Abstract
The CDKN2A locus, which contains the tumor suppressor gene p16INK4a, is associated with an increased risk of age-related inflammatory diseases, such as cardiovascular disease and type 2 diabetes, in which macrophages play a crucial role. Monocytes can polarize toward classically (CAMφ) or alternatively (AAMφ) activated macrophages. However, the molecular mechanisms underlying the acquisition of these phenotypes are not well defined. Here, we show that p16INK4a deficiency (p16−/−) modulates the macrophage phenotype. Transcriptome analysis revealed that p16−/− BM-derived macrophages (BMDMs) exhibit a phenotype resembling IL-4–induced macrophage polarization. In line with this observation, p16−/− BMDMs displayed a decreased response to classically polarizing IFNγ and LPS and an increased sensitivity to alternative polarization by IL-4. Furthermore, mice transplanted with p16−/− BM displayed higher hepatic AAMφ marker expression levels on Schistosoma mansoni infection, an in vivo model of AAMφ phenotype skewing. Surprisingly, p16−/− BMDMs did not display increased IL-4–induced STAT6 signaling, but decreased IFNγ-induced STAT1 and lipopolysaccharide (LPS)–induced IKKα,β phosphorylation. This decrease correlated with decreased JAK2 phosphorylation and with higher levels of inhibitory acetylation of STAT1 and IKKα,β. These findings identify p16INK4a as a modulator of macrophage activation and polarization via the JAK2-STAT1 pathway with possible roles in inflammatory diseases.
Introduction
The tumor suppressor p16INK4a is encoded by the CDKN2A locus on the human chromosome 9p21 and on the murine chromosome 4. p16INK4a belongs to the INK4 family of cyclin-dependent kinase (CDK) inhibitors, also including p15INK4b, p18INK4c, and p19INK4d.1-5 p16INK4a inhibits cell-cycle progression by preventing cyclin D–CDK 4/6 complex formation. As a consequence, pRb hyperphosphorylation and its association with E2F, which induces transcription of S-phase genes, are inhibited. p16INK4a inactivation by deletion, point mutation, or promoter methylation, occurs frequently in most tumors.6
Besides its role in cancer as an inhibitor of cell-cycle progression, p16INK4a plays a crucial role in senescence and aging.7,8 Indeed, expression of p16INK4a increases with age in various tissues from several species.9-11 A genome-wide association study has shown association of the CDKN2A locus with an increased risk of the age-related frailty syndrome.12 In addition, increased p16INK4a expression causes the age-dependent decline in proliferation of self-renewing cellular compartments such as HSCs,13 which give rise to immune cells.
Although the role of p16INK4a in mature immune cells has not yet been investigated, several studies has shown that the CDKN2A locus is associated with an increased risk for coronary heart disease,14 atherosclerosis,15 and type 2 diabetes (T2D).16 In these pathologies, immune cells, such as macrophages, play a crucial role. Besides their pleiotropic immune functions, macrophages also play a role in the development and homeostasis of several tissues, such as adipose tissue17 and liver.18 Depending on the cytokine environment, macrophages differentiate into distinct subclasses with specific characteristics. Classically activated macrophages (CAMφ) differentiate in presence of Th1 cytokines, such as IFNγ, or in presence of bacterial products such as lipopolysaccharide (LPS). CAMφ trigger proinflammatory responses required to kill intracellular pathogens.19 Alternatively activated macrophages (AAMφ), induced by Th2 cytokines such as IL-4 and IL-13, are associated with Th2-type immune responses as seen in helminth parasite infections.19 During inflammation, AAMφ play a key role in protecting the organism against tissue damage.20 However, little is known about the mechanisms underlying the acquisition of the AAMφ phenotype.
In the present study, we investigated whether p16INK4a deficiency influences macrophage activation in vitro, using BM-derived macrophages (BMDMs), and in vivo by infection with the parasite Schistosoma mansoni. BMDMs from p16INK4a-deficient (p16−/−) mice exhibit a phenotype resembling IL-4–induced macrophage polarization and an enhanced response to the Th2 cytokine IL-4 compared with BMDMs from wild-type (p16+/+) mice. By contrast, their response to Th1 stimuli is diminished. Moreover, S mansoni infection of mice transplanted with p16−/− BM resulted in an increased hepatic AAMφ signature in vivo. Finally, we show that p16INK4a deficiency does not influence the IL-4–induced STAT6 pathway. By contrast, the lower response to classic activation stimuli likely occurs through an inhibition of the phosphorylation of both STAT1 and IKKα,β, which correlated with decreased JAK2 phosphorylation and increased acetylation of STAT1 and IKKα,β.
Methods
Mice
C57BL/6J p16INK4a-deficient and littermate control mice on a C67BL/6J background (> 97%) were kindly provided by P. Krimpenfort (Netherlands Cancer Institute, Amsterdam). p16−/− mice showed neither abnormalities in biochemical parameters nor spontaneous tumor development at the studied age. Flow cytometric analysis of splenic T and B cells or macrophages showed no differences between p16−/− and p16+/+ mice (data not shown). All protocols were conducted with the approval of the Pasteur Institute ethical review board (Lille, France).
BM chimeras
Eight-week-old C57BL/6J mice were lethally irradiated (8 Gy) and injected in the tail vein the next day with 10 × 106 BM cells isolated from p16−/− or p16+/+ donor mice. Mice were supplied with autoclaved acidified water (pH = 2) supplemented with neomycin 100 mg/L (Sigma-Aldrich), polymyxin B sulfate 60 000 U/L (Invitrogen) 1 week before and 4 weeks after transplantation. Mice were studied 6 weeks posttransplantation to allow repopulation by the donor BM. To ensure that donor BM replaced the resident blood cell population, DNA was extracted from whole blood with an Illustra blood kit (GE Healthcare). PCR was performed with forward 5′-GCA GTG TTG CAG TTT GAA CCC-3′ and reverse 5′-TGT GGC AAC TGA TTC AGT TTG-3′ primer sets yielding products of different lengths, which were separated on a 1.5% agarose gel and quantified with the Gel Doc XR system (Bio-Rad). More than 95% of host blood cells were from donor origin.
Primary cell isolation and culture
LPS-elicited neutrophils from air pouches were isolated as described.21 Splenic CD4+ T cells and CD43− B cells were isolated from female mice, respectively using Dynal (Invitrogen), biotin-conjugated anti-CD43 (BD Biosciences) and streptavidin conjugated to magnetic beads (Miltenyi Biotec) according to the manufacturer's instructions. BM-derived dendritic cells (DCs) were prepared as described.22 BMDMs were obtained from femoral and tibial BM suspensions, plated at 10 × 106 cells in 10-cm plates and differentiated in BMDM medium (RPMI 1640 containing HEPES 25mM supplemented with 10% low endotoxin FBS, 15% L929-conditioned medium, 2mM l-glutamine, 1mM gentamycine). Stimulations and microarray analysis were carried out at day 8. IL-4 (15 ng/mL) and IFNγ (2.5 ng/mL and 10 ng/mL) were obtained from PeproTech. LPS (100 ng/mL) from Escherichia coli serotype 0111B4 was from Sigma-Aldrich. CINK4 (no. 219492) was obtained from Merck. For indirect coculture experiments, conditioned medium of alternatively differentiated p16+/+ and p16−/− macrophages was added to cultures of BMDMs 24 hours before LPS (100 ng/mL) stimulation. After 4 hours, CAMφ were washed and cultured for 24 hours in fresh medium. The resulting media were assayed for cytokine secretion by ELISA.
Flow cytometry
During in vitro differentiation, asynchronously growing cultures of BMDMs were gently scraped every 2 days with 1× PBS/10mM EDTA/2% trypsine, washed with PBS and collected by centrifugation (5 minutes, 146g). For protein analysis, nonspecific staining was avoided by incubating cells with purified rat anti–mouse CD16/CD32 (BD Pharmingen) for 20 minutes at 4°C and labeled with anti–mouse F4/80-FITC (Caltag Laboratories) for 30 minutes at 4°C. Cells were analyzed using a Coulter EPICS XL4-MCL Flow cytometer (Beckman Coulter) and Expo32 software (Applied Cytometry Systems). For cell sorting, cells were incubated with anti–mouse F4/80-FITC Abs (11-4801; eBioscience) and rat IgG2a K isotype (11-4321; eBioscience) and sorted on an EPICS-ALTRA cell sorter (Beckman Coulter). For FACS-based cell-cycle analysis, cells were fixed with 70% ethanol for 1 hour at −20°C and labeled with 50 μg/mL propidium iodide containing 50 μg/mL RNase A for 30 minutes at room temperature. The percentage of cell-cycle distribution was obtained using the Multicycle software (Phoenix Flow Systems).
Microarray studies
Mouse genome MOE 430 2.0 gene chips (Affymetrix) were used in triplicate for each condition. Sample preparation, hybridization, and scanning were performed according the one-cycle protocol (Affymetrix). Only genes significantly regulated (P < .05) were analyzed further with Genomatix Bibliosphere. Microarray data have been deposited to http://www.ebi.ac.uk/arrayexpress under accession number E-MEXP-3216.
RNA extraction and analysis
Total RNA was obtained from BMDMs using EXTRACT-ALL, DNase-I treated, and cDNA was generated using cDNA reverse transcription kit (Applied Biosystems). Real-time quantitative PCR (QPCR) detection was conducted on a Stratagene Mx3005P (Agilent technologies) using Brilliant II SYBR Green reagent (Agilent Technologies) and specific primers (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). mRNA levels were normalized to 36B4 mRNA and fold induction was calculated using the 2−ΔΔCt method.
Cytokine production measurement
IL-6, TNF, IL-1Rn, IL-4, IL-10, and IFNγ levels in culture supernatants were measured with ELISA kits according to the manufacturer's instructions (R&D Systems). Plasma IgE from infected animals was determined by ELISA using purified rat anti–mouse IgE (BD Pharmingen) as a capture Ab, and biotin rat anti–mouse IgE as a detection Ab (BD Pharmingen). Standards were prepared using purified mouse IgE κ isotype control (BD Pharmingen).
Protein extraction and Western blotting
BMDM lysates were prepared using cell lysis buffer (Cell Signaling Technology) supplemented with 1mM PMSF. Lysates were resolved in 16% or 10% SDS-PAGE and analyzed by Western blot. Anti-p16 (sc-1207), -actin (sc-1616), and -p38 (sc-535) Abs were purchased from Santa Cruz Biotechnology. Anti–phospho-STAT6 and -STAT6 (nos. 9361 and 9362), anti–phospho-STAT1 and -STAT1 (nos. 9171 and 9172), anti–phospho-p38 (no. 9211), anti–phospho-IKKα,β (no. 2681), anti-IκBα (no. 9242), and anti–phospho-JAK2 and -JAK2 (nos. 3771 and 3230) Abs were purchased from Cell Signaling Technology. Hybridizations were done 1:1000 in 5% BSA, 1× TBS, 0.1% Tween 20 overnight at 4°C. Anti–rabbit IgG peroxidase-conjugate secondary Ab 1:10 000 (Sigma-Aldrich) was added for 1 hour at room temperature. Signals were visualized using the enhanced chemiluminescence Western blot detection kit (Amersham ECL plus Western blotting Detection System; GE Healthcare). Membranes were stripped with Reblot Plus Strong Antibody Stripping Solution (Millipore) and reblotted with the indicated Abs.
Immunoprecipitation assay
Five hundred micrograms of BMDM lysates were prepared using lysis buffer supplemented with trichostatin A 1μM (Sigma-Aldrich). Lysates were mixed with 100 μL of protein G/protein A agarose (Millipore) and rotated for 1 hour at 4°C. After centrifugation for 2 minutes at 900g, anti-acetyl-Lysine Ab 1:100 (ab21623; Abcam) was added. Samples were rotated overnight at 4°C. The protein G/protein A agarose mix (100 μL) was added to the samples and incubated for 1 hour at 4°C. The beads were washed 3 times and the pellets dissolved in 40 μL of 2× Laemmli buffer. Proteins were analyzed by Western blotting as previously described.
S mansoni infection
Chimeric mice were anesthetized and percutaneously infected with 40 S mansoni cercariae and killed 9 weeks later. The liver was perfused and adult worms were recovered and quantified. A piece of liver was weighed and dissolved in a KOH solution (5%, 37°C during 16 hours) and eggs were counted. Organs were snap-frozen or fixed in paraformaldehyde for further analysis. Resin-embedded livers were cut at 5 μm and Mallory trichrome staining was performed to determine granuloma size and hepatic collagen. Immunostaining was performed using Abs directed to Moma-2 (sc59332; Santa Cruz Biotechnology), Arginase1 (Arg-I; ab60176-100), Fizz1 (ab39626-50), and Ym1 (ab93034) from Abcam. May-Grunwald-Giemsa staining was performed and eosinophils were counted per granuloma area. Granulomas were isolated using an Arcturus laser capture microdissection system from 14-μm sections of snap-frozen liver samples. For ex vivo restimulation, spleens were homogenized, filtered (70 μm) and erythrocytes were lysed. Cells were cultured in RPMI 1640 (Invitrogen) supplemented with 10% low endotoxin FBS and 1mM gentamycin. Cells (5 × 105) were stimulated with 10 μg/mL soluble egg Ag (SEA) or anti-CD3 (no. 555274; BD Pharmingen) Abs during 4 days after which medium was collected for cytokine determination by ELISA.
Statistical analysis
Groups were compared using 2-tailed nonpaired t tests using GraphPad Prism software. In vitro data are expressed as means ± SD and in vivo data are expressed as means ± SEM.
Results
p16INK4a expression increases during macrophage maturation, but p16INK4a deficiency does not modulate macrophage cell-cycle progression
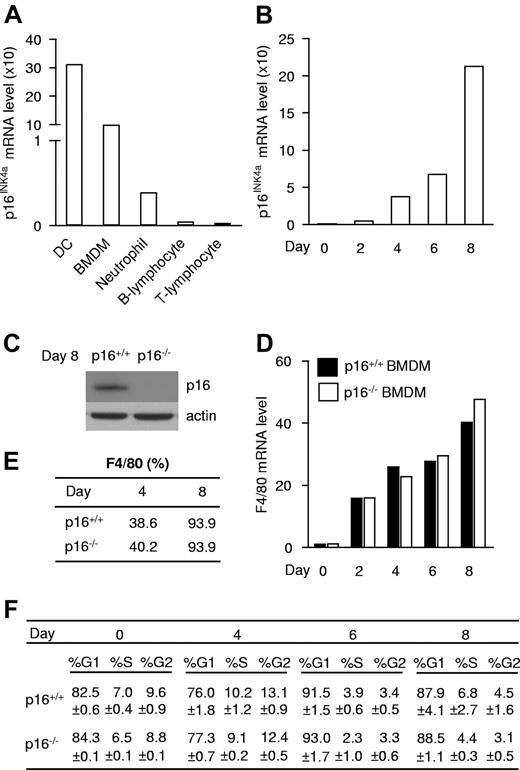
To assess whether and in which mature immune cell type p16INK4a is expressed, p16INK4a mRNA was measured in DCs, BMDMs, and neutrophils, all of myeloid origin, and in B and T lymphocytes. p16INK4a was highly expressed in BMDMs and DCs, while its expression was markedly lower in neutrophils and virtually absent in lymphocytes (Figure 1A). Because the CDKN2A locus has been associated with diseases with inflammatory components in which macrophages play a central role, we focused on the role of p16INK4a in these cells. BMDMs displayed a gradual increase in p16INK4a mRNA expression during in vitro differentiation (Figure 1B). Cell-sorting analysis showed that p16INK4a mRNA expression increased specifically in F4/80-positive cells (macrophages; supplemental Figure 1). Together, these data show that increased expression occurs in a higher number of macrophages which also express higher p16INK4a levels. In line, p16INK4a protein was expressed in differentiated macrophages (Figure 1C). Absence of p16INK4a (Figure 1C) did not influence macrophage maturation because it affected neither mRNA (Figure 1D) nor protein (Figure 1E) expression of the macrophage surface marker F4/80 during differentiation. Interestingly, absence of p16INK4a did not affect cell-cycle progression at any time point of in vitro macrophage differentiation (Figure 1F). Expression analysis of the INK4 family members p15INK4b, p18INK4c, p19INK4d, and p19ARF, another product of the INK4A/ARF locus which inhibits the p53 pathway, as well as the KIP/CIP family members p21WAF1 and p27KIP1, revealed increased expression of p15INK4b in p16INK4a-deficient (p16−/−) BMDMs (supplemental Figure 2). Together with the observation that expression of Cyclin D, a direct E2F target gene, is similar in both genotypes (supplemental Figure 3), these data suggest that the induction of p15INK4b may compensate for p16INK4a deficiency in the control of the cell cycle. These data indicate that p16INK4a expression increases during macrophage differentiation and that p16INK4a deficiency influences neither maturation nor the cell cycle in BMDMs.
p16INK4a is expressed, but does not influence maturation or the cell cycle in macrophages. (A) p16INK4a mRNA expression was measured in immune cells isolated from mice: dendritic cells (DC), BM-derived macrophages (BMDMs), neutrophils, B lymphocytes, and T lymphocytes. (B) p16INK4a mRNA expression was measured at different stages during differentiation of BMDM isolated from p16+/+ mice. Because p16INK4a fold induction differs between experiments, data are from 1 of 3 distinct experiments. (C) p16INK4a protein expression in p16+/+ and p16−/− BMDM was analyzed by Western blot with an anti-p16INK4a Ab. Anti-actin Ab was used as loading control. (D-E) Increase of the macrophage marker F4/80 mRNA expression (D) and protein expression (E) measured by, respectively, QPCR and flow cytometry in p16+/+ and p16−/− BMDM. Data are shown from 1 of 3 independent experiments. (F) Cell-cycle analysis by propidium iodide staining of p16+/+ and p16−/− BMDM during differentiation expressed in % of events ± SD.
p16INK4a is expressed, but does not influence maturation or the cell cycle in macrophages. (A) p16INK4a mRNA expression was measured in immune cells isolated from mice: dendritic cells (DC), BM-derived macrophages (BMDMs), neutrophils, B lymphocytes, and T lymphocytes. (B) p16INK4a mRNA expression was measured at different stages during differentiation of BMDM isolated from p16+/+ mice. Because p16INK4a fold induction differs between experiments, data are from 1 of 3 distinct experiments. (C) p16INK4a protein expression in p16+/+ and p16−/− BMDM was analyzed by Western blot with an anti-p16INK4a Ab. Anti-actin Ab was used as loading control. (D-E) Increase of the macrophage marker F4/80 mRNA expression (D) and protein expression (E) measured by, respectively, QPCR and flow cytometry in p16+/+ and p16−/− BMDM. Data are shown from 1 of 3 independent experiments. (F) Cell-cycle analysis by propidium iodide staining of p16+/+ and p16−/− BMDM during differentiation expressed in % of events ± SD.
Macrophage p16INK4a deficiency leads to a phenotype resembling IL-4–induced macrophage polarization
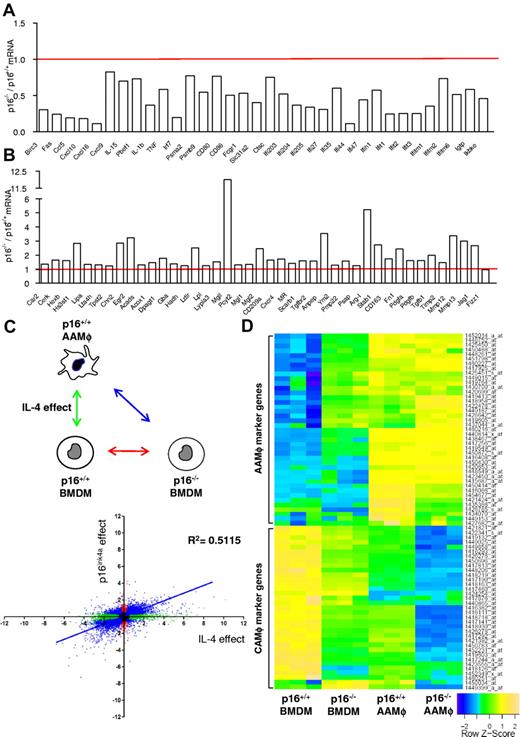
Microarray analysis of p16−/− BMDMs revealed that the expression of a large number of inflammatory genes was significantly lower compared with wild-type (p16+/+) BMDMs under basal conditions (Figure 2A). Several of these genes are typically associated with a CAMφ phenotype.23-27 By contrast, several AAMφ-associated genes, linked to IL-4–induced STAT6 activation,23-27 were significantly higher expressed in p16−/− BMDMs (Figure 2B). However, the AAMφ markers Ym1/2 and Fizz1 were not spontaneously induced by p16INK4a deficiency. The differential expression of several genes was confirmed by QPCR (supplemental Table 2).
p16INK4a deficiency induces a gene expression profile resembling IL-4–induced macrophage polarization. Microarray analysis using mRNA from p16−/− BMDM compared with p16+/+ BMDM showed (A) decreased mRNA expression of classically activated macrophage-associated genes and (B) increased mRNA expression of alternatively activated macrophage-associated genes. Data are expressed as fold change relative to p16+/+ BMDM. (C) Differential gene expression in p16−/− BMDM relative to p16+/+ BMDM was correlated with the changes induced in IL-4–induced p16+/+ AAMφ. The figure shows 2 log values of the probesets significantly (P < .05) regulated only in p16−/− BMDM (red dots), only in IL-4–polarized p16+/+ AAMφ (green dots) and by both conditions (blue dots), compared with p16+/+ BMDM. The x-axis represents differences in gene expression induced by IL-4, whereas the y-axis represents the effect of p16INKa deficiency. These comparisons are depicted in the schematic representation of the protocol in the corresponding colors. Pearson correlation analysis was done for probesets differentially expressed by both conditions (blue). (D) Heat map of p16+/+ BMDM, p16−/− BMDM, IL-4–polarized p16+/+, and p16−/− AAMφ gene expression profiles. Colors fluctuate from blue (poorly expressed) to green (intermediate expression) and yellow (high expression). Additional information regarding gene description, fold induction, and P value can be found in supplemental Table 3.
p16INK4a deficiency induces a gene expression profile resembling IL-4–induced macrophage polarization. Microarray analysis using mRNA from p16−/− BMDM compared with p16+/+ BMDM showed (A) decreased mRNA expression of classically activated macrophage-associated genes and (B) increased mRNA expression of alternatively activated macrophage-associated genes. Data are expressed as fold change relative to p16+/+ BMDM. (C) Differential gene expression in p16−/− BMDM relative to p16+/+ BMDM was correlated with the changes induced in IL-4–induced p16+/+ AAMφ. The figure shows 2 log values of the probesets significantly (P < .05) regulated only in p16−/− BMDM (red dots), only in IL-4–polarized p16+/+ AAMφ (green dots) and by both conditions (blue dots), compared with p16+/+ BMDM. The x-axis represents differences in gene expression induced by IL-4, whereas the y-axis represents the effect of p16INKa deficiency. These comparisons are depicted in the schematic representation of the protocol in the corresponding colors. Pearson correlation analysis was done for probesets differentially expressed by both conditions (blue). (D) Heat map of p16+/+ BMDM, p16−/− BMDM, IL-4–polarized p16+/+, and p16−/− AAMφ gene expression profiles. Colors fluctuate from blue (poorly expressed) to green (intermediate expression) and yellow (high expression). Additional information regarding gene description, fold induction, and P value can be found in supplemental Table 3.
To further characterize this IL-4–like phenotype of p16−/− BMDMs, the gene signature of these cells was compared with the gene signature of p16+/+ BMDMs polarized in vitro by addition of IL-4 from day 0 of differentiation, generating p16+/+ AAMφ.28 Compared with p16+/+ BMDMs, 14 311 probesets were differentially expressed in p16+/+ AAMφ (Figure 2C green dots), whereas 7605 probesets were regulated in p16−/− BMDMs (Figure 2C red dots), of which the majority (78%) was also differentially expressed in p16+/+ AAMφ (Figure 2C blue dots). In addition, a strong correlation (P < .0001; Pearson R = 0.7; R2 = 0.5) was found between the 2 experimental conditions (Figure 2C), as also illustrated by a heat map representation of a selection of AAMφ and CAMφ marker genes (Figure 2D, supplemental Table 3).
Because p16−/− BMDMs displayed lower expression levels of inflammatory genes (Figure 2A,D), the secretion of inflammatory cytokines produced in p16−/− BMDMs compared with p16+/+ BMDMs was measured. Under basal conditions, secretion of the pro-inflammatory cytokines IL-6 and TNF was lower (Figure 3A-B) in p16−/− BMDMs. By contrast, secretion of anti-inflammatory IL-1Rn was, albeit modestly, higher (Figure 3C). These observations are in line with the microarray data and suggest that p16−/− BMDMs are functionally different from their wild-type counterparts.
p16INK4a-deficient macrophages phenotypically and functionally resemble IL-4–polarized alternatively activated macrophages. Protein secretion of IL-6 (A), TNF (B), and IL-1Rn (C) was measured in the culture medium of p16+/+ and p16−/− BMDM by ELISA. (D-F) QPCR analysis of Arg-I (D), Ym1/2 (E), and Mgl2 (F) of p16+/+ and p16−/− AAMφ polarized by addition of 15 ng/mL IL-4 from day 0 of differentiation. (G) Schematic representation of the indirect coculture experiment. (H-I) LPS-induced secretion of IL-6 (H), and TNF (I) was determined by ELISA in p16+/+ BMDM supernatants resulting from indirect coculture. Statistically significant differences are indicated (t test; ***P < .001, **P < .01, and *P < .05).
p16INK4a-deficient macrophages phenotypically and functionally resemble IL-4–polarized alternatively activated macrophages. Protein secretion of IL-6 (A), TNF (B), and IL-1Rn (C) was measured in the culture medium of p16+/+ and p16−/− BMDM by ELISA. (D-F) QPCR analysis of Arg-I (D), Ym1/2 (E), and Mgl2 (F) of p16+/+ and p16−/− AAMφ polarized by addition of 15 ng/mL IL-4 from day 0 of differentiation. (G) Schematic representation of the indirect coculture experiment. (H-I) LPS-induced secretion of IL-6 (H), and TNF (I) was determined by ELISA in p16+/+ BMDM supernatants resulting from indirect coculture. Statistically significant differences are indicated (t test; ***P < .001, **P < .01, and *P < .05).
Next, we compared the response of p16−/− vs p16+/+ BMDMs to polarization into AAMφ induced by IL-4 added at the start (day 0) of differentiation. Induction of the AAMφ marker genes Arg-I (Figure 3D) and Ym1/2 (Figure 3E) was higher in p16−/− AAMφ. However, induction of Mgl2 (Figure 3F) was not different in these cells, suggesting that the IL-4 responsiveness of certain, but not all, genes is dependent on p16INK4a. This heterogeneity in regulation of AAMφ marker genes in IL-4–polarized p16−/− AAMφ confirms the microarray results (Figure 2D).
Because p16−/− macrophages display increased AAMφ marker expression after IL-4–induced polarization and because AAMφ exert paracrine anti-inflammatory effects,29 the functional effects of p16INK4a deficiency on IL-4–induced macrophage polarization were analyzed by indirect coculture experiments. Hereto, conditioned medium from IL-4–polarized p16−/− or p16+/+ AAMφ was added to p16+/+ BMDMs and their response to LPS was measured (Figure 3G). Conditioned medium from p16−/− AAMφ resulted in a more pronounced inhibition of LPS-induced secretion of IL-6 and TNF (Figure 3H-I). Thus, medium from p16−/− AAMφ more potently inhibits proinflammatory responses.
Collectively, these results evidence that p16INK4a deficiency results in a phenotype partially resembling IL-4–induced macrophage polarization. Moreover, IL-4–polarized p16−/− macrophages display a more pronounced AAMφ phenotype.
p16INK4a-deficient macrophages are more responsive to IL-4, while they respond less to IFNγ or LPS
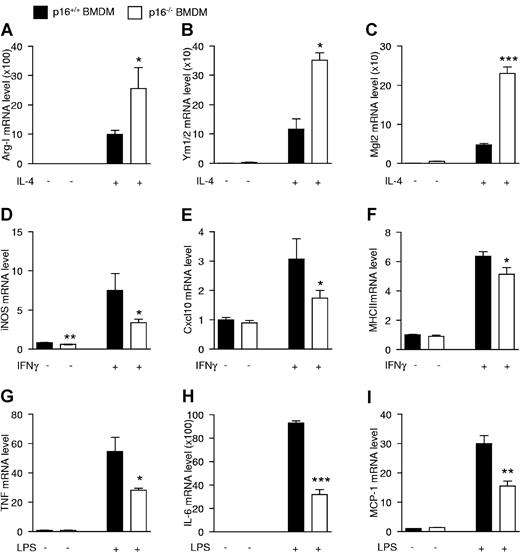
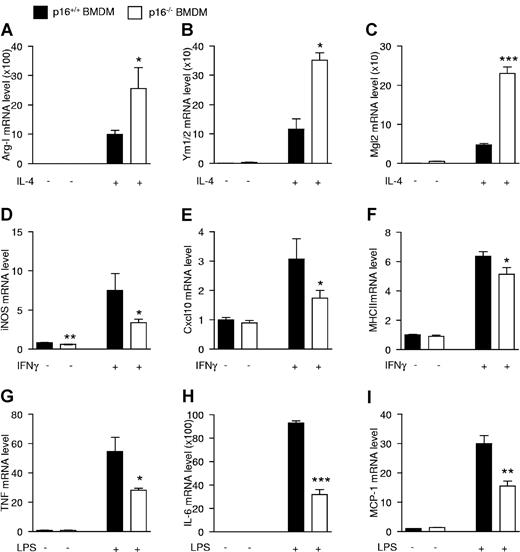
Because p16−/− BMDMs resemble IL-4–polarized macrophages and because p16−/− AAMφ show more potent anti-inflammatory effects than wild-type macrophages, the response of p16−/− BMDMs to acute (24 hours) IL-4 activation was assessed. Interestingly, IL-4 treatment resulted in a larger increase of Arg-I, Ym1/2, and Mgl2 mRNA in p16−/− compared with p16+/+ BMDMs (Figure 4A-C).
p16INK4a deficiency modulates macrophage responses to alternatively and classically polarizing stimuli. mRNA expression from p16+/+ and p16−/− BMDM activated with 15 ng/mL IL-4 during 24 hours (A-C), 10 ng/mL IFNγ during 12 hours (D-F), or 100 ng/mL LPS during 2 hours (G) or 24 hours (H-I). mRNA expression of the IL-4–induced target genes Arg-I (A), Ym1/2 (B), and Mgl2 (C); IFNγ-induced target genes iNOS (D), Cxcl10 (E), and MHCII (F); and LPS-induced target genes TNF (G), IL-6 (H), and MCP-1 (I) was quantified by QPCR and expressed as fold increase compared with respective controls. Statistically significant differences are indicated (t test; ***P < .001 **P < .01, and *P < .05).
p16INK4a deficiency modulates macrophage responses to alternatively and classically polarizing stimuli. mRNA expression from p16+/+ and p16−/− BMDM activated with 15 ng/mL IL-4 during 24 hours (A-C), 10 ng/mL IFNγ during 12 hours (D-F), or 100 ng/mL LPS during 2 hours (G) or 24 hours (H-I). mRNA expression of the IL-4–induced target genes Arg-I (A), Ym1/2 (B), and Mgl2 (C); IFNγ-induced target genes iNOS (D), Cxcl10 (E), and MHCII (F); and LPS-induced target genes TNF (G), IL-6 (H), and MCP-1 (I) was quantified by QPCR and expressed as fold increase compared with respective controls. Statistically significant differences are indicated (t test; ***P < .001 **P < .01, and *P < .05).
AAMφ are known to be less sensitive to inflammatory stimuli.19 Therefore, the effects of IFNγ and LPS, 2 proinflammatory stimuli, were tested on these cells. Interestingly, p16−/− BMDMs were less responsive to IFNγ, shown by the lower induction of the IFNγ target genes iNOS, Cxcl10, and MHCII (Figure 4D-F), as well as to LPS, shown by the decreased response of the LPS-regulated genes TNF, IL-6, and MCP-1 (Figure 4G-I). Expression levels of the cognate receptors for IL-4, IFNγ, and LPS was similar in both genotypes (supplemental Figure 4A-C), suggesting that the altered responses were not because of differences in receptor expression.
Collectively, these data show that p16−/− BMDMs are more responsive to alternative activation by IL-4 and less responsive to the classic macrophage activators IFNγ and LPS.
Hematopoietic p16INK4a deficiency increases hepatic expression of alternatively activated macrophage markers on S mansoni infection
Because helminth parasite infections induce a strong Th2 immune response resulting in AAMφ differentiation,20,30 the influence of p16INK4a deficiency on macrophage polarization in vivo was investigated during S mansoni infection. To assess the role of p16INK4a specifically in hematopoietic cells, p16+/+ and p16−/− BM was transplanted in lethally irradiated wild-type recipients. Mice were killed 9 weeks after infection, corresponding to the peak of the Th2 immune response, and the hepatic pathology was subsequently characterized.
Expression of Th2 and Th1 cytokines in livers (supplemental Figure 5A-D) and spleens (supplemental Figure 6A-C), hepatic infection parameters (Table 1), plasma IgE levels (supplemental Figure 7), eosinophil infiltration (Table 1), as well as mortality and weight loss (data not shown) did not differ between p16−/− and p16+/+ transplanted animals. These results indicate that there are no differences in the lymphocyte-dependent Th2 response, in line with the very low expression of p16INK4a in T cells (Figure 1A).
Because macrophages express significant levels of p16INK4a (Figure 1A) and because these cells are abundant in S mansoni–induced granulomas,20 macrophage markers were measured in the livers of infected mice. Hepatic mRNA levels of F4/80, CD68, and CD14 were equally induced on infection (Figure 5A-C), indicating that macrophage numbers were not modified. However, the induction of the AAMφ markers Ym1/2 (Figure 5D), Fizz1 (Figure 5E), and Mgl2 (Figure 5F) was higher in mice transplanted with p16−/− versus p16+/+ BM. Moreover, expression of the tissue remodeling markers αSMA and Timp1 (supplemental Figure 5E-F) was higher in mice transplanted with p16−/− BM, consistent with the tissue remodeling function of AAMφ.31
Hematopoietic deficiency of p16INK4a exacerbates hepatic alternative macrophage activation of S mansoni–infected mice. Hepatic mRNA expression from mice transplanted with either p16+/+ or p16−/− BM which were not infected (naive) or infected with S mansoni, 9 weeks postinfection. mRNA expression of the macrophage markers F4/80 (A), CD68 (B), and CD14 (C); AAMφ markers Ym1/2 (D), Fizz1 (E), and Mgl2 (F) was quantified by QPCR and expressed as fold increase compared with their respective naive controls. Data were obtained from n = 10 naive and n = 14 infected mice per genotype. Hepatic granulomas were isolated by laser-capture microdissection and mRNA expression of the AAMφ markers Ym1/2 (G), Fizz1 (H), and Mgl2 (I) was quantified by QPCR. Statistically significant differences are indicated (t test; **P < .01 and *P < .05).
Hematopoietic deficiency of p16INK4a exacerbates hepatic alternative macrophage activation of S mansoni–infected mice. Hepatic mRNA expression from mice transplanted with either p16+/+ or p16−/− BM which were not infected (naive) or infected with S mansoni, 9 weeks postinfection. mRNA expression of the macrophage markers F4/80 (A), CD68 (B), and CD14 (C); AAMφ markers Ym1/2 (D), Fizz1 (E), and Mgl2 (F) was quantified by QPCR and expressed as fold increase compared with their respective naive controls. Data were obtained from n = 10 naive and n = 14 infected mice per genotype. Hepatic granulomas were isolated by laser-capture microdissection and mRNA expression of the AAMφ markers Ym1/2 (G), Fizz1 (H), and Mgl2 (I) was quantified by QPCR. Statistically significant differences are indicated (t test; **P < .01 and *P < .05).
Immunohistochemical analysis showed that the parasite-induced granulomas are highly macrophage-enriched (supplemental Figure 8). Moreover, Arg-I, Fizz1, and Ym1 colocalized with the general macrophage marker Moma-2. Surprisingly, the macrophage population within the granulomas appeared heterogeneous because these markers did not entirely colocalize (supplemental Figure 8). Nevertheless, these data suggest that the increase in hepatic AAMφ marker expression occurs mainly in macrophages. To unequivocally demonstrate this, the macrophage-positive areas of the granulomas were isolated by laser-capture microdissection from p16−/− and p16+/+ BM transplanted mice and mRNA levels were measured. p16−/− granulomas displayed increased Mgl2 and Fizz1 mRNA levels, whereas Ym1/2 mRNA tended to be increased (Figure 5G-I). F4/80, CD68, and CD14 expression were identical, indicating that macrophage numbers did not differ between genotypes (data not shown).
Because macrophage polarization influences T-cell activation,20,32 the ex vivo response of splenocytes from mice transplanted with p16+/+ or p16−/− BM to SEA was measured. Ag-independent stimulation of naive splenocytes by anti-CD3 showed similar IL-4 production (data not shown) suggesting an equal ability of T cells to synthesize Th2 cytokines, regardless of macrophages within the splenocyte preparation. By contrast, Ag-dependent restimulation with SEA showed a lower production of both IL-4 and IFNγ in splenocytes from p16−/− transplanted mice (supplemental Figure 6D-E), while IL-10 production was similar (supplemental Figure 6F), hence resulting in an unchanged IL-4/IFNγ balance.33 Collectively, these results provide evidence for a role of p16INK4a in AAMφ skewing in vivo.
p16INK4 deficiency in macrophages decreases STAT1 and IKKα,β signaling without altering STAT6 phosphorylation
To unravel the mechanisms underlying the phenotype induced by p16INK4a deficiency, the microarray data were analyzed to identify which pathways are affected. Interestingly, many genes with a decreased expression in p16−/− BMDMs respond to activation of the IFNγ and NF-κB inflammatory signaling pathways (supplemental Figure 9), of which STAT1 and IKKα,β, respectively, are crucial components. Detailed examination of these gene expression patterns revealed that expression levels were comparable (Cxcl10, ifi27, ifi203, ifi47, ifit1, Ccl5, Irak3, Birc3, and Ly6e) or even lower (ifi44, CD55, and NFκBia) in p16−/− BMDMs under basal conditions compared with IL-4–polarized p16+/+ AAMφ (Figure 6). Moreover, most inflammatory genes were more strongly inhibited in IL-4–induced p16−/− AAMφ than in p16+/+ AAMφ (Figures 6 and 2D), suggesting an additive effect of p16INK4a deficiency on IL-4 treatment. By contrast, such a synergism could not be observed with respect to the AAMφ marker genes because many of these AAMφ marker genes responded similarly in IL-4–induced p16−/− and p16+/+ AAMφ (supplemental Table 4, Figure 2D). Collectively, these data show that the pathways controlling IFNγ, NF-κB, and IL-4 signaling are affected by p16INK4a deficiency in macrophages.
p16INK4a deficiency results in an alteration of STAT1 and NF-κB signaling. Representation of the relative microarray intensity values from a selection of down-regulated genes in p16+/+ and p16−/− BMDM with or without polarization (AAMφ) by 15 ng/mL IL-4 from day 0 of differentiation. Statistically significant differences are indicated (a: P < .05 compared with p16+/+ BMDM; b: P < .05 compared with p16−/− BMDM; c: P < .05 compared with p16+/+ AAMφ).
p16INK4a deficiency results in an alteration of STAT1 and NF-κB signaling. Representation of the relative microarray intensity values from a selection of down-regulated genes in p16+/+ and p16−/− BMDM with or without polarization (AAMφ) by 15 ng/mL IL-4 from day 0 of differentiation. Statistically significant differences are indicated (a: P < .05 compared with p16+/+ BMDM; b: P < .05 compared with p16−/− BMDM; c: P < .05 compared with p16+/+ AAMφ).
To determine whether the observed differences were related to indirect effects through pRb, BMDMs were treated with CINK4, a potent inhibitor of pRb hyperphosphorylation.34 CINK4 treatment resulted in a dose-dependent decrease of cyclin D mRNA levels both in p16+/+ and p16−/− BMDMs (supplemental Figure 3). Furthermore, treatment with CINK4 in combination with IFNγ did not influence the different phenotype between the 2 genotypes (supplemental Figure 10), thus excluding a role for pRb therein.
STAT6 is the major transcription factor activated by IL-4. Surprisingly, IL-4 treatment induced STAT6 phosphorylation to a similar extent in p16−/− and p16+/+ BMDMs (Figure 7A). By contrast, IFNγ-induced STAT1 phosphorylation (Figure 7B) and LPS-induced IKKα,β phosphorylation were markedly diminished (Figure 7C) in p16−/− versus p16+/+ BMDMs. Interestingly, IκBα protein levels were increased in p16−/− BMDMs under basal conditions (Figure 7C). However, phosphorylation of p38, another downstream target of TLR-4, was not different between p16−/− and p16+/+ BMDMs (Figure 7C), suggesting that p16INK4a deficiency specifically alters the NF-κB response.
p16INK4a deficiency diminishes JAK2-STAT1 and NF-κB signaling and increases acetylation of STAT1 and IKKα,β. Total protein extracts of p16+/+ and p16−/− BMDM treated with 15 ng/mL IL-4 (A), 2.5 ng/mL IFNγ (B) and (E), and 100 ng/mL LPS (C) for the times indicated. Western blots were performed with the indicated Abs. (D) Total protein extracts from p16+/+ and p16−/− BMDM treated with 2.5 ng/mL IFNγ for 5 hours were then immunoprecipitated (IP) with an anti-Acetyl-Lys Ab and then immunoblotted with an anti-STAT1 Ab or with an anti-IKKα,β Ab, as indicated. Input corresponds to 5% of total protein extract used for immunoprecipitation.
p16INK4a deficiency diminishes JAK2-STAT1 and NF-κB signaling and increases acetylation of STAT1 and IKKα,β. Total protein extracts of p16+/+ and p16−/− BMDM treated with 15 ng/mL IL-4 (A), 2.5 ng/mL IFNγ (B) and (E), and 100 ng/mL LPS (C) for the times indicated. Western blots were performed with the indicated Abs. (D) Total protein extracts from p16+/+ and p16−/− BMDM treated with 2.5 ng/mL IFNγ for 5 hours were then immunoprecipitated (IP) with an anti-Acetyl-Lys Ab and then immunoblotted with an anti-STAT1 Ab or with an anti-IKKα,β Ab, as indicated. Input corresponds to 5% of total protein extract used for immunoprecipitation.
Several genes coding for proteins with acetyl transferase activity were induced in p16−/− BMDMs (Table 2). Acetylation of STAT1 and IKKα,β inhibits their phosphorylation and thereby their transcriptional activity.35,36 Immunoprecipitation of acetylated proteins revealed that STAT1 and IKKα,β were acetylated to a higher extent in p16−/− than in p16+/+ BMDMs (Figure 7D). To identify whether STAT1 acetylation is the cause or the consequence of decreased STAT1 phosphorylation, JAK2 phosphorylation, which is upstream in this pathway, was assessed. JAK2 phosphorylation was less pronounced under basal conditions and after activation with IFNγ in p16−/− BMDMs (Figure 7E). Because acetylation of STAT1 can interfere with NF-κB signaling36 and both pathways act synergistically,37 the decreased STAT1 and NF-κB signaling and their increased acetylation probably results from decreased JAK2 phosphorylation.
Discussion
We identify a novel role for the tumor suppressor protein p16INK4a in the regulation of the macrophage phenotype in vitro and in vivo. p16INK4a deficiency promotes a STAT6-independent IL-4–induced macrophage polarization, whereas LPS and IFNγ responses are lower. These actions of p16INK4a deficiency likely occur through modification of the JAK2-STAT1 and NF-κB pathways.
Although p16INK4a is mainly known as a cell-cycle inhibitor, cell cycle and macrophage maturation were unaffected in the absence of p16INK4a, and an E2F target gene signature,38,39 which would be expected to be increased by p16INK4a deficiency, could not be identified in p16−/− BMDMs. Because p15INK4b is a critical tumor suppressor in the absence of p16INK4a40 and because an increase of p15INK4b expression was observed in p16−/− BMDMs, a compensatory mechanism between these CDKI might have taken place with respect to cell-cycle control.
Interestingly, p16INK4a has been previously shown to modulate transcriptional activities of the proinflammatory transcription factors NF-κB and AP-1. Via its ankyrin motifs, p16INK4a binds c-jun41 and NF-κB,42 inhibiting their activity in several immortalized cell lines. However, this mechanism does not appear to play a role in primary murine macrophages, where the loss of p16INK4a has anti-inflammatory effects. Another CDKI, p21WAF1/CIP1, was also reported to modulate inflammation without altering cell cycle or differentiation. However, in contrast with p16INK4a deficiency, p21WAF1/CIP1 deficiency in macrophages resulted in a higher sensitivity to inflammatory stimuli.43,44 In addition, p21WAF1/CIP1 expression was unchanged in p16−/− BMDMs.
In a model of AAMφ differentiation in vivo, infection with the helminth S mansoni resulted in equal Th2-associated responses in p16−/− and p16+/+ BM transplanted mice. These observations indicate unaltered T-lymphocyte function, in line with the low expression levels of p16INK4a in these cells. By contrast, AAMφ markers were significantly increased in the livers and in hepatic granulomas of p16−/− BM transplanted animals, probably because of a cell-autonomous activity of p16INK4a in macrophages. Because total BM was transplanted, a contribution of other hematopoietic cell types to this cell-autonomous macrophage response cannot be excluded. However, because expression of the measured markers was also up-regulated in the macrophage-rich granulomas and because AAMφ markers colocalized with macrophages in the granuloma, the effects are, at least in part, because of p16INK4a deficiency in the macrophages. Surprisingly, different AAMφ markers did not strictly colocalize, suggesting the existence of different macrophage populations in defined regions of the granuloma, which could be because of different gradients of stimuli within the granuloma.
Microarray analysis showed that p16−/− BMDMs under basal conditions display a gene signature resembling that of IL-4–induced macrophage polarization,27 with the exception of the AAMφ marker genes Ym1/2 and Fizz1. Moreover, p16−/− BMDMs were more responsive to IL-4 activation and displayed weaker proinflammatory responses to IFNγ and LPS. Importantly, our data show that p16INK4a deficiency does not influence pRb-mediated activation of the E2F transcription factor. The decreased IFNγ-induced phosphorylation of STAT1 and the LPS-induced IKKα,β phosphorylation in p16−/− BMDMs probably account for the attenuated inflammatory responses. Decreased IKKα,β phosphorylation combined with increased IκBα protein levels correlated with the profound decrease of NF-κB target gene expression in p16−/− BMDMs. Because a cluster of genes coding for proteins with acetyl transferase activity was induced in p16−/− BMDMs and because STAT136 and IKKα,β35 acetylation reduces their ability to be phosphorylated, the decrease in STAT1 and IKKα,β phosphorylation may result from increased acetylation of these proteins in p16−/− BMDMs. The acetylation-phosphorylation balance may be influenced by alterations upstream in the signaling pathway. Because phosphorylation of JAK2, the first component to be phosphorylated on receptor dimerization,45 was decreased, it appears that the p16INK4a deficiency-induced defect in JAK2 phosphorylation may be implicated in the reduced Stat1 phosphorylation. The increased acetylation status may be a consequence of this mechanism, contributing to the phenotype of p16−/− BMDMs. Moreover, JAK2 phosphorylation was less pronounced already under basal culture conditions, in line with the observation that p16INK4a deficiency confers a lower basal inflammatory phenotype, which is further exacerbated on stimulation. Because p16INK4a deficiency alters the IFNγ response early in the pathway and TLR4 signaling only at the level of NF-κB, but not, for example, at the level of p38, p16INK4a deficiency thus likely acts directly on the IFNγ and indirectly on the NF-κB signaling pathway.
Surprisingly, STAT6 phosphorylation, the major transcription factor for IL-4–mediated signaling response, was comparable in BMDMs of both genotypes, although IL-4–responsive genes were increased in p16−/− BMDMs, suggesting that other pathways are involved. It has been shown that STAT1 activation antagonizes expression of STAT6-induced AAMφ marker genes46,47 and that decreased STAT1 phosphorylation can result in an increase of AAMφ marker genes48 by several molecular mechanisms. First, IL-4 action is inhibited by STAT1 activation through inhibition of STAT6 phosphorylation and its subsequent nuclear translocation in human monocytes.49 However, p16INK4a deficiency did not modify STAT6 phosphorylation. Second, IFNγ inhibits STAT6 activation by inducing the expression of the suppressor of cytokine signaling 1 (SOCS1) in macrophages.46 Although SOCS1 mRNA levels were not different between p16−/− and p16+/+ BMDMs (data not shown), we cannot exclude its involvement via posttranscriptional mechanisms. Third, STAT1 and STAT6 have been shown to compete for binding the same promoters47 because decreased STAT1 activity increases binding of STAT6 to the promoters of several of its target genes. To our knowledge, an effect of NF-κB signaling on STAT6-dependent gene expression has not yet been demonstrated. Therefore, it is possible that the AAMφ-like phenotype of p16−/− BMDMs is secondary to the decreased CAMφ-associated response via JAK2-STAT1.
In conclusion, our results identify p16INK4a as a novel modulator of macrophage polarization and show that p16INK4a deficiency skews macrophages toward an IL-4–like phenotype. Because the CDKN2A locus is predictive for the risk of T2D and CVD,14,15 in which macrophage polarization plays a role,28,50 the role of p16INK4a in the control of macrophage function could be one mechanism contributing to the association of this locus with these diseases.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
P. Krimpenfort provided p16INK4a-deficient mice. Discussions with G. Raes, J. Vanginderachter, and M. de Winther are greatly acknowledged. The authors thank E. Vallez for mouse breeding, J. Brozek (Genfit SA, Loos, France) for microarray raw data analysis, and T. Coevoet, N. Jouy, and A. Lucas for technical assistance.
This work was supported by the Fondation pour la Recherche Médicale (DCV20070409276, B.S.), the EFSD/GSK Program 2009, the Cost Action (BM0602), and the Conseil régional Nord Pas-de-Calais and FEDER. C.C. was supported by a doctoral fellowship from the Nouvelle Société Française d'Athérosclérose/Schering-Plough/MSD. K.W. was supported by a European FP7 Marie Curie grant (PIEF-GA-2009-235221) and a European Atherosclerosis Society grant.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authorship
Contribution: C.C. and K.W. designed and performed experiments, analyzed data, and wrote the manuscript. L.F., S.A.H., C.P., K.B., E.B., J.V., S.F., and P.R. contributed to some of the experiments; A.T. and G.C. contributed to data interpretation; and D.D., B.S., and R.P. contributed to data interpretation, manuscript preparation, and project supervision.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bart Staels, Inserm UR 1011, Institut Pasteur de Lille, 1 rue du Professeur Calmette, BP 245, Lille 59019, France; e-mail: bart.staels@pasteur-lille.fr.
References
Author notes
C.C. and K.W. contributed equally to this work.