Abstract
Most antiangiogenic therapies currently being evaluated in clinical trials target the vascular endothelial growth factor pathway; however, the tumor vasculature can acquire resistance to vascular endothelial growth factor-targeted therapy by shifting to other angiogenesis mechanisms. Insulin-like growth factor binding protein-3 (IGFBP-3) has been reported to suppress tumor growth and angiogenesis by both IGF-dependent and IGF-independent mechanisms; however, understanding of its IGF-independent mechanisms is limited. We observed that IGFBP-3 blocked tumor angiogenesis and growth in non–small cell lung cancer and head and neck squamous cell carcinoma. Conditioned media from an IGFBP-3–treated non–small cell lung cancer cell line displayed a significantly decreased capacity to induce HUVEC proliferation and aortic sprouting. In cancer cells, IGFBP-3 directly interacted with Erk1/2, leading to inactivation of Erk1/2 and Elk-1, and suppressed transcription of early growth response protein 1 and its target genes, basic fibroblast growth factor and platelet-derived growth factor. These data suggest that IGF-independent Erk1/2 inactivation and decreased IGFBP-3–induced Egr-1 expression block the autocrine and paracrine loops of angiogenic factors in vascular endothelial and cancer cells. Together, these findings provide a molecular framework of IGFBP-3's IGF-independent antiangiogenic antitumor activities. Future studies are needed for development of IGFBP-3 as a new line of antiangiogengic cancer drug.
Introduction
Angiogenesis, the formation of new capillaries from existing blood vessels, is essential to carcinogenic processes, including solid tumor formation, growth, invasion, and metastasis.1 Most tumors can stimulate angiogenesis by switching on the production of numerous cytokines and growth factors, including fibroblast growth factors (FGFs), vascular endothelial growth factors (VEGFs), and platelet-derived growth factors (PDGFs).2 Several antiangiogenic agents are in various phases of clinical trials for human cancer; however, most of these agents target the VEGF signaling pathway.3 Therefore, other potential therapeutic agents that block non-VEGF angiogenic pathways need to be evaluated.
Insulin-like growth factor-binding protein-3 (IGFBP-3), a member of a family of 6 IGFBPs, has demonstrated antiproliferative, proapoptotic, antiangiogenic, and antimetastatic activity in a variety of cancer cells.4-8 It may also have IGF-independent antitumor activities through cell-surface or intracellular protein interaction, its nuclear translocation, or its transcriptional regulation.7,9-12 However, the mechanisms that mediate IGFBP-3's IGF-independent antitumor activity have not been clearly defined.
The 82-kDa phosphoprotein transcription factor early growth response protein 1 (Egr-1), an immediate early gene product, has been implicated in multiple cellular processes, including cell growth, apoptosis, wound healing, and angiogenesis. Mitogenic stimuli, including serum, PDGF, peptide growth factors, and B-Raf, and nonmitogenic stresses, including γ-irradiation and hypoxia, activate Egr-1 expression through serum response elements (SREs) in the Egr-1 promoter, where serum response factor (SRF) and ternary complex factors form transcriptionally active ternary complexes.13 Once activated, Egr-1 binds to GC-rich, cis-acting promoter elements and controls the expression of multiple genes that encode growth factors, cytokines, adhesion molecules, and proteases, including IGF-1, IGF-2, TGF-β1, fibronectin, urokinase-type plasminogen activator, VEGF-1R, VEGF, PDGF-A and -B, and basic FGF (bFGF), which are believed to have important functions in cancer cell survival, apoptosis, angiogenesis, invasion, and metastasis.14,15 The association between Egr-1 and tumor angiogenesis has been observed in various tumor types.16-18 A DNA-based enzyme and a siRNA that target Egr-1 suppress bFGF expression and tumor angiogenesis and growth in various cancer cell types.18-21
In this study, we determined the mechanisms by which IGFBP-3 exerts its IGF-independent antiangiogenic antitumor activity. Our findings reveal that IGFBP-3's engagement of Erk1/2 inactivates Erk1/2 and Elk1 in an IGF-independent manner, resulting in inhibited binding of Elk1 to SRE sites in the Egr-1 upstream promoter and reduced transcription of Egr-1 and its target genes, including PDGF and bFGF. Egr-1, bFGF, and PDGF proteins were highly down-regulated in our non–small cell lung cancer (NSCLC) mouse models; we also observed angiogenesis suppression. These data suggest that increase in IGFBP-3 level could lead to inhibition of tumor angiogenesis.
Methods
Cell culture, animals, and other reagents
H460, H1299, A549 (NSCLC cell lines), UMSCC38 (head and neck squamous cell carcinoma [HNSCC] cell lines), and HUVEC (Cambrex Bio Science) cell lines were cultured as previously described.8 NSCLC cell lines (H1299, A549, and H460) and HUVEC were cultured in RPMI 1640 with 10% FBS and in EGM (Lonza Walkersville Inc), respectively, in a humidified environment with 5% CO2. Six-week-old female athymic nude mice (10 mice for each group) and chick eggs (Harlan-Sprague-Dawley and Charles River laboratories) were maintained in a defined pathogen-free environment. All animal procedures were performed in accordance with a protocol approved by the M. D. Anderson Institutional Animal Care and Usage Committee.
Tumor xenograft model and immunohistochemical analysis
Ad-BP-3's antiangiogenic effects on established H1299 subcutaneous or UMSCC38 HNSCC orthotopic tumor models were determined as described elsewhere.8,11,22 In brief, after the H1299 xenograft tumor volume reached ∼ 75 mm3, mice (n = 5) were given single intratumoral injections (1 × 1010 particles) of IGFBP-3–expressing (Ad-BP-3) or empty viruses (Ad-EV). Tumors embedded in paraffin were subjected to immunohistochemical staining by the use of an ABC staining kit (Vector Laboratories) with anti-bFGF (1:400 dilution), anti-PDGF (1:400 dilution), and anti–Egr-1 (1:400 dilution) antibodies. For CD31 staining, frozen tumor tissue sections were stained with anti-CD31 antibody (1:100 dilution).
Matrigel plug assay
The in vivo mouse Matrigel plug assay was performed with A549 NSCLC cells infected with Ad-BP-3 (50 pfu/cell) or Ad-EV (50 pfu/cell). Each treatment group included 10 mice. The number of blood vessels per field was analyzed by microscopy at 10× magnification.
Cell proliferation, migration, invasion, and tube-formation assay
The in vitro migration, invasion, and tube-formation assays were performed as described elsewhere.8,23,24 In brief, in the coculture assay with H460 cells, HUVECs (4 × 104) were seeded onto the cell culture inserts (1-μm pore size; Becton Dickinson) and H460 cells (2 × 105) transfected with pBP-3, pBP-3-ggg, or pEgr1 were transferred to the bottom of a 12-well plate. Three days later, HUVEC proliferation was assessed by use of the MTT assay. To determine IGFBP-3's effects on NSCLC cells' angiogenesis-stimulating effects, we infected H1299, H460, and A549 cells with Ad-BP-3 (50 pfu/cell) or Ad-EV or treated them with rBP-3 or control vehicle, as previously described.25 Conditioned media (CM) were collected from NSCLC cells and added to HUVECs for cell proliferation or tube formation or to check aortas for endothelial cell sprouting, as previously described.8,24 HUVEC tube formation was scored after 8 hours, and cell proliferation was analyzed by the MTT assay after 3 days, as previously described.26 Each condition was tested in 6 wells. The details are described in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Transcription analysis
Quantitative RT-PCR was performed as described elsewhere.27 The primer sequences used are described in supplemental Methods. To avoid amplification of genomic DNA, each gene primer was chosen from different exons. To analyze the bFGF promoter, we amplified the 5′-flanking region (−590 to +26) of the human bFGF gene (GenBank accession number: NM_002006) from human genomic DNA (Sigma-Aldrich) and cloned into the SmaI site of the luciferase reporter vector pGL3-Basic (Promega). The detailed methods used to construct the mutant Egr-1 promoter vectors, transfect plasmids and siRNAs, and perform the luciferase reporter assay are described in supplemental Methods.
ChIP
A ChIP assay was performed with H460 cells infected with Ad-BP-3 or Ad-EV or treated with rBP-3 or control vehicle. Extracts from equal numbers of cells were immunoprecipitated with antibodies against Elk-1, SRF, IGFBP-3, or preimmune serum as a negative control. PCR was performed with the use of primers encompassing the SRE elements, as depicted in Figure 5A, and an exon 1 primer was used as a negative control. All PCR primers and conditions are described in the supplemental Methods. An aliquot of the whole-cell protein-DNA complex (2% of the immunoprecipitated volume) was subjected to PCR analysis to confirm the protein-bound DNA sequence.
Immunoprecipitation, in vitro pull, and Western blot analyses
H1299 cells were transfected with 5 μg of control (EV) or pCMV6-IGFBP-3-Flag (pBP-Flag). After they were starved of serum, the cells were stimulated with 10% FBS for 0 and 20 minutes. Whole-cell lysates were prepared, and Western blotting was performed as described elsewhere.28 For immunoprecipitation, total cell lysates were precleared with the appropriate protein-G or protein-A beads and then incubated with goat antibody (1 μg) against Erk1/2 or rabbit antibody against p38α and protein-G or protein-A beads. IGFBP-3-Flag binding was detected with a mouse anti-Flag antibody. For the in vitro pulldown assay, rBP-3 (1 μg) conjugated to NTA-agarose beads (50 μL, 50% slurry) was incubated with H1299 cell lysate (150, 300, and 450 μg) and then washed with PBS and 10mM imidazole. The beads were then boiled in 2× sample buffer for 10 minutes and used for the Western blot analysis. For the negative control, blank BSA-coated NTA beads were incubated with 450 μg of cell lysate.
Immunofluorescence staining
H1299 cells were seeded on cover slips (Deckgläser; Menzel-Gläser) coated with rat tail collagen type I (100 μg/mL; BD Biosciences). After being incubated overnight, the cells were serum-starved for 3 hours and treated with rhIGFBP-3 at 10 μg/mL in a serum-free medium (RPMI 1640) and fixed at 0, 10, 30, and 60 minutes in 4% paraformaldehyde in PBS for 3 minutes. After being thoroughly washed with PBS, the cells were treated with methanol on ice and washed with PBS. After blocking with 5% BSA, primary rabbit anti-IGFBP-3 (Santa Cruz Biotechnology) and mouse anti-Erk1/2 (Cell Signaling Technology) antibodies were added to the cells overnight at 4°C. After being washed in PBS, the cells were stained with donkey anti–rabbit antibodies conjugated with Alexa Fluor 568 and donkey anti–mouse antibodies conjugated with Alexa Fluor 488. During the washing steps, the cells were stained with Hoechst 33342. The cover slips were mounted and imaged with the use of an Olympus IX71 FV 500 laser confocal microscope with Fluoview Version 5.0 software. All images were obtained with a 60× objective lens with 2× optical zoom. The colocalization analysis was performed by use of the colocalization finder function of ImageJ software (NIH).
Statistical analysis
Data are given as the mean ± SD. To determine the statistical significance between groups, we used paired Student t tests and 95% confidence intervals. In all statistical analyses, 2-sided P values < .05 were considered statistically significant.
Results
IGFBP-3 inhibits NSCLC and HNSCC tumor growth and angiogenesis
We previously demonstrated that IGFBP-3 overexpression inhibits the growth of H1299 NSCLC xenografts in nude mice.11 To determine whether IGFBP-3's antiangiogenic activity contributes to its antitumor activities, we performed a series of experiments by using adenoviral (Ad-BP-3) and recombinant (rBP-3) IGFBP-3.
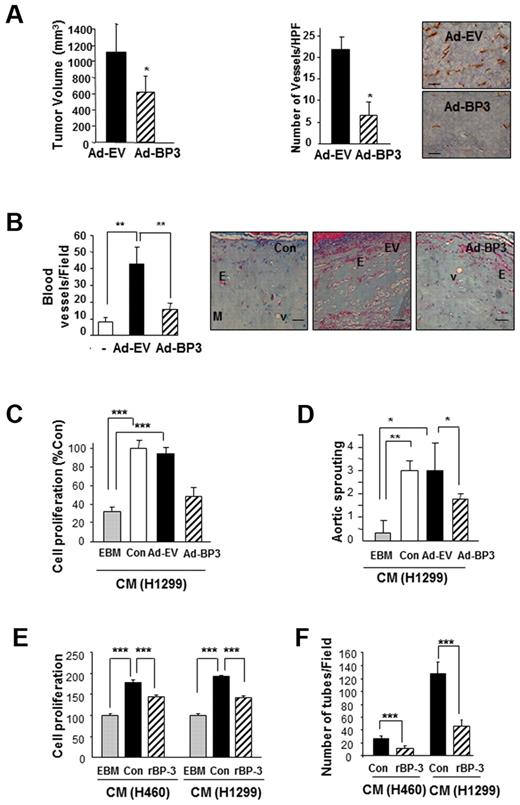
We first determined Ad-BP-3's effect on tumor growth and angiogenesis in 1299 NSCLC xenograft tumors established in athymic nude mice. Ad-BP-3 induced significant decreases in H1299 NSCLC xenograft tumor growth (Figure 1A). Anti-CD31 staining of the tumor tissues injected with Ad-BP-3 revealed significantly decreased tumor vascularization compared with those injected with control viruses (AD-EV; Figure 1A right). The Matrigel plugs that contained Ad-BP-3–infected A549 cells had significantly fewer blood vessels than did those containing Ad-EV cells (P < .01; Figure 1B). These findings suggest that IGFBP-3 has antiangiogenic and antitumor activity in NSCLC.
IGFBP-3 suppresses tumor growth and angiogenesis in NSCLC xenografts and vascular endothelial cells. (A) H1299 xenograft tumor growth (left) 10 days after injection with IGFBP-3–expressing adenoviruses (Ad-BP-3) or empty viruses (Ad-EV). Tumor growth is expressed as the mean ± SEM. An immunohistochemical analysis of CD31 (right) was performed in xenograft tissues, and the number of CD31-immunoreactive vessels per high-power field was counted. The results represent the mean calculated from 5 mice (bars, SDs). *P < .05 compared with the control group. Representative CD31 immunostaining in H1299 xenograft tissues is included. (B) Matrigel plug assay with A549 cells. Gross observed results of blood vessels are expressed as the mean of 5 tumors ± SEM, **P < .01. (C-D) Effect of CM from indicated NSCLC cell lines that had been infected with Ad-EV or Ad-BP-3 (C-D) or treated with rBP-3 (E-F) on HUVEC proliferation (C,E), chick aortic sprouting (D), and HUVEC tube formation (F). The results represent the means (bars, SDs) of 5 identical wells. *P < .05; **P < .01; ***P < .001.
IGFBP-3 suppresses tumor growth and angiogenesis in NSCLC xenografts and vascular endothelial cells. (A) H1299 xenograft tumor growth (left) 10 days after injection with IGFBP-3–expressing adenoviruses (Ad-BP-3) or empty viruses (Ad-EV). Tumor growth is expressed as the mean ± SEM. An immunohistochemical analysis of CD31 (right) was performed in xenograft tissues, and the number of CD31-immunoreactive vessels per high-power field was counted. The results represent the mean calculated from 5 mice (bars, SDs). *P < .05 compared with the control group. Representative CD31 immunostaining in H1299 xenograft tissues is included. (B) Matrigel plug assay with A549 cells. Gross observed results of blood vessels are expressed as the mean of 5 tumors ± SEM, **P < .01. (C-D) Effect of CM from indicated NSCLC cell lines that had been infected with Ad-EV or Ad-BP-3 (C-D) or treated with rBP-3 (E-F) on HUVEC proliferation (C,E), chick aortic sprouting (D), and HUVEC tube formation (F). The results represent the means (bars, SDs) of 5 identical wells. *P < .05; **P < .01; ***P < .001.
Tumor angiogenesis is partly mediated by tumor-secreted angiogenic growth factors that interact with their receptors expressed on endothelial cells.29 To determine whether IGFBP-3 suppresses the secretion of angiogenic factors from NSCLC, we collected CM from Ad-EV– or Ad-BP-3–infected H1299 cells after incubating them in a serum-free medium for 1 day. As shown in Figure 1C, HUVECs that had been treated with CM from untreated (Con) or Ad-EV–infected cells, but not from Ad-BP-3–infected cells, demonstrated significantly greater proliferation than did those treated with endothelial cell basal medium only. The ex vivo chick aortic ring arch assay showed similar results; the CM from untreated or Ad-EV–infected cells, but not the CM from Ad-BP-3–infected cells, significantly stimulated the formation of endothelial cell sprouts (Figure 1D).
Because protein expression induced by adenoviruses can be much greater than that seen under real-life conditions, we further determined the effects of exogenously added recombinant IGFBP-3 (rBP-3), which has a cytosolic half-life of 3 hours.28 We performed a Western blot analysis to confirm that no residual rBP-3 was present in the CM (data not shown). HUVECs that had been treated with the CM from BP-3–pretreated cancer cells also showed significantly reduced proliferation (Figure 1E) and tube formation (Figure 1F) compared with those treated with CM from untreated NSCLC cells. These findings suggest that IGFBP-3 has antiangiogenic and antitumor activity in NSCLC, at least partly because of its effects on NSCLC cells' secretion of angiogenic factors.
IGFBP-3 induces down-regulation of the bFGF-Egr-1 transcription loop
We assessed the angiogenic factors that are regulated by IGFBP-3. Consistent with previous findings in which IGFBP-3 was found to mediate the antiangiogenic action of the farnesyl transferase inhibitor SCH66336, which suppresses VEGF expression in NSCLC and HNSCC cells,8,30 Ad-BP-3 reduced VEGF expression in H1299 and A549 NSCLC cells (unpublished data). We determined IGFBP-3's effects on bFGF expression. Ad-BP-3 led to decreased bFGF expression in H1299, A549, and H460 NSCLC cells and UMSCC38 HNSCC cells (Figure 2A) over the range of doses used in the cell-based experiments described in Figure 1. IGFBP-3's ability to reduce bFGF protein expression was further confirmed by a Western blot analysis of rBP-3–treated H1299 cells (supplemental Figure 1). A CM analysis from UMSCC38 cells revealed that rBP-3 reduced bFGF secretion (Figure 2B). Ad-BP-3 negatively regulated bFGF transcription in H460 cells (Figure 2C). Conversely, IGFBP-3 knockdown by siRNA led to increased bFGF mRNA levels. We then determined IGFBP-3's effects on bFGF promoter activity by transiently transfecting H460 cells with a pGL3-basic luciferase reporter construct that contained the 5′-flanking promoter region of human bFGF (−589 to +26). bFGF promoter activity was significantly reduced after Ad-BP-3 infection or rBP-3 treatment but increased after IGFBP-3 siRNA transfection (Figure 2D). Together, these data demonstrate the inhibitory effects of IGFBP-3 on bFGF transcription.
IGFBP-3 down-regulates bFGF transcription. (A) Western blot analysis of IGFBP-3 and bFGF expression in NSCLC and HNSCC cells 2 days after infection with Ad-EV or Ad-BP-3. (B) Reduced bFGF levels in the CM from rBP-3–treated UMSCC38 cells. N.S. indicates nonspecific bands. (C) Semiquantitative RT-PCR analysis of bFGF expression in H460 cells infected with Ad-EV or Ad-BP-3 (left) or transfected with scrambled (Con) or IGFBP-3 (BP-3) siRNA (right). (D) Luciferase assay to determine the effect of IGFBP-3 on bFGF promoter activity in H460 cells transiently transfected with bFGF-Luc in association with Ad-BP-3 or Ad-EV infection at the indicated doses (left), rBP-3 treatment (middle), or scrambled (Con) or IGFBP-3 (BP-3) siRNA cotransfection (right). The results represent the means (bars, SDs) of triplicate results. *P < .05; **P < .01; ***P < .001.
IGFBP-3 down-regulates bFGF transcription. (A) Western blot analysis of IGFBP-3 and bFGF expression in NSCLC and HNSCC cells 2 days after infection with Ad-EV or Ad-BP-3. (B) Reduced bFGF levels in the CM from rBP-3–treated UMSCC38 cells. N.S. indicates nonspecific bands. (C) Semiquantitative RT-PCR analysis of bFGF expression in H460 cells infected with Ad-EV or Ad-BP-3 (left) or transfected with scrambled (Con) or IGFBP-3 (BP-3) siRNA (right). (D) Luciferase assay to determine the effect of IGFBP-3 on bFGF promoter activity in H460 cells transiently transfected with bFGF-Luc in association with Ad-BP-3 or Ad-EV infection at the indicated doses (left), rBP-3 treatment (middle), or scrambled (Con) or IGFBP-3 (BP-3) siRNA cotransfection (right). The results represent the means (bars, SDs) of triplicate results. *P < .05; **P < .01; ***P < .001.
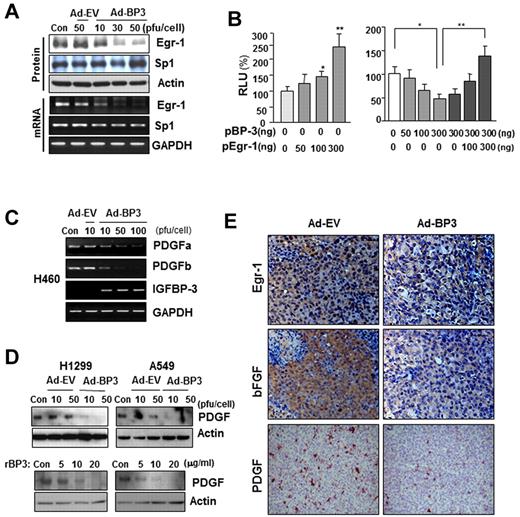
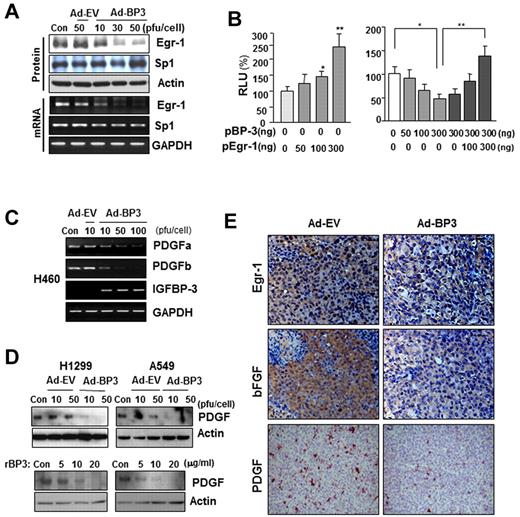
We next determined which transcription factors are involved in IGFBP-3–regulated bFGF promoter activity. The Egr-1 and Sp-1 transcription factors have long been recognized as MAPK-dependent activators of bFGF transcription.13,31,32 Therefore, we determined IGFBP-3's effects on Egr-1 and Sp-1 expression. Infection of H460 cells with Ad-BP-3 dramatically reduced Egr-1 expression at the protein and mRNA levels, with no detectable changes in Sp-1 expression of (Figure 3A). RT-PCR (supplemental Figure 2A) and Northern blot (supplemental Figure 2B) analyses further confirmed rBP-3's and Ad-BP-3's ability to reduce bFGF and Egr-1 mRNA levels. A band shift analysis revealed a reduction in nuclear complexes bound to the Egr-1 gene promoter in Ad-BP-3–infected H460 cells compared with in Ad-EV–infected cells (supplemental Figure 2C). These findings suggest that IGFBP-3 inhibits Egr-1 expression, leading to reduced bFGF promoter activity and attenuated bFGF expression.
IGFBP-3 down-regulates bFGF expression promoter activity by regulating Egr-1 transcription. (A) Western blot (top) and semiquantitative RT-PCR (bottom) analyses of IGFBP-3's effect on Egr-1 and Sp-1 protein and mRNA expression in H460 cells that had been infected with Ad-BP-3 or Ad-EV for 2 days. (B) Luciferase assay to determine Egr-1's effect on bFGF expression. H460 cells transiently transfected with bFGF-Luc and pEgr-1, pBP-3, or both; *P < .05, **P < .01. (C-D) RT-PCR analysis of PDGFa and PDGFb mRNA expression in H460 cells (C) and Western blot analysis of PDGF protein expression in H1299 and A549 cells (D) after infection with Ad-EV or Ad-BP-3 or treatment with rBP-3. (E) Immunohistochemical analysis of Egr-1, bFGF, and PDGF expression in H1299 xenografts 10 days after injection with Ad-EV or Ad-BP-3.
IGFBP-3 down-regulates bFGF expression promoter activity by regulating Egr-1 transcription. (A) Western blot (top) and semiquantitative RT-PCR (bottom) analyses of IGFBP-3's effect on Egr-1 and Sp-1 protein and mRNA expression in H460 cells that had been infected with Ad-BP-3 or Ad-EV for 2 days. (B) Luciferase assay to determine Egr-1's effect on bFGF expression. H460 cells transiently transfected with bFGF-Luc and pEgr-1, pBP-3, or both; *P < .05, **P < .01. (C-D) RT-PCR analysis of PDGFa and PDGFb mRNA expression in H460 cells (C) and Western blot analysis of PDGF protein expression in H1299 and A549 cells (D) after infection with Ad-EV or Ad-BP-3 or treatment with rBP-3. (E) Immunohistochemical analysis of Egr-1, bFGF, and PDGF expression in H1299 xenografts 10 days after injection with Ad-EV or Ad-BP-3.
To determine Egr-1's effect on IGFBP-3–mediated antiangiogenic activity in NSCLC cells, we transiently cotransfected H460 cells with the bFGF luciferase reporter construct and an Egr-1 expression vector, an IGFBP-3 expression vector (pBP-3), or both. As shown in Figure 3B, Egr-1 expression led to a dose-dependent increase in bFGF promoter activity (left) and attenuated IGFBP-3's inhibitory effects on promoter activity (right). These results indicate that Egr-1 plays a specific role in IGFBP-3–mediated suppression of bFGF transcription. IGFBP-3 also inhibited the expression of PDGF, another Egr-1–regulated angiogenic factor, as shown by the results of an RT-PCR analysis of PDGF-a and -b mRNA levels in H460 cells (Figure 3C) and a Western blot analysis of PDGF protein expression in H1299 and A549 cells (Figure 3D) that had been infected with Ad-BP-3 or treated with rBP-3. An immunohistochemical analysis of H1299 xenografts injected with Ad-BP-3 (Figure 1A) also demonstrated reduced Egr-1, bFGF, and PDGF staining (Figure 3E). Collectively, these results demonstrate that IGFBP-3–mediated Egr-1 suppression results in decreased bFGF and PDGF expression.
IGFBP-3 suppresses Egr-1 promoter activity in an IGF-independent manner
The conventional function of IGFBP-3 is to regulate cell growth and promote apoptosis by sequestering free IGFs. However, studies have shown that IGFBP-3 also has IGF-independent antitumor activity.4,33 To determine whether Egr-1 transcription regulation by IGFBP-3 is IGF dependent, we cotransfected H460 cells with a human Egr-1 reporter construct and expression vectors carrying K-ras (V12) or H-ras (V12), along with pBP-3 or an empty vector control.
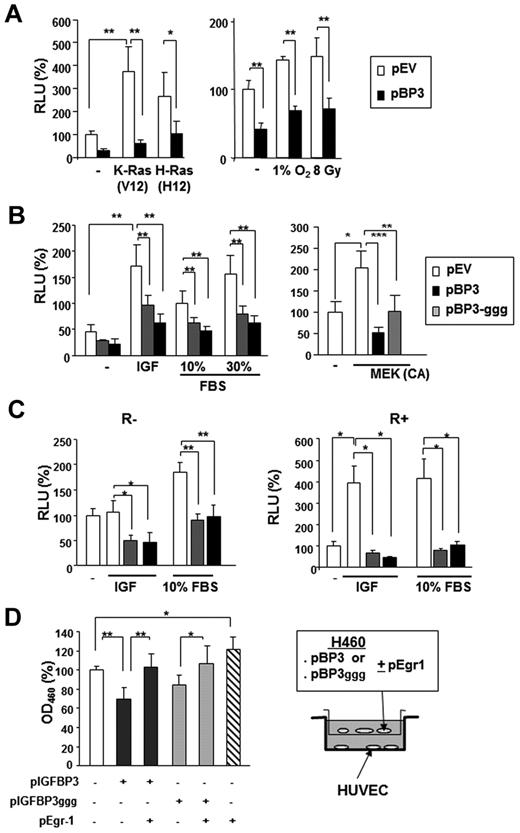
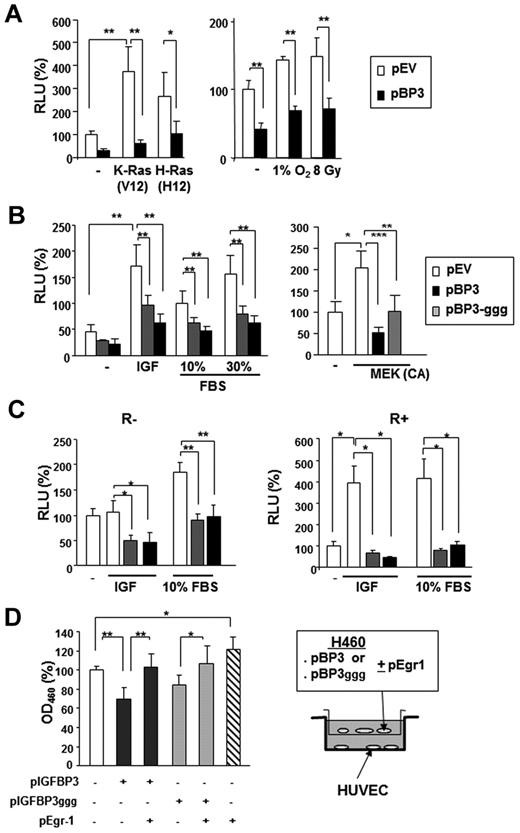
Transfection with pBP-3 resulted in substantial loss of the Egr-1 promoter activity that had been stimulated by K-ras (V12) and H-ras (V12; Figure 4A). Egr-1 transcription is known to be activated by environmental stresses, such as hypoxia and radiation.34,35 Egr-1 promoter activity, stimulated by γ-radiation (8 Gy) or incubation in 1% O2, was significantly suppressed by pBP-3 (Figure 4A right). We also found that pBP-3-ggg, a mutant IGFBP-3 with a substitution of 3 glycine residues (Gly56Gly80Gly81) that are critical for the IGF binding domain,33 inhibited the Egr-1 promoter activity stimulated by IGF-1, FBS, or constitutively active MAPK kinase (MEK; ΔN3/S218E/S222D) (Figure 4B).36 pBP-3 and pBP-3-ggg reduced IGF-1– and 10% FBS-stimulated Egr-1 promoter activity in both R− (mouse embryonic fibroblasts from an IGF-1R–null mouse) and R+ (R− cells transfected with IGF-1R) cells (Figure 4C).37 These findings suggest that the effects of IGFBP-3 on Egr-1 promoter activity are IGF-1 independent.
IGFBP-3 inhibits Egr-1 expression independently IGF-1. (A-B) The wild-type 1.2-kb Egr-1 promoter reporter construct (Egr1-A-Luc) was transiently transfected, with or without pBP-3 or pBP-3-ggg, into NSCLC H460 cells. (A) Cells were stimulated by cotransfection of plasmids containing mutants of K-Ras (V12) or H-Ras (V12) or by exposure to hypoxia (1% O2) or γ-radiation (8 Gy). (B) Cells were stimulated by IGF-1 (50 ng/mL) or FBS (10% and 30%) for 24 hours or cotransfected with plasmids expressing CA MEK. (C) R− (IGF-1R null mouse fibroblasts) and R+ (R− cells transfected with IGF-1R) cell lines were cotransfected with Egr1-A-Luc and pBP-3 or pIGFBP-3-ggg and then stimulated by IGF-1 (50 ng/mL) or FBS (10%) for 24 hours. The data are the mean ± SD from 3 independent experiments, with 4 replicates per experiment. *P < .05, **P < .01. (D) In vitro evaluation of the antiangiogenic potential of IGFBP-3. pBP-3–transfected H460 cells show less stimulatory activity for HUVEC proliferation than untransfected H460 cells in a coculture assay system. The values are the mean ± SD from 2 separate experiments, with 3 replicates per experiment. *P < .05, **P < .01, ***P < .001.
IGFBP-3 inhibits Egr-1 expression independently IGF-1. (A-B) The wild-type 1.2-kb Egr-1 promoter reporter construct (Egr1-A-Luc) was transiently transfected, with or without pBP-3 or pBP-3-ggg, into NSCLC H460 cells. (A) Cells were stimulated by cotransfection of plasmids containing mutants of K-Ras (V12) or H-Ras (V12) or by exposure to hypoxia (1% O2) or γ-radiation (8 Gy). (B) Cells were stimulated by IGF-1 (50 ng/mL) or FBS (10% and 30%) for 24 hours or cotransfected with plasmids expressing CA MEK. (C) R− (IGF-1R null mouse fibroblasts) and R+ (R− cells transfected with IGF-1R) cell lines were cotransfected with Egr1-A-Luc and pBP-3 or pIGFBP-3-ggg and then stimulated by IGF-1 (50 ng/mL) or FBS (10%) for 24 hours. The data are the mean ± SD from 3 independent experiments, with 4 replicates per experiment. *P < .05, **P < .01. (D) In vitro evaluation of the antiangiogenic potential of IGFBP-3. pBP-3–transfected H460 cells show less stimulatory activity for HUVEC proliferation than untransfected H460 cells in a coculture assay system. The values are the mean ± SD from 2 separate experiments, with 3 replicates per experiment. *P < .05, **P < .01, ***P < .001.
We then determined whether this IGFBP-3–induced decrease in Egr-1 expression affected IGFBP-3–mediated antiangiogenic activity. To this end, we cocultured HUVECs with H460 cells cotransfected with pEgr-1 and pBP-3 or pBP-3-ggg. As shown in Figure 4D, HUVECs that had been cocultured with H460 cells transfected with Egr-1 and pBP-3 or pBP-3ggg had significantly reduced proliferation compared with those cultured with H460 cells transfected with pBP-3 or pBP-3-ggg alone, indicating attenuation of the antiangiogenic effects of pBP-3 and pBP-3ggg by Egr-1 expression. Taken together, these results demonstrate that IGFBP-3 has antiangiogenic activity in NSCLC cells by inhibiting Egr-1 promoter activity through IGF-1–independent pathways.
IGFBP-3 inhibits Erk phosphorylation and subsequent Elk-1 activation, leading to a reduction in Elk-1 binding to SREs in the Egr-1 promoter
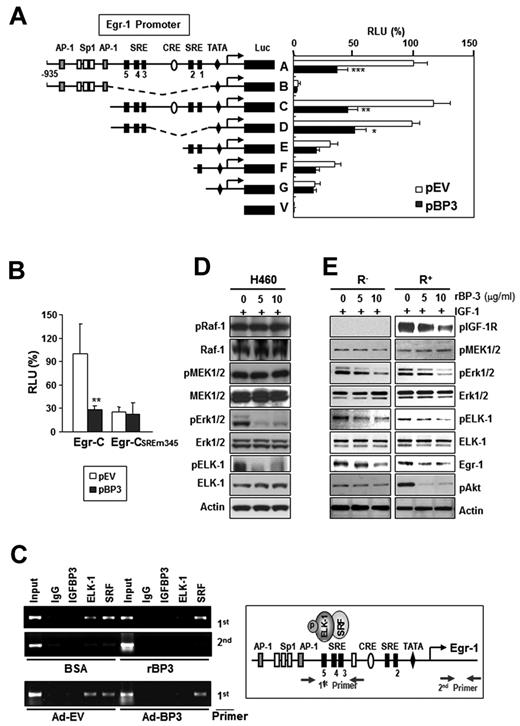
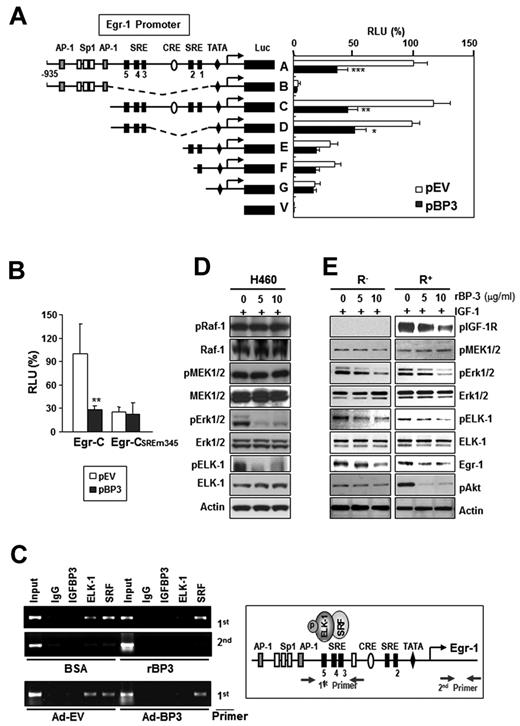
The promoter region (−935 to +12) of Egr-1 contains AP-1, Sp-1, cAMP response element, and SREs and their adjacent Ets binding sites.13 To identify the promoter elements that are involved in IGFBP-3–regulated Egr-1 promoter activity, we assessed a 1.2-kb Egr-1 promoter construct (A) and several 5′-truncated deletion mutant constructs (B to V)13 for promoter activity in H460 cells that had been transiently cotransfected with pBP-3. Deletion of the 2 AP-1 binding sites and 3 Sp-1 binding sites (C construct) or the CRE sites and two 3′-SREs (SRE 1 and 2; D construct) had little effect on promoter activity compared with the A construct, and the promoter activities of these 3 constructs (A, C, and D) were strongly inhibited by cotransfection with pBP-3 (Figure 5A). In contrast, removal of regions upstream of the SRE 1 or 2 sites (E and F constructs) led to a dramatic reduction in Egr-1 promoter's activity and response to IGFBP-3 expression. Loss of all 5 SRE sites and 2 CRE sites (B construct) led to a total elimination of Egr-1 promoter activity. In contrast, pBP-3 had no effect on the activity of the Egr-1 promoter that carried mutations in the 3 5′-SRE sites (SRE 3, 4, and 5; CSREm345 construct; Figure 5B).
IGFBP-3 inhibits Egr-1 transcription by inactivating Erk-Elk1 and Elk1 binding to SRE sites in the Egr-1 promoter. (A-B) IGFBP-3's effects on Egr-1 promoter activity. H460 cells were transiently cotransfected with Egr-1 Luc constructs (A) or an Egr-1 Luc construct carrying mutations in the 3 5′-SRE sites (SRE 3, 4, and 5; B), along with an empty vector (pEV) or pBP-3. The important genetic elements in the Egr-1 regulatory region are shown, including the SRE sites, CRE sites, GC, and TATA boxes. The data are the mean ± SD from 3 independent experiments, with 4 replications per experiment. *P < .05, **P < .01, ***P < .001 compared with pEV-transfected cells. (C) IGFBP-3 reduces in vivo binding of Elk-1 to the Egr-1 promoter. H460 cells, treated with rBP-3 or bovine serum albumin or infected with Ad-BP-3 or Ad-EV, cross-linked and immunoprecipitated with antibodies specific for IGFBP-3, Elk-1, SRF, or a normal serum control antibody (immunoglobulin). The second primer denotes PCR samples using a pair of negative control primers corresponding to the exon 1 sequence of the Egr-1 gene. (D-E) Western blot analysis for the indicated proteins in H460 cells (D) and R− and R+ cells (E) treated with the indicated concentrations of rBP-3 for 2 days and stimulated with IGF-1 for 15 minutes.
IGFBP-3 inhibits Egr-1 transcription by inactivating Erk-Elk1 and Elk1 binding to SRE sites in the Egr-1 promoter. (A-B) IGFBP-3's effects on Egr-1 promoter activity. H460 cells were transiently cotransfected with Egr-1 Luc constructs (A) or an Egr-1 Luc construct carrying mutations in the 3 5′-SRE sites (SRE 3, 4, and 5; B), along with an empty vector (pEV) or pBP-3. The important genetic elements in the Egr-1 regulatory region are shown, including the SRE sites, CRE sites, GC, and TATA boxes. The data are the mean ± SD from 3 independent experiments, with 4 replications per experiment. *P < .05, **P < .01, ***P < .001 compared with pEV-transfected cells. (C) IGFBP-3 reduces in vivo binding of Elk-1 to the Egr-1 promoter. H460 cells, treated with rBP-3 or bovine serum albumin or infected with Ad-BP-3 or Ad-EV, cross-linked and immunoprecipitated with antibodies specific for IGFBP-3, Elk-1, SRF, or a normal serum control antibody (immunoglobulin). The second primer denotes PCR samples using a pair of negative control primers corresponding to the exon 1 sequence of the Egr-1 gene. (D-E) Western blot analysis for the indicated proteins in H460 cells (D) and R− and R+ cells (E) treated with the indicated concentrations of rBP-3 for 2 days and stimulated with IGF-1 for 15 minutes.
These findings suggest that the regions upstream of SRE 1 and 2, especially the 3 5′-SRE sites (SRE 3, 4, and 5), contain promoter elements that are regulated by IGFBP-3. The Egr-1 promoter sequence that encompasses SRE sites 3-5 contains a cluster of Ets motifs (GGA sequence),38 and these SRE sites and adjacent Ets motifs are continuously occupied by ternary complex factors transcription factors.39-41 The authors of previous studies have shown that Elk-1, in combination with SRF, participates in the initiation of Egr-1 transcription.42 Hence, we hypothesized that IGFBP-3 treatment would inhibit the binding of Elk-1 or SRF to the Egr-1 promoter, leading to reduced Egr-1 transcription. We performed a ChIP assay by using 2 primer sets that encompassed the 5′-SRE sites or the first exon (as a negative control) and H460 cells treated with Ad-BP-3 or rBP-3. IGFBP-3 did not interact directly with the SRE sites and did not affect SRF's binding to these sites (Figure 5C). In contrast, IGFBP-3 markedly reduced Elk-1's binding to the 5′-SRE sites. The immunoprecipitates with preimmune serum (immunoglobulin) showed no significant binding to these sites. These results suggest that IGFBP-3 regulates Egr-1 transcription by indirectly inhibiting Elk-1's binding to the 5′-SRE sites in the Egr-1 promoter.
The authors of previous studies have indicated that transcriptional Egr-1 activation is regulated by Elk-1 phosphorylation through the MEK/Erk signaling cascade,20 which is required to recruit SRF to the SREs in the Egr-1 promoter.43 Thus, we determined whether IGFBP-3 interfered with the Raf-MAPK-Elk phosphorylation cascade in H460 cells. A Western blot analysis revealed that phosphorylated Erk1/2 (pErk1/2) and phosphorylated Elk-1 (pElk-1) expression was dramatically reduced by IGFBP-3 treatment, whereas total and phosphorylated Raf and MEK1/2 levels were not affected (Figure 5D). Likewise, pBP-3 transfection led to reduced pErk1/2 expression, with no change in pMEK1/2 levels (supplemental Figure 3). An additional experiment using R− and R+ cells treated with rBP-3 and stimulated with IGF-1 resulted in reduced pErk1/2, pElk-1, and Egr-1 levels, with no change in pMEK1/2 levels in both cell lines (Figure 5E). In contrast, neither pIGF-1R nor pAkt levels were affected in rBP-3–treated R− cells. Together, these findings suggest that IGFBP-3 inhibits Erk1/2 and Elk-1 activation through IGF-1–independent mechanisms, leading to reduced binding of Elk-1 to SREs in the Egr-1 promoter and Egr-1 transcription suppression.
IGFBP-3 binds to and inactivates Erk1/2
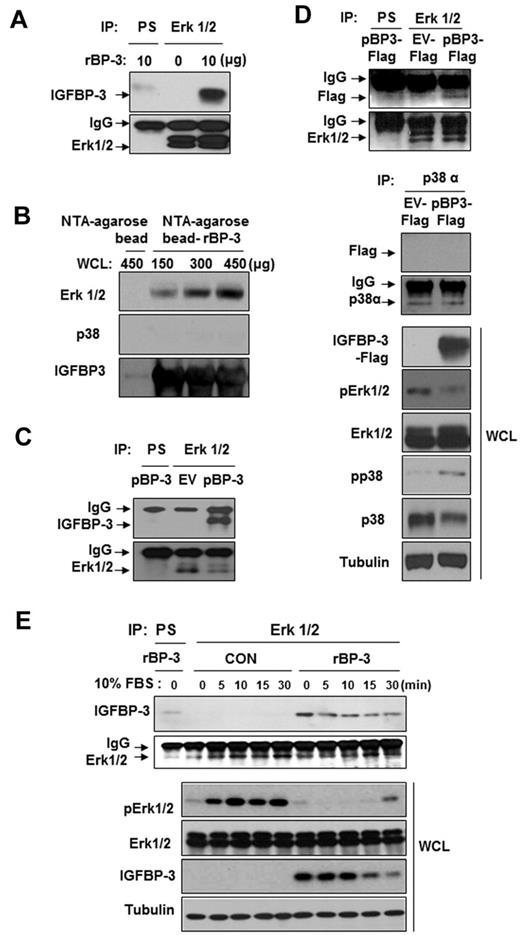
We determined the IGF-independent mechanisms by which IGFBP-3 may deregulate Erk1/2 activation. IGFBP-3 has been shown to act independently of the IGF system by binding to its own receptors or other cellular proteins, although the mechanism by which this occurs has not been clearly identified. Therefore, we determined whether IGFBP-3 interacted with Erk1/2 using H1299 cells that do not express IGFBP-3.44 First, we performed coimmunoprecipitation assays by using H1299 cell lysates that had been pretreated with rBP-3. H1299 cell immunoprecipitation with the use of an anti-Erk1/2 antibody, followed by Western blotting for IGFBP-3, revealed an association between Erk1/2 and rBP-3 (Figure 6A). Second, we determined in vitro whether IGFBP-3 directly interacts with Erk1/2. rBP-3 protein conjugated to NTA-agarose beads was incubated with H1299 cell lysates, washed, electrophoresed, and immunoblotted for Erk1/2 and p38. We found an interaction between IGFBP-3 and Erk1/2 that increased with the amount of the cell lysate added (Figure 6B). However, we did not detect an interaction between p38α and IGFBP-3.
IGFBP-3 binds to and inactivates Erk. (A,C-D) H1299 cells were pretreated with rBP-3 for 3 hours (A) or transiently transfected with pEV or pBP-3 (C), or with pEV-Flag or pBP-3-Flag (D) for 2 days. Lysates from H1299 cells were used for immunoprecipitation with rabbit anti-Erk1/2 antibody. Erk1/2, Flag, IGFBP-3, and p38α were detected by Western blotting. Whole cell lysates (WCLs) were subjected to a Western blot assay for phospho-Erk and phospho-p38 to determine specific inhibition of Erk by IGFBP-3. EV: empty vector. pBP-3-Flag: pCMV6-IGFBP-3-Flag. (B) Ni-NTA bead-bound his-tagged rBP-3 was incubated with H1299 whole cell lysate (150, 300, or 450 μg) for 5 hours before being washed and subjected to Western blot analysis for Erk1/2, p38α, and IGFBP-3. (E) H1299 cells were incubated with rBP-3 for 3 hours and stimulated with 10% FBS; samples were removed at different time points, as shown. The cell lysates were used for immunoprecipitation with rabbit anti-Erk antibody. Western blotting was used to detect IGFBP-3 and Erk from immunoprecipitation or whole cell lysates.
IGFBP-3 binds to and inactivates Erk. (A,C-D) H1299 cells were pretreated with rBP-3 for 3 hours (A) or transiently transfected with pEV or pBP-3 (C), or with pEV-Flag or pBP-3-Flag (D) for 2 days. Lysates from H1299 cells were used for immunoprecipitation with rabbit anti-Erk1/2 antibody. Erk1/2, Flag, IGFBP-3, and p38α were detected by Western blotting. Whole cell lysates (WCLs) were subjected to a Western blot assay for phospho-Erk and phospho-p38 to determine specific inhibition of Erk by IGFBP-3. EV: empty vector. pBP-3-Flag: pCMV6-IGFBP-3-Flag. (B) Ni-NTA bead-bound his-tagged rBP-3 was incubated with H1299 whole cell lysate (150, 300, or 450 μg) for 5 hours before being washed and subjected to Western blot analysis for Erk1/2, p38α, and IGFBP-3. (E) H1299 cells were incubated with rBP-3 for 3 hours and stimulated with 10% FBS; samples were removed at different time points, as shown. The cell lysates were used for immunoprecipitation with rabbit anti-Erk antibody. Western blotting was used to detect IGFBP-3 and Erk from immunoprecipitation or whole cell lysates.
Because large amounts of recombinant protein may cause coimmunoprecipitation artifacts and binding of cellular extracts to IGFBP-3 conjugated to affinity beads may not establish that IGFBP-3 interacts with Erk1/2 in vivo, we performed coimmunoprecipitation assays by using H1299 cell lysates that had been transfected with expression vectors carrying IGFBP-3 (pBP-3; Figure 6C) or Flag-tagged IGFBP-3 (pBP-3-Flag; Figure 6D). We found an association between Erk1/2 and IGFBP-3 in these cells, further confirming an Erk1/2-IGFBP-3 interaction. In contrast, a control immunoprecipitation using an anti-p38α antibody revealed that p38α was not associated with IGFBP-3 (Figure 6D bottom). IGFBP-3-Flag expression induced a marked decrease in Erk phosphorylation, with a slight increase in p38α phosphorylation. These findings suggest a direct and specific interaction between Erk1/2 and IGFBP-3. Thus, the interaction between IGFBP-3 and Erk does not appear to be an artifact of the high IGFBP-3 concentration.
We then correlated Erk1/2 activation with the association between IGFBP-3 and Erk1/2 by incubating H1299 cells with rBP-3 in serum-free conditions and analyzing samples obtained at several time points up to 30 minutes after stimulation with 10% FBS. IGFBP-3 was strongly associated with Erk1/2 under the serum-starved condition (Figure 6E). We found that 10% FBS stimulated Erk1/2 activation in control cells; however, Erk1/2 remained inactive in rBP-3–treated cells in which IGFBP-3 associated with Erk1/2. IGFBP-3 levels in H1299 cells gradually decreased 15 minutes after FBS stimulation, and Erk1/2 was rephosphorylated. These findings suggest that IGFBP-3 binds to Erk1/2, obstructing its activation and downstream signaling cascades.
Transcolocalization of IGFBP-3 with Erk in H1299 cells
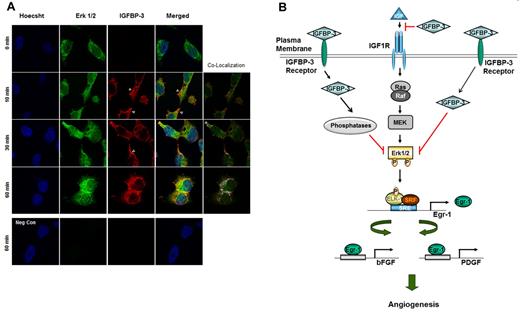
We performed confocal microscopy to determine whether exogenously supplied recombinant IGFBP-3 protein entered the cytosol and interacted with Erk. Serum-starved H1299 cells were treated with rBP-3 (10 μg/mL) for 1 hour; the cells were collected after 0, 10, 30, and 60 minutes and subjected to immunofluorescence staining with antibodies against IGFBP-3 or Erk1/2. As shown in Figure 7A, IGFBP-3 was localized near the membrane and in the cytosol 10 minutes after treatment and translocalized more to the cytosol by 60 minutes. Furthermore, IGFBP-3 and Erk1/2 colocalization at the cytoplasmic and perinuclear regions was markedly increased after 60 minutes. Interestingly, nuclear colocalization of IGFBP-3 and Erk1/2 was not detected, suggesting that IGFBP-3 binds and inactivates Erk1/2, resulting in suppression of Erk1/2 nuclear localization.
Translocalization and colocalization of rBP-3 with Erk in H1299 cells. (A) H1299 cells on cover slips were treated with rhIGFBP-3 (10 μg/mL) and fixed at 0, 10, 30, and 60 minutes after being washed 3 times in PBS. Nuclei (blue) were stained with Hoechst 33342 (1 μg/mL). Erk1/2 (green) was stained with mouse anti-Erk antibody and secondary antibodies conjugated with Alexa Fluor 488. IGFBP-3 (red) was stained with rabbit anti-IGFBP-3 antibody and secondary antibodies conjugated with Alexa Fluor 568. For the negative control, cells treated with rhIGFBP-3 for 60 minutes were stained with secondary but not primary antibodies. Neg Con: negative control. Colocalization map: schematic plot of colocalization; white dots represent the colocalization of Erk and IGFBP-3. The empty arrowhead indicates IGFBP-3 accumulation near the cell membrane. (B) Schematic model of IGFBP-3's angiogenesis inhibition. IGFBP-3 inhibits tumor angiogenesis by IGF-dependent and -independent mechanisms. In the IGF-independent mechanism, IGFBP-3 directly binds to and inactivates Erk1/2, prevents Elk-1 activation and binding between activated Elk-1 and Egr-1 promoter, and inhibits expression of Egr-1 and its target genes, including bFGF and PDGF, resulting in suppression of angiogenesis and tumor growth.
Translocalization and colocalization of rBP-3 with Erk in H1299 cells. (A) H1299 cells on cover slips were treated with rhIGFBP-3 (10 μg/mL) and fixed at 0, 10, 30, and 60 minutes after being washed 3 times in PBS. Nuclei (blue) were stained with Hoechst 33342 (1 μg/mL). Erk1/2 (green) was stained with mouse anti-Erk antibody and secondary antibodies conjugated with Alexa Fluor 488. IGFBP-3 (red) was stained with rabbit anti-IGFBP-3 antibody and secondary antibodies conjugated with Alexa Fluor 568. For the negative control, cells treated with rhIGFBP-3 for 60 minutes were stained with secondary but not primary antibodies. Neg Con: negative control. Colocalization map: schematic plot of colocalization; white dots represent the colocalization of Erk and IGFBP-3. The empty arrowhead indicates IGFBP-3 accumulation near the cell membrane. (B) Schematic model of IGFBP-3's angiogenesis inhibition. IGFBP-3 inhibits tumor angiogenesis by IGF-dependent and -independent mechanisms. In the IGF-independent mechanism, IGFBP-3 directly binds to and inactivates Erk1/2, prevents Elk-1 activation and binding between activated Elk-1 and Egr-1 promoter, and inhibits expression of Egr-1 and its target genes, including bFGF and PDGF, resulting in suppression of angiogenesis and tumor growth.
Discussion
IGFBP-3 is known to have IGF-independent antiangiogenic antitumor activities; however, mechanisms by which the IGF-independent activities of IGFBP-3 are mediated are not clearly understood. In this article, we have demonstrated a novel principal mechanism by which IGFBP-3 blocks tumor angiogenesis induced by NSCLC cells, including blocking the autocrine and paracrine loops of angiogenic factors by inhibiting the production of angiogenic factors such as bFGF and PDGF. IGFBP-3 appears to bind to and inactivate Erk1/2, which results in Elk-1 inactivation and Elk-1–SRE binding suppression in the Egr-1 promoter. Ultimately, this inhibits the transcription of Egr-1 and its target genes, including bFGF and PDGF.
Because IGFBP-3 has been shown to inhibit vascular endothelial cell survival45,46 and induce tumor vasculature normalization,47 it is believed to have antiangiogenic properties. In support of this notion, IGFBP-3–mediated antitumor activities have been found to involve angiogenesis suppression in various cancer types.7,8,45 However, to date, the mechanisms by which IGFBP-3 regulates angiogenesis are not understood.
We investigated the mechanisms that mediate IGFBP-3's antiangiogenic activity and found that it regulates the expression of bFGF and PDGF, potent angiogenic factors, in NSCLC, HNSCC, and vascular endothelial cells. Because the Egr-1 transcription factor plays a role in regulating these angiogenic factors' expression by binding to the G/C-rich consensus element in their promoters, we determined effects of IGFBP-3 on Egr-1 expression. As expected, IGFBP-3 decreased Egr-1 promoter activity and expression; it also led to reduced transcription of Egr-1 target genes, including bFGF and PDGF. Furthermore, Egr-1 overexpression restored the bFGF promoter and angiogenic activities that had been suppressed by IGFBP-3. Therefore, IGFBP-3 may regulate tumor angiogenesis through mechanisms that involve Egr-1, a transcription factor for bFGF and PDGF expression.
IGFBP-3 leads to decreased clusterin expression, which is induced by stress-induced Egr-1 transactivation through IGF-1R-Src-MEK-Erk signal transduction cascade activation.48 Hence, the inhibitory effect of IGFBP-3 on Egr-1 expression and the resultant reduction in bFGF and PDGF transcription may have occurred through IGF-dependent mechanisms. However, Egr-1 regulation by IGFBP-3 appears to include novel IGF-independent mechanisms for the following reasons: (1) both wild-type IGFBP-3 and the non-IGF–binding IGFBP-3-ggg mutant suppressed the Egr-1 promoter activity that had been stimulated by mitogenic and nonmitogenic stimuli in H1299, IGF-1R–null (R−), and IGF-1R–positive (R+) cells; and (2) Egr-1 overexpression abrogated the antiangiogenic activity of wild-type IGFBP-3 and the IGFBP-3-ggg mutant in NSCLC cells. Our gross promoter deletion and mutation analyses revealed that the SREs and their adjacent Ets sites are critical to the IGFBP-3–mediated reduction in Egr-1 transcription. Importantly, our ChIP assay results showed that Elk-1's binding to SRE sites 3-5 was inhibited by IGFBP-3 treatment, whereas SRF binding was not affected. This pattern of constitutive binding by SRF without ternary complex formation is consistent with some49,50 SRE-mediated transcription models. One possible mechanism of the IGFBP-3–mediated down-regulation of Elk-1 binding is inactivation of the Ras-Raf-MEK1/2-Erk1/2 signaling cascade, which phosphorylates and activates Elk-1 and regulates SRE-Ets–mediated Egr-1 transcription.49 The Ras-Raf-MEK1/2-Erk1/2 pathway regulates endothelial cell behavior during angiogenesis by stimulating cell proliferation, survival, migration, and invasion.51,52 MEK1 knockout mice have defects in angiogenesis in the placenta, resulting in embryonic lethality.53 Surprisingly, our biochemical analysis revealed that rBP-3 treatment resulted in Erk1/2 inactivation and suppression of Elk-1 and Egr-1 expression without concordant inactivation of MEK1/2 in NSCLC, R+, and R− cells. These findings demonstrate that IGFBP-3 can inactivate Erk/Elk-1 independent of MEK-1.
Scaffolding proteins, such as Ras kinase suppressor, MAPK organizer-1, and MEK partner-1, are known as regulators of the Ras-Raf-MEK signaling pathway.54 This knowledge led us to begin investigating the potential involvement of these scaffolding proteins in IGFBP-3–mediated Erk1/2 inactivation. However, we detected no interactions among these scaffolding proteins and IGFBP-3 in H1299 or H460 cells that had been transfected with pBP-3 or infected with Ad-BP-3 (data not shown). Furthermore, interaction between these scaffolding proteins and signaling components of Ras-Raf-MEK-Erk signaling was not appreciably changed by the IGFBP-3 expression. These findings suggest that IGFBP-3 inactivates Erk1/2 through mechanisms that are independent of these scaffolding proteins. Surprisingly, our data clearly demonstrated that IGFBP-3 can directly interact with Erk1/2, leading to Erk1/2 activity suppression. This interaction appeared to be Erk1/2-specific because the high level of IGFBP-3 obtained after treatment with recombinant protein or transient transfection with mammalian expression vector did not induce p38 binding.
In summary, our data provide a model of an IGF-independent mechanism through which IGFBP-3 inhibits the Erk1/2-Elk-1 activation loop and thus reduces Egr-1 expression. The intracellular translocalization of IGFBP-3 may have been mediated by membrane receptors, as proposed in previous studies.55-57 In this study, recombinant IGFBP-3 translocated through the membranes into the cytoplasm. Additional mechanisms underlying the IGFBP-3–mediated inactivation of Erk1/2 should be explored (such as whether IGFBP-3's binding to surface receptors triggers phosphatases to dephosphorylate Erk1/2), our current data provide a schematic model in which IGFBP-3 directly binds to Erk1/2 and inhibits its activation, thereby preventing Elk-1 phosphorylation. IGFBP-3's dissociation from Erk1/2 during mitogen stimulation allows Erk1/2 activation, leading to Elk-1–induced expression of Egr-1, a master transcription factor that regulates genes that are strongly implicated in tumor growth and angiogenesis. Ultimately, this activation stimulates the Egr-1–mediated transcriptional events observed in cancer cells (Figure 7B).
Antiangiogenic therapies currently in clinical trials induce acquired resistance in cells by causing them to shift to other growth factor-induced angiogenesis mechanisms. Therefore, IGFBP-3's ability to regulate multiple potent angiogenic factors makes it an attractive antiangiogenic, antineoplastic agent. However, because reduced tumor vascularization after antiangiogenic therapy could induce hypoxia and thus promote the spread of cancer cells toward a more oxygenated environment,58 extensive studies are needed before clinical trials in which IGFBP-3 overexpression is induced are considered. In this work, we did not determine the exact mechanisms by which IGFBP-3 enters cells and by which serum stimulation induces IGFBP-3 dissociation from Erk1/2, although serum stimulation may activate proteases, including various MMPs, that cause IGFBP-3's degradation.59-61 Further studies are warranted to understand the molecular mechanisms of IGFBP-3-Erk1/2 interaction.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Warren Davis (Medical University of South Carolina) and David M. Cohen (Oregon Health and Science University) for generously donating plasmids.
This work was supported by National Institutes of Health grants R01 CA100816 and R01 CA109520 (H.-Y.L.) and P50 CA907007 (S.M.L.). This research is supported in part by the National Institutes of Health through M. D. Anderson's Cancer Center Support Grant CA016672. Specifically, this study was performed by use of the microscopy equipment that is part of the Flow Cytometry and Cellular Imaging Core Facility at M. D. Anderson.
National Institutes of Health
Authorship
Contribution: J.-H.K., D.S.C., O.-H.L., and S.-H.O. performed the in vitro experiments; S.-H.O. contributed to the animal experiments; S.M.L. provided clinical input and financial support; and J.-H.K. and H.-Y.L. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for O.-H.L. is the Severance Integrative Research Institute for Cerebral & Cardiovascular Diseases, Yonsei University Health System, Seoul, Republic of Korea. The current affiliation for S.-H.O is the Division of Cancer Biology, National Cancer Center, Goyang-si, Republic of Korea.
Correspondence: Ho-Young Lee, PhD, Research Institute of Pharmaceutical Sciences, College of Pharmacy, Seoul National University, 599 Gwanak-ro, Gwanak-gu, Seoul 151-742, Republic of Korea; e-mail: hylee135@snu.ac.kr.
References
Author notes
J.-H.K. and D.S.C. contributed equally to this work.