Abstract
The FOXO transcription factors control proliferation and apoptosis in different cell types. Their activity is regulated by posttranslational modifications, mainly by the PI3K-PKB pathway, which controls nuclear export and degradation. We show that FOXO1 is highly expressed in normal germinal center B cells as well as in non-Hodgkin lymphomas, including follicular lymphoma, diffuse large B-cell lymphoma, mucosa-associated lymphoid tissue non-Hodgkin lymphoma, B-cell chronic lymphocytic leukemia, and mantle cell lymphoma. In contrast, in 31 of 32 classical Hodgkin lymphoma (cHL) cases, Hodgkin and Reed-Sternberg cells were FOXO1 negative. Neoplastic cells of nodular lymphocyte-predominant Hodgkin lymphoma were negative in 14 of 20 cases. FOXO1 was down-regulated in cHL cell lines, whereas it was expressed in non-Hodgkin lymphoma cell lines at levels comparable with normal B cells. Ectopic expression of a constitutively active FOXO1 induced apoptosis in cHL cell lines and blocked proliferation, accompanied with cell-cycle arrest in the G0/G1 phase. We found that, in cHL cell lines, FOXO1 is inactivated by multiple mechanisms, including constitutive activation of AKT/PKB and MAPK/ERK kinases and up-regulation of microRNAs miR-96, miR-182, and miR-183. These results suggest that FOXO1 repression contributes to cHL lymphomagenesis.
Introduction
FOXO1 belongs to the subgroup O of forkhead transcription factors (FOX), which share the highly conserved forkhead DNA-binding domain. This O subgroup consists of the 4 members, FOXO1, FOXO3, FOXO4, and FOXO6.1 FOXO transcription factors control different cellular processes, such as stress response, proliferation, apoptosis, and cell differentiation.2 FOXO target genes include the cell-cycle regulators CDKN1A and CDKN1B, proapoptotic genes BIM, PMAIP1/NOXA, and FASL as well as oxidative stress protectors SOD2 and CAT.1,3,4 FOXO1 is of particular interest in B cells because it is highly expressed (http://biogps.gnf.org) and plays a nonredundant role in B-cell differentiation by activating recombination activating genes Rag1 and Rag2 and germinal center (GC) genes Bcl6 and Aicda.5–7 In addition, FOXO1 was reported to be a regulator of B-cell death, and its inactivation by B-cell receptor signaling to AKT/PKB kinase was found to be critical for survival of mature B cells.8 Several other kinases, including the IκB kinase9 and ERK,10 have been identified to phosphorylate and facilitate nuclear export of FOXO proteins.
It is well documented that the oncogenic program of B-cell lymphomas (BCLs) is tightly related to the survival program of their nontransformed precursor cells. In the majority of non-Hodgkin BCLs, the oncogenic program is established as an error of normal GC processes (ie, somatic hypermutation and class switch recombination), which facilitate mutations of tumor suppressor genes and translocation of oncogenes.11 Execution of the oncogenic program in these lymphomas requires the maintenance of the GC program and the prevention of post-GC differentiation steps (eg, by BCL6 and PAX5 translocation)12 and probably by deregulation of other transcription factors regulating GC phenotype and terminal differentiation of B cells.13 Unlike other types of BCLs, neoplastic cells of classical Hodgkin lymphoma (cHL) lose most of their B-cell phenotype.14 At the same time, Hodgkin and Reed-Sternberg (HRS) cells express many genes of activated B cells, indicating that the oncogenic program of cHL may arise in the process of deregulation of mechanisms, controlling activity of NF-κB,15 ERK,16 and JAK/STAT17 pathways in activated B cells. In addition, AKT and ERK kinases are frequently constitutively activated in different B-lymphoma entities, including follicular lymphoma (FL),18 diffuse large BCL (DLBCL),19 Burkitt lymphoma (BL),20 and cHL.21,22 Therefore, it has been hypothesized that FOXO1 inactivation might be a common event contributing to lymphomagenesis in several lymphoma entities.8
Given the proposed critical role of inactivation of FOXO1 function in BCLs, we investigated FOXO1 expression in different BCL entities. Unexpectedly, whereas FOXO1 expression was maintained in majority of non-Hodgkin lymphomas (NHLs), down-regulation was observed in cHL and lymphocyte-predominant Hodgkin lymphoma (LPHL). We found that re-expression of FOXO1 inhibited proliferation and induced apoptosis in all cHL cell lines. We also found that the depletion of FOXO1 in cHL is brought about by complex mechanisms, including constitutive AKT and ERK activation, up-regulation of miR-96, miR-182, and miR-183, and chromosomal aberrations.
Methods
Cell lines, chemicals, and treatments
cHL cell lines of B-cell origin KM-H2, L1236, L428, SUP-HD1 and BL cell lines Namalwa, Daudi, and Ramos, FL cell lines DoHH-2 and WSU-NHL, and DLBCL cell lines Karpas-422 and SU-DHL-5 were cultured in RPMI 1640 medium (Life Technologies) supplemented with 10% FCS (PAN Biotech), l-glutamine, 100 U/mL of penicillin, 100 μg/mL of streptomycin, and 50μM 2-mercaptoethanol at 37°C and 5% CO2. The cHL cell line U-HO123 was cultured in RPMI 1640/Iscove medium (1:4) supplemented with 10% FCS, glutamine, and antibiotics. The clones of cHL cell lines, stably expressing FOXO1(A3)ER, were created by transfection with pcDNA-FOXO1(A3)ER vector, using a nucleofector device (Lonza Germany) followed by selection with 1 mg/mL of G418 sulfate, or by infection with pCFG5-FOXO1(A3)ER vector followed by selection with 100 μg of zeocin (Calbiochem), respectively. As control, we used cell lines transfected with empty vectors.
A pcDNA-FOXO1(A3)ER vector was created by recloning of FOXO1(A3)ER from pBABE- FOXO1(A3)ER into the BamH1/XhoI site of pcDNA3.1(+) vector. The pBABE- FOXO1(A3)ER vector expressing the constitutively active form of human FOXO1 in-frame with the modified tamoxifen-specific version of the murine estrogen receptor-α ligand-binding domain24 was provided by T. G. Unterman (Chicago, Illinois). The pCFG5-FOXO1(A3)ER vector was constructed by cloning of FOXO1(A3)ER into BamHI/EcoRV sites of the pCFG5-IEGZ vector. FOXO1-ER constructs were induced by 4-hydroxytamoxifen (4-OHT; Calbiochem) at a final concentration of 200nM. MEK1/2 inhibitor U0126 was obtained from Cell Signaling, and AKT inhibitor KP372-1 was from MoBiTec GmbH.
FISH and array comparative genomic hybridization
KM-H2, L428, L1236, SUP-HD1, and U-HO1 cells were prepared for metaphase cytogenetics and FISH following routine cytogenetics protocols as previously described.25 To investigate copy number changes of the FOXO1 locus on chromosome 13q14.11 in these cell lines, we used the following locus-specific and reference FISH probes: LSI FOXO1 Dual Color, Break Apart rearrangement probe, LSI D13S319 (13q14.2), and LSI 13q34 Spectrum Green (Abbott Molecular). For each cell line, 100 interphase nuclei and 20 metaphase cells were captured and scored. Because of cell-to-cell variation, only the most common signal constellations in representative metaphases for each cell line were reported. Furthermore, array comparative genomic hybridization profiles of microdissected HRS cells from 53 cHL patients that had been reported in a previous study were specifically reanalyzed for copy number changes of the FOXO1 gene locus.25
DNA methylation and mutational analysis
The analysis of the methylation status of FOXO1 promoter CpG island and mutational status of FOXO1 coding exons is described in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Apoptosis, cell-cycle analysis, and viability tests
Apoptosis was assessed by annexin V staining PE-conjugated (BioVision). A total of 1 × 105 cells were washed in PBS and resuspended in 500 μL of binding buffer, containing 1 μL of the annexin V–PE reagent and 2 μg/mL of 7-amino-actinomycin D. After 5 minutes of incubation at room temperature, fluorescence was recorded using a FACSCalibur flow cytometer (BD Biosciences). The results were calculated as specific apoptosis (SA) using the formula: SA(%) = 100 × (AE − AC)/(100 − AC), where AE equals percent of apoptotic cells in the experiment group and AC equals percent of apoptotic cells in the control group. For cell-cycle analysis, cells were washed with PBS, fixed in cold 70% ethanol overnight, spun down, and incubated for 1 hour at room temperature with 40 μg/mL propidium iodide and 100 μg/mL RNAse A in PBS. The cell-cycle distribution was measured by flow cytometry and analyzed with help of ModFit LT Version 2.0 software (Verify Software House).
Cell viability MTT assay was done as described.26 The IC50 was calculated using BioDataFit Version 1.02 software (http://www.changbioscience.com/stat/ec50.html).
Quantitative RT-PCR
Total RNA was isolated from 1 × 106 cells using the High Pure RNA Isolation kit or from frozen tissues using the High Pure RNA Tissue kit (Roche Applied Science) according to the manufacturer's instructions. A total of 2 mg of RNA and 0.5 mg of dT 18-mer primer were heated for 5 minutes at 70°C. After cooling on ice, the first strand of cDNA was synthesized by M-MLV reverse transcriptase (Promega). Quantitative RT-PCR was performed using QuantiTect SYBR Green PCR Kit (QIAGEN) by a LightCycler 480 real-time PCR instrument (Roche Diagnostics). The primers sequences are listed in supplemental Table 1. Primer sequences were identified using Genscript online software (www.genscript.com). All oligonucleotides were synthesized by biomers.net (Ulm, Germany).
Western immunoblot
Immunoblot was done as described earlier.27 The proteins were detected either by rabbit monoclonal antibodies against FOXO1(C29H4); AKT(pan)(C67E7); phospho-MAPK1/2(Thr202/204) #4370; MAP kinase (137F5) #4695; or by rabbit polyclonal antibodies against phospho-AKT(Ser473) #9271; phospho-AKT(Thr308) #9272 (Cell Signaling). Actin expression was detected by anti-actin rabbit antibody A5060 (Sigma-Aldrich). For detection of bound rabbit Ig, we used goat anti–rabbit IgG-HRP (sc-2004; Santa Cruz Biotechnology). Signals were visualized using the SuperSignal West Dura extended-duration substrate (Thermo Scientific).
Human material, immunohistochemistry
Thirty-two cases of cHL and 20 cases of LPHL were included in this study. Twenty of the cHL cases belonged to the nodular sclerosis type, 6 cases were mixed cellularity type, 4 were lymphocyte-rich, and 2 were cHL not otherwise specified because of small size of the tissue sample. Twenty-eight cases of cHL were analyzed for EBV infection, 22 cases were EBV latent membrane protein (LMP) negative and in 6 cases HRS cells were LMP positive. NHL samples included 21 cases of FL, 13 cases of DLBCL, 6 cases of BL, 13 cases of mucosa-associated lymphoid tissue (MALT) NHL, 13 cases of B-cell chronic lymphocytic leukemia (B-CLL), and 13 cases of mantle-cell lymphoma (MCL). Lymphoma diagnosis was in accordance with the current World Health Organization classification.28 As control, we used 5 samples of nonneoplastic tonsils. All samples were drawn from our archive of formalin-fixed, paraffin-embedded tissues and pseudonymized to comply with the German law for ethical usage of archival tissue for clinical research (Deutsches Ärzteblatt 2003; 100 A1632). Approval for these studies was obtained from the University of Ulm ethics board. The CD19+ tonsillar cells and B-cell subtypes were isolated as described earlier.29 For immunostaining, deparaffinized tissue sections were heat-denatured in a pressure cooker and incubated with rabbit monoclonal antibody against FOXO1 used for immunoblot (1:25 dilution). Bound antibody was labeled using EnVision System (Dako Germany). Peroxidase activity was visualized by the substrate 3-amino-9-ethylcarbazole (0.1 mg/mL in 0.17M sodium acetate, pH 5.2 plus 0.01% H2O2). Nuclei were counterstained with hemalaun. LMP was detected by mouse anti-LMP antibody CS1-4 (M0897, Dako Germany) with help of REAL Detection Systems kit (Dako Germany). Picture acquisition was performed applying a Zeiss Axiophot microscope equipped with a KY-F75U digital camera (JVC) and the DISKUS Version 4.5 software (C.H. Hilgers Technisches Büro). Images were processed with Adobe Photoshop CS2 Version 9.0 software (Adobe systems).
miRNA assays
RNA was isolated using Pure-Link miRNA Isolation Kit (Invitrogen), and cDNA was synthesized using miScript Reverse Transcription Kit (QIAGEN). The expression of specific miRNAs was detected using miScript Primer Assay and miScript SYBR Green PCR kit (QIAGEN). U6 was used as reference gene. For primer sequence, see supplemental Table 1. To clarify the role of the miR-96, miR-182, and miR-183 in FOXO1 regulation, we used the psi-CHECK-2 dual luciferase reporter construct, containing sequences identical to FOXO1 3′-untranslated region (UTR) wild-type binding sites for the miR-96/182 (2122-2146 of FOXO1 3′-UTR) and to the miR-183 binding sites (2187-2102 of FOXO1 3′-UTR; supplemental Table 2). The sites were found with the help of miRanda online software (http://www.microrna.org, release August 2010) and scored as “Conserved miRNA with good mirSVR score.” As control, we used a construct with scrambled complementary nucleotides inside the miRNA binding sites. The constructs were created on the basis of the reporter vectors kindly provided by I. Guttilla and B. White (University of Connecticut Health Center, Farmington, CT) as described.30 To inhibit function of the miRNAs, we used miScript miRNA inhibitors (QIAGEN) specific for miR-96, miR-182, and miR-183. L428 cells were transfected with 1μM of the specific miRNA inhibitor or with the miScript miRNA negative control RNA by nucleofection (Lonza). The cells were harvested for protein isolation 48 hours after transfection.
Results
FOXO1 expression is low in cHL
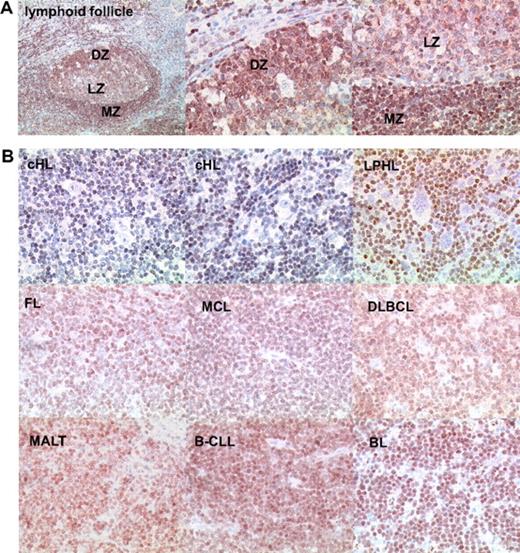
Of the different members of the FOXO family, FOXO1 is highly expressed in the B-cell lineage (http://biogps.gnf.org). This could be confirmed by immunohistochemistry of tonsil sections (Figure 1A). In tonsils, there was strong staining in the dark zone of the GC, where centroblasts can be found at high density, as well as in the follicular mantle zone. Slightly weaker staining was seen in the GC light zone that harbors centrocytes as the dominant cell population. Costaining of FOXO1 and proliferation marker Ki-67 using immunofluorescence corroborated expression of FOXO1 in proliferating GC cells (supplemental Figure 1). We then characterized B-cell derived primary tumors representing different BCLs (Figure 1B). FOXO1 was expressed in multiple non-Hodgkin BCL subtypes at levels comparable with normal B cells, although the expression levels were somewhat variable. B-CLL and BL demonstrated the highest levels of FOXO1 expression, with strong specific staining observed in 12 of 13, and 5 of 6 cases, respectively. Strong FOXO1 staining was also detected in 9 of 13 cases of MALT-NHL, 11 of 21 cases of FL, 10 of 13 cases of MCL, and 7 of 13 cases of DLBCL. However, in 31 of 32 cHL cases analyzed, HRS cells were FOXO1-negative (Figure 1B; Table 1). Only one cHL sample showed weak FOXO1 staining in HRS cells. As an internal control, intense FOXO1 expression was found in a substantial fraction of the reactive bystander population surrounding the HRS cells. In addition, 14 of 20 LPHL cases were negative for FOXO1. Thus, high FOXO1 expression characteristic for normal B cells is generally retained in a majority of NHLs, whereas in cHL and LPHL FOXO1 expression is typically lost.
FOXO1 expression in lymphoid FL, HL, and NHL. (A) FOXO1 expression in tonsillar lymphoid follicle. Low magnification image represents immunohistochemical staining of FOXO1 distribution in the whole follicle (original magnification ×50). High-magnification images show FOXO1 expression in cells of dark zone containing centroblasts (DZ) and light and marginal zone containing centrocytes and resting B cells, respectively (LZ and MZ; original magnification ×200). (B) FOXO1 expression in BCLs. Neoplastic cells of cHL and LPHL are FOXO1-negative (original magnification ×200). Objective: Plan-Neofluar 10×/0.3 NA for the low magnification of normal lymphoid follicle and Plan-Neofluar 40×/0.75 NA objective for the high magnification normal lymphoid follicle and lymphoma images.
FOXO1 expression in lymphoid FL, HL, and NHL. (A) FOXO1 expression in tonsillar lymphoid follicle. Low magnification image represents immunohistochemical staining of FOXO1 distribution in the whole follicle (original magnification ×50). High-magnification images show FOXO1 expression in cells of dark zone containing centroblasts (DZ) and light and marginal zone containing centrocytes and resting B cells, respectively (LZ and MZ; original magnification ×200). (B) FOXO1 expression in BCLs. Neoplastic cells of cHL and LPHL are FOXO1-negative (original magnification ×200). Objective: Plan-Neofluar 10×/0.3 NA for the low magnification of normal lymphoid follicle and Plan-Neofluar 40×/0.75 NA objective for the high magnification normal lymphoid follicle and lymphoma images.
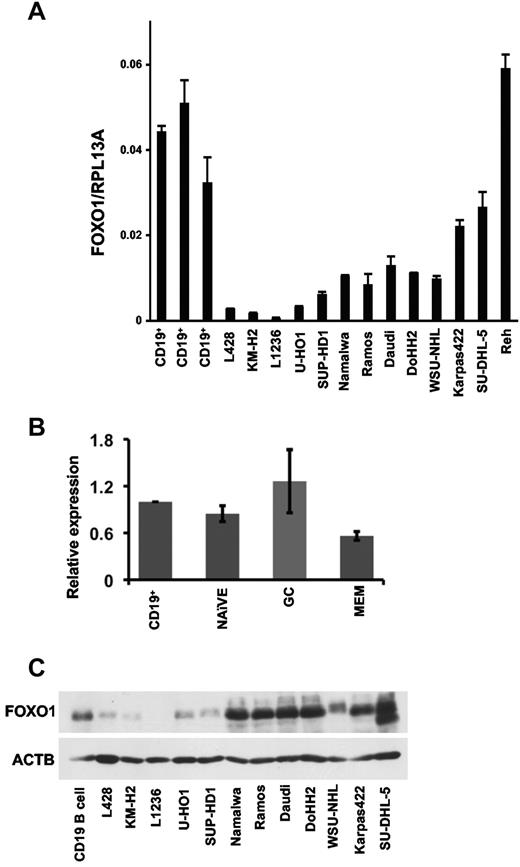
We then asked whether the low protein expression determined by immunohistochemistry would also be reflected at the RNA level. To this end, we analyzed FOXO1 expression in CD19+ tonsillar B cells, cHL, BL, DLBCL, FL, and pre-BCL cell lines using quantitative RT-PCR (Figure 2A). Expression of FOXO1 mRNA was significantly higher in CD19+ tonsillar cells than in BCL cell lines, with the exception of the Reh pre-B-lymphoma cell line. Remarkably, cHL cell lines had the lowest levels of FOXO1 mRNA expression (Figure 2A), thus supporting the immunohistochemistry data obtained in clinical samples. Immunohistochemistry stainings had revealed some differences in FOXO1 expression among subsets of B cells. We therefore compared FOXO1 mRNA expression in GC, which are bona fide normal counterparts of many BCL entities, memory, and naive B cells. FOXO1 expression in these B-cell subtypes did not differ significantly from total tonsillar CD19+ cells (Figure 2B). We also assessed FOXO1 protein expression in normal tonsillar CD19+ B cells and lymphoma cell lines by immunoblot (Figure 2C). FOXO1 protein expression in normal B cells was significantly higher than in cHL cell lines. In contrast, the BL cell lines Namalwa, Ramos, and Daudi, the DLBCL cell lines DoHH2, WSU-NHL, and SU-DHL-5, and the FL cell line Karpas-422 expressed FOXO1 at levels comparable with CD19+ cells.
FOXO1 expression in B-cell subsets and in BCL cell lines. (A) Expression of FOXO1 mRNA in tonsillar CD19+ cells, cHL, BL, FL, DLBCL, and pre-BCL cell lines was assessed by quantitative RT-PCR as described in “Quantitative RT-PCR.” Data are mean ± SD of target gene to reference gene (FOXO1/RPL13A) ratio. (B) Expression of FOXO1 mRNA in B-cell subsets. The naive, GC, memory cells (MEM), and CD19+ cells were isolated from the fresh surgical material by magnetic cell sorting. The FOXO1 mRNA expression was measured by quantitative RT-PCR, and the data were analyzed by comparative Ct method. The figure represents mean of relative expression ± SD. (C) Expression of FOXO1 protein in human tonsillar CD19+ cells and in cHL cell lines. The proteins from samples of CD19+ cells and cultured lymphoma cell lines were separated by PAGE, and FOXO1 expression was assessed by immunoblot.
FOXO1 expression in B-cell subsets and in BCL cell lines. (A) Expression of FOXO1 mRNA in tonsillar CD19+ cells, cHL, BL, FL, DLBCL, and pre-BCL cell lines was assessed by quantitative RT-PCR as described in “Quantitative RT-PCR.” Data are mean ± SD of target gene to reference gene (FOXO1/RPL13A) ratio. (B) Expression of FOXO1 mRNA in B-cell subsets. The naive, GC, memory cells (MEM), and CD19+ cells were isolated from the fresh surgical material by magnetic cell sorting. The FOXO1 mRNA expression was measured by quantitative RT-PCR, and the data were analyzed by comparative Ct method. The figure represents mean of relative expression ± SD. (C) Expression of FOXO1 protein in human tonsillar CD19+ cells and in cHL cell lines. The proteins from samples of CD19+ cells and cultured lymphoma cell lines were separated by PAGE, and FOXO1 expression was assessed by immunoblot.
FOXO1 induces growth arrest and apoptosis in cHL cell lines
Given the consistent down-regulation of FOXO1 in cHL, we asked whether FOXO1 might exert tumor suppressor function for this tumor entity. Therefore, we stably expressed in 5 cHL cell lines an inducible FOXO1(A3)ER construct encoding a constitutively active protein. In this mutated protein, the phosphorylation sites of AKT/PKB and SGK are mutated to alanines, thus preventing inactivation by these kinases. Moreover, in this construct, the FOXO1 coding sequence is fused in frame with the mutant ligand-binding domain of the estrogen receptor allowing specific activation by 4-OHT. This construct has been extensively used in previous studies to investigate FOXO1 effects in a variety of cell types, including BCL.31
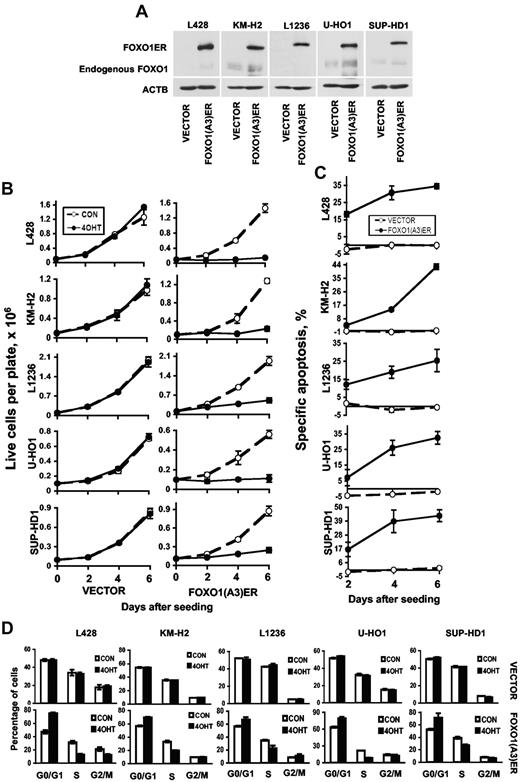
Stably transfected cHL cell lines were generated, and expression of FOXO1(A3)ER was verified by immunoblotting (Figure 3A). When FOXO1 nuclear translocation was induced by 4-OHT, we observed a significant reduction of viable cells in all cHL cell lines (Figure 3B). FOXO proteins may inhibit proliferation and/or induce cell death, depending on the cell type. We analyzed apoptosis in these cell clones and found a significant rate of apoptosis in all cHL cell lines (Figure 3C). FOXO1 activation also induced changes in cell-cycle parameters. In all cell lines, we saw reduced levels of cells in the S-phase accompanied by an increase in the G1/G0 fraction (Figure 3D). Thus, FOXO1 induces cell-cycle arrest and apoptosis in cHL cell lines.
FOXO1 induces growth arrest and apoptosis in cHL cell lines. (A) Expression of FOXO1(A3)ER in clones of cHL cell lines. Western immunoblot using antibodies specific for FOXO1 and ACTB as loading control are shown. (B) Growth-inhibitory effect of FOXO1. cHL cell lines stably expressing the empty vector or the vector containing FOXO1(A3)ER were seeded in a 6-well plate in 2 mL of complete culture medium at a density of 1 × 105 cells/well and treated with 4-OHT at a concentration of 200nM or with a vehicle. Numbers of live cells were calculated by hemacytometer using trypan blue staining (C). Apoptosis was measured by annexin V–PE/7-amino-actinomycin D staining at the same time points as cell counting. Data represent the mean ± SD of 3 independent experiments. (D) FOXO1 activation inhibits cell-cycle transition. A total of 1 × 106 cells were seeded in 10 mL of complete medium. The experimental group was treated with 200nM 4-OHT. After 24 hours of incubation with 4-OHT, cells were harvested and cell-cycle distribution was analyzed by propidium iodide. Bars represent the mean of 3 measurements ± SD. The data are representative of at least 3 independent experiments that gave similar results.
FOXO1 induces growth arrest and apoptosis in cHL cell lines. (A) Expression of FOXO1(A3)ER in clones of cHL cell lines. Western immunoblot using antibodies specific for FOXO1 and ACTB as loading control are shown. (B) Growth-inhibitory effect of FOXO1. cHL cell lines stably expressing the empty vector or the vector containing FOXO1(A3)ER were seeded in a 6-well plate in 2 mL of complete culture medium at a density of 1 × 105 cells/well and treated with 4-OHT at a concentration of 200nM or with a vehicle. Numbers of live cells were calculated by hemacytometer using trypan blue staining (C). Apoptosis was measured by annexin V–PE/7-amino-actinomycin D staining at the same time points as cell counting. Data represent the mean ± SD of 3 independent experiments. (D) FOXO1 activation inhibits cell-cycle transition. A total of 1 × 106 cells were seeded in 10 mL of complete medium. The experimental group was treated with 200nM 4-OHT. After 24 hours of incubation with 4-OHT, cells were harvested and cell-cycle distribution was analyzed by propidium iodide. Bars represent the mean of 3 measurements ± SD. The data are representative of at least 3 independent experiments that gave similar results.
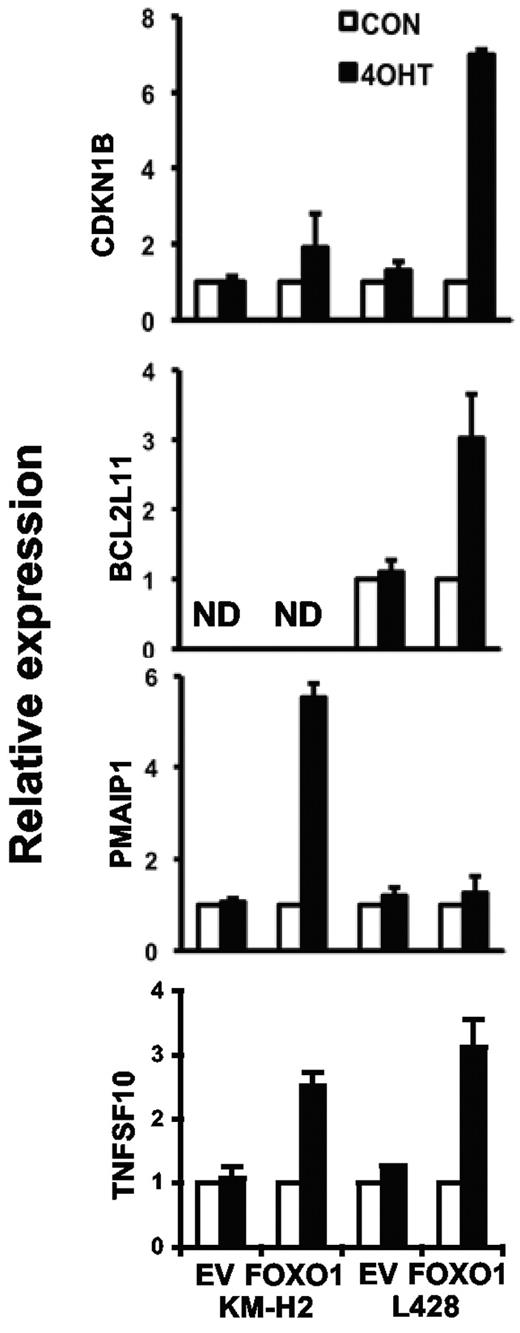
Given the phenotype of the FOXO1-expressing cells, we investigated expression of the known FOXO target genes CDKN1B/P27KIP1, PMAIP1/NOXA, BCL2L11/BIM, and TNFSF10/TRAIL in empty vector and FOXO1(A3)ER-expressing L428 and KM-H2 cell lines (Figure 4). FOXO1 activation led to 1.9- and 6.9-fold up-regulation of the cell-cycle regulator CDKN1B in KM-H2 and L428 cells, respectively. The mRNA of the proapoptotic gene BCL2L11 was not detectable and was not induced in KM-H2 cells but was enhanced 3-fold in L428 cells. In contrast, the proapoptotic gene PMAIP1 was induced in KM-H2 cells, whereas TNFSF10 was significantly induced in both cell lines. Thus, the antiproliferative and proapoptotic effects of FOXO1 in cHL correlate with the activation of its known tumor suppressor target genes.
Activation of the FOXO target genes. KM-H2 and L428 cell lines expressing FOXO1(A3)ER were treated or not treated with 200nM 4-OHT. Twenty-four hours later, cells were harvested and TNFSF10, PMAIP1, BCL2L11, and CDKN1B mRNA expression was assessed as described in Figure 2. ND indicates not detected; and EV, empty vector. Data are mean of relative expression ± SD. The measurements were done in triplicates.
Activation of the FOXO target genes. KM-H2 and L428 cell lines expressing FOXO1(A3)ER were treated or not treated with 200nM 4-OHT. Twenty-four hours later, cells were harvested and TNFSF10, PMAIP1, BCL2L11, and CDKN1B mRNA expression was assessed as described in Figure 2. ND indicates not detected; and EV, empty vector. Data are mean of relative expression ± SD. The measurements were done in triplicates.
FOXO1 locus is often deleted in HRS cells of cHL and in cHL cell lines
To identify potential mechanisms for FOXO1 down-regulation in cHL, we investigated epigenetic and genetic changes of the FOXO1 locus. As B cell–specific genes are often epigenetically silenced by CpG island promoter methylation in cHL,32 we first analyzed the methylation status of the FOXO1 core promoter in human tonsils, CD19+ tonsillar cells, centroblasts, in cHL, and in BL cell lines (supplemental Figure 2). Although a minor increase of methylation was observed in KM-H2 cells, in all other lymphoma cell lines levels of methylation did not differ from those of the normal B-cell subsets. Therefore, we conclude that methylation of promoter CpG-island does not play a major role in FOXO1 down-regulation in cHL. We also analyzed all cHL cell lines for the presence of mutations in FOXO1 coding exons. No missense or nonsense mutations were found compared with peripheral blood mononuclear cells obtained from healthy donors (data not shown).
In previous studies, conventional cytogenetic analysis revealed recurrent structural and numerical aberrations of chromosome 13, harboring the FOXO1 locus, in cHL cell lines.23,33,34 To specifically investigate copy number changes of the FOXO1 locus, we performed FISH on the cHL cell lines KM-H2, L428, L1236, SUP-HD1, and U-HO1. We identified an interstitial deletion of chromosome 13q involving FOXO1 in the diploid cell line SUP-HD1, whereas in the triploid cell lines KM-H2 and L1236, as well as in tetraploid L428, only 2 chromosomes 13 (harboring the FOXO1 locus) were present (supplemental Figure 3). Only the diploid cell line U-HO1 had 2 FOXO1 alleles. These results are consistent with relative loss of the FOXO1 locus as part of numerical and large-scale chromosome 13 alterations in all investigated cell lines. We also reanalyzed previously published copy number data of primary HRS cells25 and found 6 of 53 cHL cases (11.3%) that harbored 13q14 deletions with involvement of the FOXO1 locus. Thus, chromosomal loss of the FOXO1 locus is a recurrent finding in cHL that might contribute to decreased gene expression in some cHL cases.
Constitutively activated AKT and ERK kinases contribute to FOXO1 repression in cHL cell lines
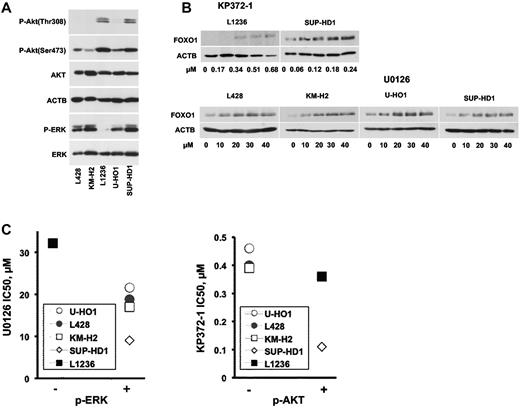
PI3K-PKB and ERK pathways regulating FOXO1 functional activity are often constitutively activated in cHL.21,22 To determine whether activation of the PI3K-PKB and/or ERK pathway might be involved in FOXO1 down-regulation in cHL, we investigated the activation status of AKT and ERK kinases, known regulators of FOXO1 activity. Among cHL lymphoma cell lines, pAKT(Thr308), a marker of AKT activation as well as phosphorylation of serine 473, which by itself does not lead to AKT activation, was detected in L1236 and SUP-HD1 cell lines (Figure 5A). Phosphorylation of ERK was found in the L428, KM-H2, U-HO1, and SUP-HD1 cHL cell lines, whereas L1236 cells showed only a weak ERK phosphorylation signal. To determine whether these signaling pathways mediate FOXO1 repression in the different cell lines, we interfered with AKT and ERK activation using the specific PDK1-AKT inhibitor KP372-1 and the MEK1/2 inhibitor U0126, respectively. We saw that the PKB/AKT inhibitor induced FOXO1 protein expression in a dose-dependent manner specifically in L1236 and SUP-HD1 cell lines. Correspondingly, U0126 increased FOXO1 expression in all cell lines except the L1236 (Figure 5B). We also investigated the effects of the inhibitors on proliferation of the cHL cell lines. We found that the efficacy of the inhibitors correlated with the activation status of their targets. The L1236 and SUP-HD1 cell lines, which were positive for pAKT (Thr308), were more sensitive to KP372-1 than the other cell lines. Similarly, the L1236 cell line was more resistant to the U0126 compared with other cHL cell lines (Figure 5C). Thus, FOXO1 repression is at least in part mediated by constitutive ERK and AKT activation in cHL.
AKT and ERK kinases inhibitors restore FOXO1 expression in cHL cell lines. (A) Constitutive activation of ERK and AKT kinases in cHL cell lines. Cell lysates of cHL cell lines were probed with antibodies specific for p-Akt(Thr308), p-Akt(Ser473), and p-ERK. The antibody to ACTB and to the total AKT and ERK were used as loading controls. (B) AKT and ERK inhibitors restore FOXO1 expression in cHL cell lines. A total of 1 × 106 cells were seeded in 3 mL of complete medium per well of a 6-well plate. The KP372-1 or U0126 kinase inhibitors were added at the same day. Twenty-four hours later, the cells were harvested and FOXO1 expression was detected by immunoblot. (C) Sensitivity of the cHL cell line to AKT inhibitor KP372-1 and to ERK inhibitor U0126. A total of 2 × 104 cells were seeded per well of a 96-well plate simultaneously with the kinase inhibitors at different concentrations. Sixty hours later, the viability of the cells was assessed by MTT staining. IC50 of U0126 and KP372-1 for p-ERK or p-AKT negative (−) and positive (+) cHL cell lines, respectively. The experiments were done in triplicate.
AKT and ERK kinases inhibitors restore FOXO1 expression in cHL cell lines. (A) Constitutive activation of ERK and AKT kinases in cHL cell lines. Cell lysates of cHL cell lines were probed with antibodies specific for p-Akt(Thr308), p-Akt(Ser473), and p-ERK. The antibody to ACTB and to the total AKT and ERK were used as loading controls. (B) AKT and ERK inhibitors restore FOXO1 expression in cHL cell lines. A total of 1 × 106 cells were seeded in 3 mL of complete medium per well of a 6-well plate. The KP372-1 or U0126 kinase inhibitors were added at the same day. Twenty-four hours later, the cells were harvested and FOXO1 expression was detected by immunoblot. (C) Sensitivity of the cHL cell line to AKT inhibitor KP372-1 and to ERK inhibitor U0126. A total of 2 × 104 cells were seeded per well of a 96-well plate simultaneously with the kinase inhibitors at different concentrations. Sixty hours later, the viability of the cells was assessed by MTT staining. IC50 of U0126 and KP372-1 for p-ERK or p-AKT negative (−) and positive (+) cHL cell lines, respectively. The experiments were done in triplicate.
The miRNAs miR-182, miR-96, and miR-183 are involved in FOXO1 repression in cHL cell lines
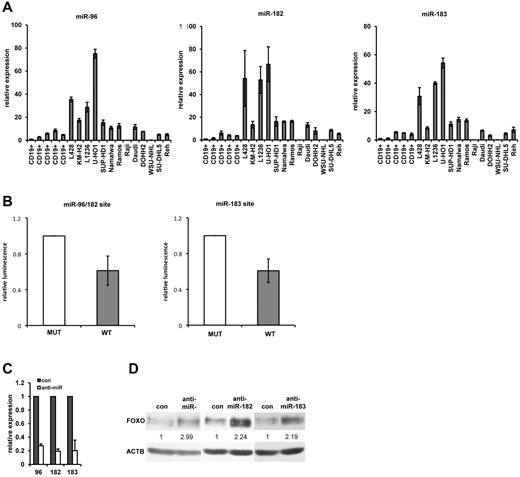
FOXO1 repression by miRNA was previously described in mammary and endometrial carcinomas. The main role in FOXO1 repression in these tumor types was attributed to miR-96, miR-183, and miR-182, which are located in the miR-183–miR-96–miR-182 cluster.30,35 We analyzed the expression of these 3 miRNAs in normal tonsillar B cells, cHL cell lines, and in different NHL cell lines by quantitative PCR (Figure 6A). Expression of all 3 miRNAs in cHL cell lines was significantly higher than in CD19+ cells. Among the cHL cell lines, the highest expression was seen in L428, L1236, and U-HO1 cells. These cHL cell lines also expressed significantly higher levels of the miRNAs than NHL cell lines. Expression of the miRNAs in KM-H2 and SUP-HD1 was higher or comparable with the NHL cell lines. To investigate the impact of the miRNAs on the repression of FOXO1, we cloned the parts of FOXO1 3′-UTR, containing the binding sites of miR-96–miR-182, and miR-183 downstream of the Renilla luciferase gene and performed reporter gene assays in L428 cells. As a control, corresponding FOXO1 3′-UTR sequences with mutated binding sites were used. The activity of constructs with wild-type binding sites was significantly lower than that of constructs with mutated binding sites, indicating involvement of these miRNAs in repression of FOXO1 (Figure 6B). To determine whether these miRNAs directly regulate FOXO1 protein levels, we interfered with their activity in L428 cells. Transfection of specific inhibitors against miR-96, miR-182, and miR-183 significantly down-regulated their levels (Figure 6C) and increased FOXO1 protein expression 2- to 3-fold compared with the negative control (Figure 6D). Thus, up-regulation of miR-96, miR-182, and miR-183 may contribute to FOXO1 repression in cHL.
The miR-183, miR-96, and miR-182 are involved in FOXO1 repression in cHL cell lines. (A) Expression of miR-183, miR-96, and miR-182. The total RNA was extracted from CD19+ tonsillar cells obtained from 5 different patients. cHL and NHL cell lines and expression of miRNAs were measured by quantitative RT-PCR. U6 mRNA was used as reference. The data were analyzed with the comparative Ct method and represent mean ± SD. (B) miRNA regulation of FOXO1 3′-UTR. miRNA activity in L428 cells was measured by electroporation of psi-CHECK-2 vector bearing either a specific wild-type target sequence (WT) common for miR-96 and miR-182, or for miR-183 sequence. The relevant mutated sequences (MUT) with mutated nucleotides in the seed region were used as control. Activity is expressed as mean ± SD of ratio of Renilla to firefly luciferase activity normalized to the mutated control. (C) Inhibition of miRNA expression by specific anti-miRNAs. L428 cells were transfected with anti-miRNAs or with the negative control RNAs. Twenty-four hours later, miRNA expression was measured by quantitative RT-PCR and analyzed and presented as described for panel A. (D) Effect of miRNA overexpression on FOXO1 levels. L428 cell line was transfected with anti-miRs or with the negative control RNAs. Forty-eight hours after transfection, the cells were harvested and FOXO1 expression was analyzed by immunoblot. ACTB was used as a loading control. The protein expression was quantified with the help of ImageJ 64 software (http://rsbweb.nih.gov/ij). All experiments were done in triplicate.
The miR-183, miR-96, and miR-182 are involved in FOXO1 repression in cHL cell lines. (A) Expression of miR-183, miR-96, and miR-182. The total RNA was extracted from CD19+ tonsillar cells obtained from 5 different patients. cHL and NHL cell lines and expression of miRNAs were measured by quantitative RT-PCR. U6 mRNA was used as reference. The data were analyzed with the comparative Ct method and represent mean ± SD. (B) miRNA regulation of FOXO1 3′-UTR. miRNA activity in L428 cells was measured by electroporation of psi-CHECK-2 vector bearing either a specific wild-type target sequence (WT) common for miR-96 and miR-182, or for miR-183 sequence. The relevant mutated sequences (MUT) with mutated nucleotides in the seed region were used as control. Activity is expressed as mean ± SD of ratio of Renilla to firefly luciferase activity normalized to the mutated control. (C) Inhibition of miRNA expression by specific anti-miRNAs. L428 cells were transfected with anti-miRNAs or with the negative control RNAs. Twenty-four hours later, miRNA expression was measured by quantitative RT-PCR and analyzed and presented as described for panel A. (D) Effect of miRNA overexpression on FOXO1 levels. L428 cell line was transfected with anti-miRs or with the negative control RNAs. Forty-eight hours after transfection, the cells were harvested and FOXO1 expression was analyzed by immunoblot. ACTB was used as a loading control. The protein expression was quantified with the help of ImageJ 64 software (http://rsbweb.nih.gov/ij). All experiments were done in triplicate.
Discussion
Here we show that FOXO1 is highly expressed in primary B cells and NHLs but is repressed in HRS cells of cHL and in cHL-derived cell lines. The mechanisms of FOXO1 repression include the activation of ERK and AKT pathways, up-regulation of miRNAs, which target FOXO1 3′-UTR, and chromosomal deletions. FOXO1 activation leads to growth arrest and apoptosis, indicating that FOXO1 repression contributes to survival and proliferation of HRS cells.
Our finding of the predominant expression of FOXO1 in the normal human follicular B cells is in line with the recently described unique role of FOXO1 in B-cell maturation. Inactivation of FOXO1 resulted in a transcriptional down-regulation of Rag1, Rag2, Il7r, Aicda, and Cd62l/Sell at different stages of B-cell development.5,6 The main regulator of GC formation BCL6 is also a FOXO target.36 Although we observed FOXO1 repression in some of the NHL cases, the strong FOXO1 expression in most of NHL entities was surprising because Foxo1 was reported to be the main mediator of physiologic B-cell death in mouse.8 Moreover, the functionally and structurally similar Foxo3 was reported to repress Myc-induced B-cell lymphomagenesis in a mouse model.37 One of the possible explanations for this might be the dependence of NHLs on the GC lineage-specific survival program, and particularly on the expression of the FOXO1 target protein BCL6,38 a GC proto-oncogene.12
The repression of FOXO1 on protein and mRNA level distinguishes HL from other BCL entities. We revealed 3 causes for this specific down-regulation: constitutive activation of AKT and ERK kinases, up-regulation of miRNAs of the miR-96–miR-182–miR-183 cluster, and chromosomal deletions. Constitutive activation of AKT and ERK kinases is typical for cHL21,39 and represents an oncogenic deregulation of a lineage-specific survival mechanism of normal activated B cells.8,40 Activation of B cells with anti-Ig antibody, CpG nucleotides, Cd40l, Il4, or LPS ultimately led to Foxo1 repression.40 This down-regulation of FOXO1 seems to be important for optimal proliferation of activated B cells.41 The activation signals facilitate FOXO1 protein phosphorylation by ERK and AKT kinases followed by its nuclear export and degradation. The FOXO1 repression can be further enhanced by disruption of the positive feedback loop in which FOXO1 activates its own promoter.42
miRNAs mediate FOXO1 repression in physiologic43 and some pathologic (mammary and endometrial carcinoma)30,35 processes. The main role in FOXO1 repression was ascribed to miR-96, miR-183, and miR-182 located in the miR-183–miR-96–miR-182 cluster.30,35 In mouse T cells, it was shown that expression of miR-182 is induced by the transcription factor STAT5.43 Therefore, our quantitative PCR data on high miR-183, miR-96, and miR-182 expression in cHL cell lines are in line with the known constitutive STAT5 activation in cHL.15 The data of miRNA arrays, showing high expression of miR-182 and miR-183 in L428 and L1236 cell lines,44 and high expression of miR-96 in HRS cells of EBV-negative nodular sclerosis cHL cases are also in agreement with our results.44
Our data on FOXO1 copy number loss in cHL cell lines are in accordance with the previously described 13q13.1-q21.32 loss in L428 and KM-H2 cell lines33 and the 13q loss in L1236 cells.45 In SUP-HD1, we found a derivative chromosome 13 with interstitial deletion similar to previous observations.23 Although none of the cell lines or HRS cells of primary cHL cases showed complete loss of both FOXO1 alleles, the deletions might be functionally relevant and further study investigating FOXO1 haploinsufficiency is warranted.46 Specifically, this phenomenon was explained by the inability of haploinsufficient cells to compensate for increased nuclear export of Foxo1, leading to a phenotype resembling complete loss of the gene. Taking into account constitutive activation of AKT or ERK promoting FOXO1 nuclear export in cHL this assumption appears to be possible. Another explanation of the haploinsufficiency phenomenon could be the deregulation of positive feedback mechanism regulating the FOXO1 promoter.23 Of note, hemizygous FOXO1 deletions were reported to be associated with FOXO1 repression in clear cell renal cell carcinoma.47
In conclusion, our data indicate that repression of FOXO1 transcription adds to autonomous growth and sustained survival of HRS cells of cHL. The mechanisms of FOXO1 repression in cHL are complex and include activation of ERK and AKT kinases, up-regulation of the miR-182, miR-96, and miR-183, and chromosome aberrations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Anita Kick, Julia Melzner, and Susana Ben Neriah for excellent technical assistance; T.G. Unterman for fruitful discussion and for providing the FOXO1(A3)ER-pBABE-puro vector; I. Guttilla and B. White for donation of the dual reporter system; and U. Schmidt-Strassburger for help with immunofluorescent staining and microscopy.
This work was supported in part by Deutsche Krebshilfe eV (grant 107547; T.W. and A.U.). C.P. was supported by Deutsche Krebshilfe eV grant 110092 (doctoral candidates progam for medical students). H.G. was supported by the China Scholarship Council. C.S. was supported by the Cancer Research Society (Steven E. Drabin fund, fellowship award).
Authorship
Contribution: L.X., A.U., H.G., C.S., J.F., C.P., and M.J.V. performed research and analyzed data; and A.U., F.L., C.S., H.J.M., R.D.G., P.M., and T.W. designed research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas Wirth, Institute of Physiological Chemistry, University of Ulm, Albert-Einstein-Allee 11, 89081 Ulm, Germany; e-mail: thomas.wirth@uni-ulm.de.
References
Author notes
L.X. and A.U. contributed equally to this study.