma and Waldenström macroglobulinemia. Circulating CLL and HCL cells had significantly higher CD160 MFIs (552.2 and 857.2, respectively) than all other sample types (76.82; P < .0001), including CLL tissue samples (318.8; range, 215.4-422.2). The processing of CLL tissue biopsies resulted in a significant decrease in MFI expression compared with peripheral blood CD160 expression, as shown by paired peripheral blood and tissue mononuclear cell suspension samples (P = .04; Figure 3B). Despite this decrease in expression, the MFI of the CLL tissue preparations was significantly higher than that of the non-CLL peripheral blood, non-CLL tissue samples, and normal polyclonal B1 cells from patients with CD5− B-LPDs (P < .0001; Figure 3B). Non-CLL/non-HCL samples had CD160 MFI measurements that were equivalent to those of normal donor B cells (P = .78). Throughout the study period, CD160 analysis was performed on several cases of reactive polyclonal B-cell lymphocytoses, and in all cases the polyclonal B cells were negative for CD160 expression.
Immunohistochemistry with CD160 in CLL, HCL, and MCL
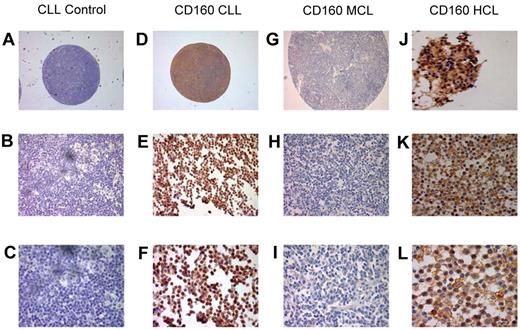
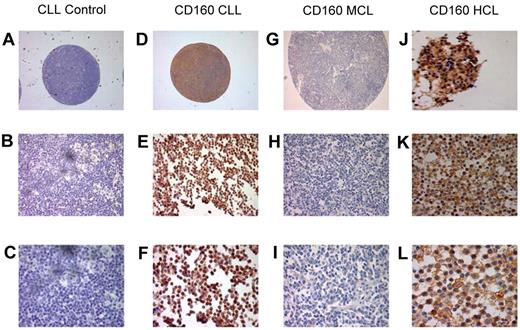
To further establish the expression of CD160 in malignant B cells, IHC staining of CD160 was performed using paraffin-embedded histological sections and TMAs (Figure 4). Of the 88 TMA CLL cases, 85 (96.5%) were positive for CD160 (Figure 4D-F). Similarly, 2 of 2 cases of HCL (a trephine biopsy, Figure 4J, and a splenic biopsy, Figure 4K-L) were positive for CD160 and showed a stronger expression than in CLL, confirming the results observed by flow cytometry. Of 97 MCL cases on TMAs, only 3 (3%) were positive for CD160 (Figure 4G-I).
Immunohistological staining of CD160 in chronic B-cell malignancies. Tissue sections and TMAs were stained with CD160 antibody (10 μg/mL), incubated overnight at 4°C, and revealed using the Super Sensitive Polymer-HRP Detection System (Biogenex). (A-C) Control: a lymph node TMA spot from a patient with CLL without the primary antibody. Magnifications were ×5 (A), ×20 (B), and ×63 (C). (D-F) Positive CD160 staining of a CLL lymph node TMA. Magnifications were ×5 (A), ×20 (B), and ×63 (C). Of the CLL TMA cases, 85 of 88 (96.5%) were CD160+. (G-I) Negative CD160 staining of an MCL lymph node on a TMA. Magnifications were ×5 (A), ×20 (B), and ×63 (C). Of the MCL TMA cases, 94 of 97 were CD160−. (J-L) Two HCL cases showing typical strong CD160 positivity, a BM trephine (J), and a splenic biopsy. Magnifications were ×43 (K) and ×63 (L).
Immunohistological staining of CD160 in chronic B-cell malignancies. Tissue sections and TMAs were stained with CD160 antibody (10 μg/mL), incubated overnight at 4°C, and revealed using the Super Sensitive Polymer-HRP Detection System (Biogenex). (A-C) Control: a lymph node TMA spot from a patient with CLL without the primary antibody. Magnifications were ×5 (A), ×20 (B), and ×63 (C). (D-F) Positive CD160 staining of a CLL lymph node TMA. Magnifications were ×5 (A), ×20 (B), and ×63 (C). Of the CLL TMA cases, 85 of 88 (96.5%) were CD160+. (G-I) Negative CD160 staining of an MCL lymph node on a TMA. Magnifications were ×5 (A), ×20 (B), and ×63 (C). Of the MCL TMA cases, 94 of 97 were CD160−. (J-L) Two HCL cases showing typical strong CD160 positivity, a BM trephine (J), and a splenic biopsy. Magnifications were ×43 (K) and ×63 (L).
Implementing the CD160FCA into clinical diagnostics
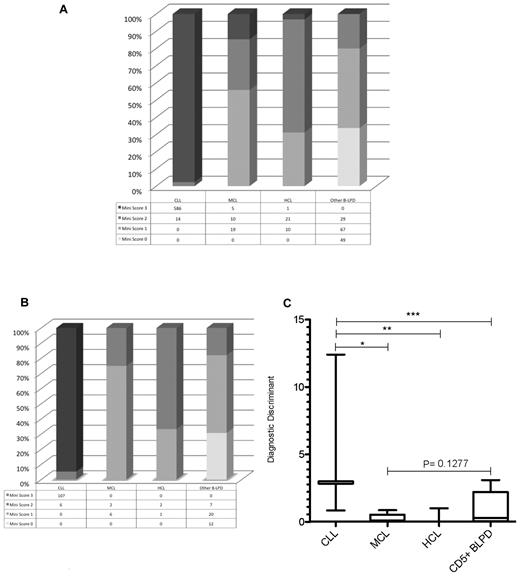
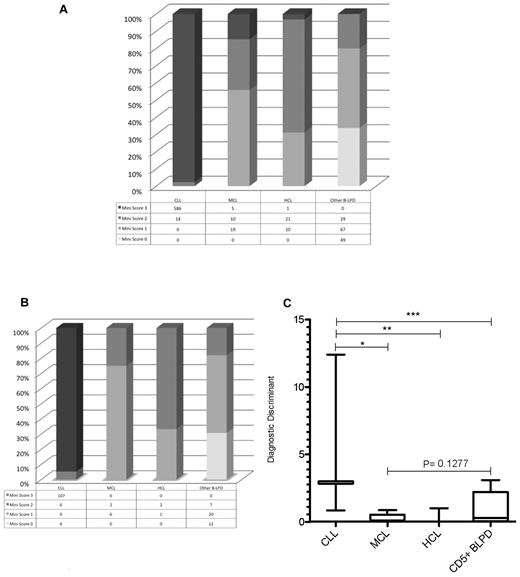
From a test cohort of 811 consecutive cases of mature B-LPD, a “mini-CLL score” was developed using the 3 most consistently expressed markers in CLL: CD5, CD23, and CD160, with each marker scoring one point. A score of 3 identified 586 of 600 cases of CLL (sensitivity = 0.98). Only 6 of 211 non-CLL cases scored 3 (false-positive rate = 0.03; Figure 5A); these included 1 case of HCL and 5 cases of CD23+CD160+ MCL in the leukemic phase. A mini-score of 3 had a very high diagnostic odds ratio of 1430 (CI: 542-3772) for CLL, with a positive predictive value of 0.99. Whereas HCL rarely causes diagnostic confusion with CLL, morphological and immunophenotypical overlap occurs between CLL and MCL in the blood. The latter is characterized by: lymphocytes with clefted nuclei (often having a “fish-mouth” appearance, and are colloquially referred to as “codocytes”), nucleoli, and pleomorphism; immunophenotypically CD5+CD23−sIgM+ (strong); and confirmed by the presence of t(11;14). Although MCL is typically CD23−, in the test cohort, 11 of 34 (32%) cases of MCL expressed CD23 (with all cases were confirmed to be MCL by G banding, FISH, and/or histology), which is similar to other reports.26,27 In contrast, 598 of 600 (> 99%) cases of CLL were CD23+. Because the percentage of CD23 positivity in MCL was lower than that in CLL, we investigated the ratio of the percentage of CD23 to CD5 expression on the malignant B cells (referred to as CD23r) as a parameter to differentiate CLL from CD23+ MCL. In CD23+ MCL cases, 87% had a ratio of 0.80 or less, with 85% of these having a ratio of 0.50 or less. In contrast, of the CLL cases analyzed (n = 431), CD23r was > 0.80 in 97% and only 0.8% had a ratio of 0.50 or less.
Mini-CLL score for B-LPDs and the generation of the diagnostic discriminant to differentiate between CLL and MCL. The mini-CLL score was derived from the CD160FCA, with one point for each marker, CD5, CD23, and CD160. (A) The mini-CLL score within the test cohort (n = 811). (B) The mini-CLL score within the validation cohort (n = 163). (C) The diagnostic discriminant was generated by multiplying the mini-CLL score and CD23r variables. CLL versus MCL (*P < .0001), HCL (**P = .0002), and CD5+ chronic B-cell malignancies (***P < .0001).
Mini-CLL score for B-LPDs and the generation of the diagnostic discriminant to differentiate between CLL and MCL. The mini-CLL score was derived from the CD160FCA, with one point for each marker, CD5, CD23, and CD160. (A) The mini-CLL score within the test cohort (n = 811). (B) The mini-CLL score within the validation cohort (n = 163). (C) The diagnostic discriminant was generated by multiplying the mini-CLL score and CD23r variables. CLL versus MCL (*P < .0001), HCL (**P = .0002), and CD5+ chronic B-cell malignancies (***P < .0001).
The disease-specific expression of the CD5/CD23/CD160 combination was also investigated using the CD160FCA methodology in a validation cohort (n = 163), which confirmed CD160 positivity in CLL (107 of 113 cases, 95%) and the utility of the mini-CLL score: 3 was diagnostic of CLL and excluded all non-CLL cases (P < .0001; sensitivity 0.95, CI: 0.89-0.98; specificity 1.00, CI: 0.93-1.00; diagnostic odds ratio of 1670, CI: 92.23-30 253); whereas a score of 0 excluded CLL, MCL, and HCL (Figure 5B). Further differentiation between B-LPDs was achieved by multiplying the mini-score by CD23r to give a single numerical value, the diagnostic discriminant (Figure 5C). The diagnostic discriminant allowed immunophenotypic separation of CLL from MCL (including leukemic CD23+ MCL), HCL, and other CD5+ B-LPDs.
Monoclonal B-cell lymphocytosis
Monoclonal B-cell lymphocytosis has been used to describe asymptomatic patients with a monoclonal lymphocytosis but < 5000 B cells/μL with a CLL phenotype.24 Within the validation cohort, 13.3% (16 of 113) of CLL cases had a lymphocyte count < 5 × 109/L, of which 15 of 16 expressed the CD160 antigen. The mean percentage of CD160 expression in these 16 cases was 59.5%, similar to that of all CLL in the validation cohort (54.6%; supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Similarly, the MFI of CD160 in the 16 cases was almost identical to that of all CLL within the validation cohort (537.6 and 552.2, respectively; P = nonsignificant; supplemental Figure 1B).
Discussion
CD160 is a cysteine-rich, glycosylphosphatidylinositol-linked membrane protein with a single Ig-like domain and weak homology to the first Ig-C2 domain of the NK receptor KIR2DL4 (22% identity and 44% similarity).5 CD160 has a broad specificity for both classic and nonclassic MHC class I molecules10 and is a ligand for herpes virus entry mediator.28 CD160 is expressed on the majority of circulating NK cells, a subset of cytotoxic T cells, most TCRγδ lymphocytes, a minor subset of circulating CD8bright+TCRαβ cells, and all intestinal intraepithelial T lymphocytes.1,2,5 In the present study, we confirm the presence of CD160 protein on circulating NK and T cells by both flow cytometry and qualitative PCR, with CD160-expressing NK and T cells representing 11% of the total lymphoid population with an MFI significantly higher than other normal lymphocytes (P < 0.0001; Figure 3A). No CD160 expression was detectable in normal B cells, regardless of developmental stage and the tissue of origin. From HSCs through to terminally differentiated plasma cells in normal donor BM as measured in immature B cells (hematogones and pre-B cells in BM), circulating mature B cells and naive CD5+ B1 cells, UCB B cells (rich in CD5+ B1 cells), and lymph node GC B cells; CD160 protein and mRNA were not detected (Figure 2). Therefore, in normal immune cells, CD160 positivity is seen in NK and T cells, but not in B cells.
To date, there has been no report on the expression of CD160 in malignant B cells of chronic B-LPDs. In the present study, we were able to show almost universal expression of CD160 in CLL and HCL in the test and validation sets totaling 974 patients, whereas there was sporadic CD160 positivity in other B-LPDs (Figure 3). The restricted expression of CD160 in malignant, as opposed to normal, B cells indicates that it is a tumor-specific marker for B-cell malignancies. In normal immune cells, CD160 triggering leads to a variety of functional effects, including cytotoxicity, cytokine secretion (IFN-γ, TNF-α, and IL-6) by circulating NK cells (which is regulated by HLA-C11,12 ), and enhancement of CD3-induced proliferation of T cells as a coreceptor in TCR signal transduction.29 Furthermore, in cytotoxic CD8+ T cells, functional cytotoxic activity is limited to those cells that express CD160.2 These observations raise the possibility that the aberrant expression of CD160 in malignant B cells may play a role in the pathophysiological process rather than simply being a marker of malignant transformation. In CLL cells, CD160 was found to mimic the functions described in normal NK and CD8+ T cells: cellular activation, improved in vitro cell survival, and cytokine production (IL-6 and IL-8).15 Moreover, PI3K/Akt signaling was required for CD160-mediated functions in CLL cells,15 similar to that described in normal immune cells, in which Syk and ERK are involved upstream and downstream of PI3K, respectively.14 We conclude that malignant B cells can potentially use aberrant CD160 expression to enhance survival and cellular activation using the same intracellular pathways as normal NK and T cells.
The “aberrant” expression of many proteins in CLL has been described previously and includes the pan T-cell marker CD5 and the TCR-signal transduction molecule ZAP-70,16,17 initially described exclusively in T cells and NK cells.18 The recruitment and phosphorylation of Syk/ZAP-70 tyrosine kinases to the TCR allows differentiation and proliferation.30,31 Similar responses were shown in ZAP-70–expressing CLL cells, with enhanced signal transduction by the BCR.32 In the B-cell lineage, ZAP-70 was initially thought to be a tumor-specific antigen, but subsequent work found that ZAP-70 is not specific to malignant B cells, but is also expressed in activated normal human B cells22 and in naive, GC, memory, cord blood, and peripheral blood B cells.23 Unlike ZAP-70, CD160 expression in malignant B cells appears to be truly “aberrant,” and we have exploited this to study its expression profile and diagnostic utility in acute and chronic B-cell malignancies. In a test cohort of 811 cases, CD160 expression was almost universal in CLL and HCL. This was confirmed in a validation cohort of 163 B-LPDs. Lymph node and splenic tissue from patients with CLL and HCL were also CD160+. Diagnostic immunophenotyping of B-LPDs requires a panel of mAbs with an ever-increasing range of target antigens proposed to help differentiate between the different diseases.33-36 However, the disease-specific CD5/CD23/CD160 combination allowed a simplified “mini-CLL score” to be derived from the single tube CD160FCA assay. A mini-score of 3 differentiated CLL from other B-LPDs, with a very high diagnostic odds ratio of 1430 (range, 542-3772) and a positive and negative predictive value of 99% and 94%, respectively; whereas a score of 0 excluded CLL, MCL, and HCL. However, biologic heterogeneity includes rare CD23− cases of CLL, whereas CD23 positivity in MCL is not uncommon.26,27,37,38 In our test and validation cohorts, 13 of 42 (31%) cases of MCL in leukemic phase were CD23+, whereas 5 MCL cases in the test cohort had a mini-CLL score of 3. The diagnostic discriminant was useful in such cases with an atypical immunophenotype by combining the information from the CD160FCA on the mini-CLL score and the level of CD23 expression (Figure 5C). In the validation cohort, the diagnostic discriminant (mini-score × CD23r) was ≥ 0.50 in all CLL cases and differentiated CLL from leukemic CD23+ MCL and other CD5+ B-LPDs (P < .0001; Figure 5C). We propose the addition of anti-CD160 mAb to existing diagnostic panels, with the diagnostic discriminant giving an additional robust, numerical summation of immunophenotypic data to aid in the diagnosis of B-LPD, particularly cases that are immunologically atypical. Our data provide a scoring system that is simpler than both historical and current systems.39,40 Other molecules have recently been reported to show CLL-restricted expression, including the tumor-associated antigen receptor tyrosine-kinase orphan receptor 1 (ROR1)41 and CD200.36,42 ROR1 mRNA and surface protein were found to be strongly expressed in MCL, as well as positivity in marginal zone lymphoma, B-ALL, and a subset of normal B-cell precursors.43,44 CD200 has an expression profile including CD19+ B cells (normal and malignant), T-cell blasts, follicular dendritic cells, thymocytes, neural tissue, and endothelium.45 Despite its expression in normal tissue, CD200 is up-regulated in CLL, resulting in a down-regulation of the Th1 immune response.42 The weak or absent of expression of CD200 in MCL has been clinically useful in the differentiation between CLL and MCL.36 The CD160 expression shown in the present study is unique compared with both ROR1 and CD200, with absent CD160 expression in normal B cells and specificity for CLL and HCL in malignant B cells.
This study demonstrates the restricted expression of the Ig-like activating NK-cell receptor CD160 in normal immune cells to NK and T cells. Neither protein nor transcript for CD160 was found in normal B cells, from immature BM precursors through to mature peripheral blood and GC B cells. In the B-cell lineage, the restriction of CD160 expression to malignant B cells indicates that it is a tumor-specific antigen and an attractive target for the assessment of minimal residual disease in CD160+ B-LPDs. The CD160 molecule is functional in malignant B cells and delivers survival and activation signals to CLL cells,15 as well as CD160+ cells of the SMZL and HCL variant (data not shown), recapitulating signaling events in normal NK and T cells. The known ligands for CD160 are expressed by both malignant B cells themselves and other cells in the lymphoid microenvironment: MHC class I,10 CD1d,46 HLA-G,10,47 and herpes virus entry mediator.6 We conclude that the interactions of CD160 with its ligands may be important in the pathophysiology of malignant B cells via autocrine, paracrine, and/or stromal cell interactions, offering new targets for therapeutic manipulation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Mr Andrew Clear for his expertise in setting up the immunohistological staining.
This study was supported by a generous donation from the Relgate Investments Ltd Charitable Foundation (Monaco) T.W.F. was funded by a Cancer Research Committee grant awarded by St Bartholomew's Hospital (London) and the London NHS Trust/Queen Mary University of London and a grant awarded by the Institute of Biomedical Science. J.G. was funded by a Research Advisory Board grant awarded by St Bartholomew's Hospital and by the London NHS Trust/Queen Mary University of London.
Authorship
Contribution: T.W.F., J.G., A.B., and S.G.A. designed the research; T.W.F., J.G.G., and S.G.A. performed the research, analyzed the data, and wrote the manuscript; F.-T.L., D.A.T., M.G.M., J.D.C., H.E.O., D.T., A.C.N., M.C., M.J., and J.G.G. performed the research and cowrote the manuscript; and A.B. provided new reagents.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Samir G. Agrawal, Senior Lecturer in Haemato-Oncology, Stem Cell Laboratory, Department of Haematology, St Bartholomew's Hospital, West Smithfield, EC1A 7BE, London, United Kingdom; e-mail: s.g.agrawal@qmul.ac.uk.