We used the opportunities afforded by the zebrafish to determine upstream pathways regulating mast cell development in vivo and identify their cellular origin. Colocalization studies demonstrated zebrafish notch receptor expression in cells expressing carboxypeptidase A5 (cpa5), a zebrafish mast cell-specific marker. Inhibition of the Notch pathway resulted in decreased cpa5 expression in mindbomb mutants and wild-type embryos treated with the γ-secretase inhibitor, Compound E. A series of morpholino knockdown studies specifically identified notch1b and gata2 as the critical factors regulating mast cell fate. Moreover, hsp70::GAL4;UAS::nicd1a transgenic embryos overexpressing an activated form of notch1, nicd1a, displayed increased cpa5, gata2, and pu.1 expression. This increase in cpa5 expression could be reversed and reduced below baseline levels in a dose-dependent manner using Compound E. Finally, evidence that cpa5 expression colocalizes with lmo2 in the absence of hematopoietic stem cells revealed that definitive mast cells initially delineate from erythromyeloid progenitors. These studies identify a master role for Notch signaling in vertebrate mast cell development and establish developmental origins of this lineage. Moreover, these findings postulate targeting the Notch pathway as a therapeutic strategy in mast cell diseases.
Introduction
Mast cells are best known for their role in acute and chronic allergic reactions; however, they also function as a crucial component in both innate and acquired immune responses1 as well as solid tumor and leukemia progression.2,3 Mast cells delineate from hematopoietic stem cells (HSCs) in the bone marrow but, unlike other blood cells, enter circulation as progenitors. They only complete maturation in resident tissues, which greatly hinders accurate lineage tracing studies in traditional mammalian models.1 We have been using the zebrafish model to study mast cell development and, specifically, the transcriptional regulation of mast cell lineage commitment. The zebrafish is a highly efficient model system for studying blood cell development.4,–6 All of the major hematopoietic cellular lineages studied to date have zebrafish counterparts, and the fundamental genetic mechanisms that control hematopoiesis are well conserved.4,7,8 We first described a mast cell counterpart in the zebrafish9 and subsequently showed conserved roles of these cells in adaptive and innate responses to inflammatory stimuli.10 Zebrafish carboxypeptidase a5 (cpa5) was identified as a zebrafish mast cell-specific marker, and gata2 and pu.1 were found to be the key transcription factors required for early mast cell lineage commitment in keeping with studies in mammalian systems.9,11,12
The Notch signaling pathway is a critical regulator of cell fate determination conserved through evolution. Aberrant Notch signaling is associated with a wide range of human disorders from developmental syndromes to cancer.13 Notch signaling is involved in the fate determination of a variety of cell types, including hematopoietic cells where it participates in differentiation, proliferation, and apoptosis.14 In mammals, the Notch pathway consists of 4 Notch genes (Notch1-4), which encode transmembrane receptor proteins. These receptors are activated by 5 ligands encoded by the Delta and Serrate/Jagged gene families: Delta-like1, (Dll1), Dll3, Dll4, Jagged 1 (Jag1), and Jag2, which are membrane-bound on neighboring cells. Ligand binding results in Notch receptor proteolysis, with the extracellular portion of Notch being endocytosed into the ligand-expressing cell. Subsequently, the intracellular portion of Notch is released from the transmembrane portion after several cleavage steps, which culminates in cleavage by the enzyme, γ-secretase.14 The liberated Notch intracellular domain (NICD) travels to the nucleus where it modulates transcription through interacting in a DNA-binding complex with CSL (CBF1/RBP-Jk, Suppressor of Hairless, Lag-1) and the Mastermind-like (MAML) proteins.15 These Notch components are highly conserved in zebrafish.16,,–19 Notch pathway activation has been most closely linked to lymphocyte development and specifically T-cell maturation20,21 but has also been more broadly implicated in myelopoiesis22,23 and more recently in mast cell development, in particular.24,,–27 Studies in mice have also suggested that critical mast cell transcription factors, Pu.122 and Gata2,28 are direct targets of the Notch pathway. To date, these links between the Notch pathway and mast cells have been identified, but a detailed interrogation of the role of Notch signaling in contributing to mast cell fate has not been previously undertaken in vivo.
We harnessed the opportunities provided by the zebrafish model system and our prior characterization and validation of cpa5 as a mast cell specific marker to conduct a comprehensive series of embryonic in vivo studies to assess the role of notch genes in vertebrate mast cell development. We incorporated a variety of approaches to inhibit zebrafish Notch pathway activation and reveal a clear dependence of the mast cell lineage on Notch signaling early in development. In addition, we found that definitive mast cells originate initially from erythromyeloid progenitor cells (EMPs). Taken together, these findings distinguish mast cells as the first blood cell lineage dependent on Notch signaling before the emergence of HSCs.16,19
Finally, in applying these findings to human disease, we demonstrate that a transgenic zebrafish line overexpressing notch1a16 recapitulates the accumulation of mast cells seen in human systemic mastocytosis, which can be abrogated through Notch pathway inhibition.
Methods
Zebrafish strains and maintenance
Zebrafish were maintained, bred, and developmentally staged according to Westerfield.29 Use of zebrafish in this study was approved by the Dalhousie University Committee on Laboratory Animals, protocol no. 11-128.
WISH
Digoxigenin- and fluorescein-labeled RNA probes were transcribed from linearized cDNA constructs according to manufacturer's protocol (Roche Diagnostics). Whole-mount in situ hybridization (WISH) assays for carboxypeptidase a5 (cpa5), gata1, gata2, pu.1, and myeloperoxidase (mpx) were performed on wild-type, mindbomb (mibta52b), Compound E (CpdE)–treated, morphant, and hsp70::GAL4;UAS::nicd1a transgenic embryos as described previously.9 Staining was performed using 5-bromo-4-chloro-3′-indolyl phosphate/nitro blue tetrazolium (Vector Laboratories).
Double WISH for colocalization of zebrafish notch homologs and cpa5
Double WISH on wild-type embryos at 28 hours post fertilization (hpf) and at 7 days post fertilization (dpf) was performed using labeled digoxigenin-cpa5 probe stained with Fast Red (Roche Diagnostics) in conjunction with fluorescein-labeled probes to zebrafish notch gene homologs: notch1a, notch1b, notch2, and notch3 (kindly provided by Dr Nathan Lawson, University of Massachusetts Medical School, Worchester, MA), stained with 5-bromo-4-chloro-3′-indolyl phosphate/nitro blue tetrazolium (Vector Laboratories).
Double fluorescent WISH for the colocalization of the zebrafish EMP marker, lmo2, and cpa5
Double WISH on wild-type embryos at 28 and 48 hpf was performed using labeled digoxigenin-cpa5 probe stained with Fast Red (Roche Diagnostics) in conjunction with fluorescein-labeled probe to the zebrafish lmo2 gene.
Notch signaling mutant, mindbomb (mibta52b)
Adult wild-type and mindbomb (mibta52b) zebrafish were incrossed, and embryos kept in 0.003% 1-phenyl-2-thiourea (Sigma-Aldrich) to prevent pigmentation for optical clarity. Embryos were dechorionated with Pronase (0.5 μg/mL; Sigma-Aldrich). Dechorionated embryos were staged and fixed with 4% paraformaldehyde (PFA; Sigma-Aldrich).
γ-secretase inhibitor CpdE treatment
A 1mM stock of Compound E (CpdE; Alexis Biochemicals) in DMSO was diluted in embryo medium and applied to dechorionated zebrafish wild-type, mibta52b, or hsp70::GAL4;UAS::nicd1a transgenic embryos at 28.5°C at final concentrations of 25, 50, and 75μM from 22 hpf to 30 hpf or 48 hpf, and then embryos were fixed with 4% PFA. Control embryo groups were treated with embryo medium with or without 0.5% DMSO.
Morpholino knockdown of zebrafish notch homologues pu.1 and HSCs
Morpholinos were purchased from Genetools LLC. Published morpholino sequences are as follows: MO notch1a 5′-GAAACGGTTCATAACTCCGCCTCGG-3′30 ; MO notch1b 5′-AATCTCAAACTGACCTCAAACCGAC-3′31 ; MO notch2 5′-AGGTGAACACTTACTTCATGCCAAA-3′32 ; MO notch3 5′-ATATCCAAAGGCTGTAATTCCCCAT-3′30 ; MO2 pu.1 5′-CCTCCATTCTGTACGGATGCAGCAT-3′9 ; MO3 runx1 5′-TGTTAAACTCACGCTGTGGCTCTC-3′16 ; and MO5 runx1 5′-AATGTGTAA-ACTCACAGTGTAAAGC-3′16
Morpholinos of notch1a, notch1b, notch2, notch3, and pu.1 were diluted to a working concentration of 1.0mM, whereas the 2 runx1 morpholinos were diluted to a working concentration of 1.3mM and combined in equal amounts to a concentration of 2.6mM. The morpholino solutions were mixed with 1% phenol red and injected into zebrafish embryos at the 1 to 4 cell stage. Dechorionated embryos were staged at appropriate developmental stage and fixed with 4% PFA.
Phenotypic rescue of morphant embryos using gata2 mRNA
gata2 mRNA was synthesized according to high-yield capped RNA transcription mMESSAGE mMACHINE Kit (Applied Biosystems). For notch1b morphant embryos, 12.5 ng/μL of gata2 mRNA was injected into embryos at the 1 to 4 cell stage and for pu.1 morphant embryos, 25 ng/μL of gata2 mRNA was injected into embryos at the 1 to 4 cell stage. Dechorionated embryos were staged at 28 hpf and fixed with 4% PFA.
Genotyping and generation of nicd1a overexpressing transgenic zebrafish
Transgenic hsp70::GAL4 and UAS::nicd1a adult fish were anesthetized with Tricaine (0.003%), and a 2- to 5-mg tail clip was taken for genotyping. Samples were partially digested using the REDExtract-N-Amp tissue PCR kit according to manufacturer's protocol (Sigma-Aldrich). A 20-μL PCR was conducted to amplify hsp70::GAL4 and UAS::nicd1a according to published literature.33 nicd1a forward and reverse PCR primers were 5′-CATCGCGTCTCAGCCTCAC-3′ and 5′-CGGAATCGTTTATTGGTGTCG-3′, respectively. GAL4 forward and reverse PCR primers were 5′-CGGGCATTTACTTTATGTTGC-3′ and 5′-GCCTTGATTCCACTTCTGG-3′, respectively.
Double-transgenic embryos (hsp70::GAL4;UAS::nicd1a) were produced from breeding positive adult hsp70::GAL4 with positive UAS::nicd1a fish. The double transgenic embryos were heat-shocked at 37°C at 13 to 15 hpf for 45 minutes. Embryos were raised at 28.5°C to the appropriate developmental stage and fixed with 4% PFA (both transgenic lines kindly provided by the Zon Laboratory, Boston, MA).
WISH assay analysis and quantification
Morpholino and WISH experiments were performed on average 2 or 3 times. Total number of embryos used in each specific WISH assay is indicated in the respective figure legends. Quantification of cpa5, gata2, pu.1, gata1, and mpx expression was accomplished by categorizing embryos based on gene expression levels (low, normal, and high) compared with wild-type expression levels. These data are presented as bar graphs following the respective representative WISH images in each figure and in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
BrdU incorporation assay
Dechorionated zebrafish wild-type or hsp70::GAL4;UAS::nicd1a transgenic embryos at 28 hpf were treated with a 10mM working concentration of Bromodeoxyuridine (BrdU; BD Biosciences) in embryo medium. The assay was carried out as described previously.34
Results
Zebrafish mast cells express notch receptor genes
To determine whether Notch receptors are expressed in zebrafish mast cells, we took advantage of our previously identified zebrafish MC-specific marker, cpa5, and performed colocalization studies, using double WISH with antisense RNA probes for cpa5 and each of zebrafish notch1a, notch1b, notch2, and notch3 genes. Coexpression of each notch gene and cpa5 was overlapping in mast cells at 28 hpf in wild-type embryos and observed in a proportion of cpa5-positive mast cells at 7 dpf in the anterior lateral mesoderm (ALM; supplemental Figure 1).
Genetic and pharmacologic inhibition of Notch signaling ablates mast cell development
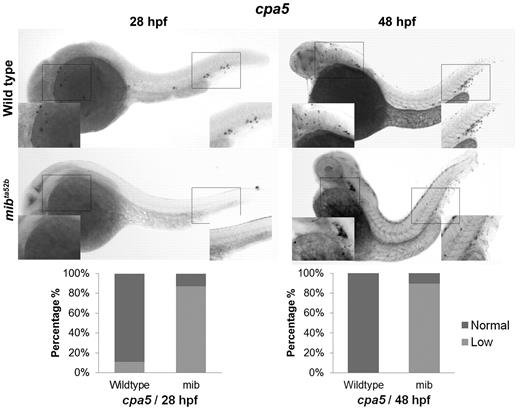
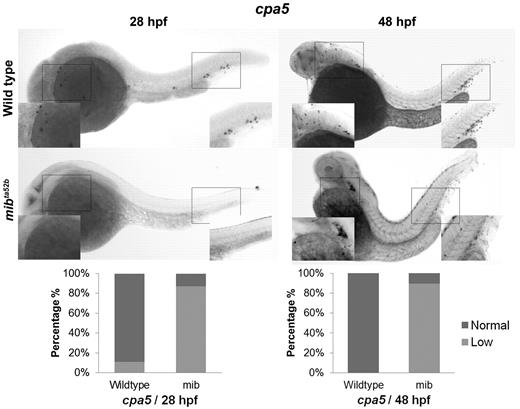
Given evidence for expression of Notch receptors on zebrafish mast cells, we examined the impact of Notch pathway inhibition on mast cell development. WISH analysis of cpa5 was performed in wild-type and mindbomb (mibta52b) mutant embryos, which are deficient in Notch signaling because of a selective defect in the processing of Notch ligands.13 In these experiments, mibta52b mutants demonstrated evidence of minimal cpa5 expression compared with wild-type embryos at 28 hpf. At 48 hpf, cpa5 expression was reduced in the ALM but was completely absent in the region of the caudal hematopoietic tissue (CHT; Figure 1). In keeping with previous studies,16 other myeloid markers in mibta52b mutants were maintained at wild-type levels, although gata1 expression was found to be significantly reduced (supplemental Figure 2). These results suggest that Notch inhibition through mindbomb results in delayed onset of definitive mastopoiesis as evident by minimal cpa5 expression at 28 hpf and reduced anterior expression at 48 hpf. At either time point, no cpa5 expression was observed posteriorly in the region of the posterior blood island/CHT, which suggests that this anatomic subpopulation is particularly sensitive to Notch regulation.
The zebrafish notch signaling mutant mindbomb (mibta52b) displays decreased cpa5 expression. WISH using a digoxigenin-labeled RNA antisense probe to zebrafish cpa5 was performed at 28 and 48 hpf. Bar graphs represent the percent of embryos categorized by WISH expression levels. At 28 hpf, the mibta52b mutant (n = 107) shows a dramatic decrease in cpa5 expression compared with wild-type embryos (n = 96), whereas at 48 hpf, the mibta52b mutant (n = 19) displays reduced cpa5 expression in the ALM and absent cpa5 expression in the CHT compared with wild-type (n = 12). Insets: Magnified view of the area of interest (5× objective, MZ6 microscope; Leica).
The zebrafish notch signaling mutant mindbomb (mibta52b) displays decreased cpa5 expression. WISH using a digoxigenin-labeled RNA antisense probe to zebrafish cpa5 was performed at 28 and 48 hpf. Bar graphs represent the percent of embryos categorized by WISH expression levels. At 28 hpf, the mibta52b mutant (n = 107) shows a dramatic decrease in cpa5 expression compared with wild-type embryos (n = 96), whereas at 48 hpf, the mibta52b mutant (n = 19) displays reduced cpa5 expression in the ALM and absent cpa5 expression in the CHT compared with wild-type (n = 12). Insets: Magnified view of the area of interest (5× objective, MZ6 microscope; Leica).
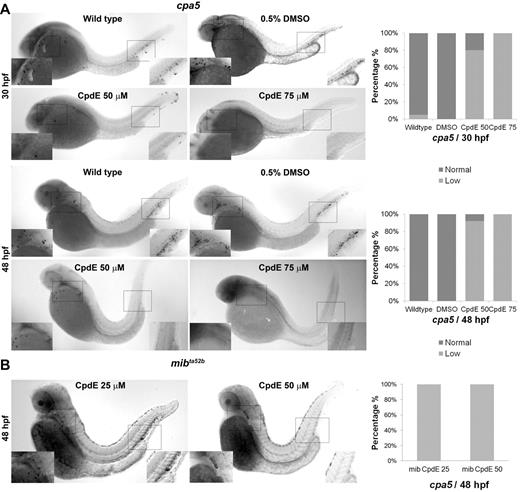
Next, we inhibited Notch signaling pharmacologically using CpdE, a highly active specific γ-secretase inhibitor. Treatment of wild-type zebrafish embryos with CpdE (50-75μM) induced a dose-dependent decrease in cpa5 expression at 30 and 48 hpf (Figure 2A), which was similarly observed at lower doses (25-50μM) in mibta52b mutants (Figure 2B). Interestingly, in wild-type embryos at the lower dose of 50μM CpdE, gata2 expression was also found to be decreased, whereas gata1 and pu.1 expression was maintained. However, at the higher dose of 75μM CpdE, expression of gata1 as well as pu.1 and mpx was found to be reduced (supplemental Figure 3). These results suggest that Notch inhibition may directly affect gata2 expression with specific impact on mast cell specification. By contrast, at higher doses of CpdE, gata2 expression is more severely reduced, thereby impacting other downstream hematopoietic markers.
Notch pathway inhibition of wild-type embryos by the γ-secretase inhibitor CpdE similarly displays decreased cpa5 expression. WISH using a digoxigenin-labeled RNA antisense probe to zebrafish cpa5 was performed at 30 and 48 hpf. Bar graphs represent the percent of embryos categorized by WISH expression levels. (A) Treatment with 50μM CpdE results in a decrease in cpa5 expression at 30 hpf (n = 35), whereas treatment with 75μM CpdE shows absent cpa5 expression (n = 10) compared with untreated controls (n = 44). Similarly, at 48 hpf, treatment with CpdE shows a dose-dependent decrease in cpa5 expression (n = 25 for 50μM CpdE, n = 30 for 75μM CpdE, n = 24 for wild-type embryos). DMSO 0.5% was used as a control, and from this group 24 and 23 embryos were analyzed at 30 and 48 hpf, respectively. (B) Treatment with 25 or 50μM CpdE results in a dose-dependent decrease of cpa5 expression in mibta52b mutants at 48 hpf, both anteriorly and posteriorly (n = 12 for 25μM, n = 11 for 50μM CpdE). Insets: Magnified view of the area of interest (5× objective, MZ6 microscope; Leica).
Notch pathway inhibition of wild-type embryos by the γ-secretase inhibitor CpdE similarly displays decreased cpa5 expression. WISH using a digoxigenin-labeled RNA antisense probe to zebrafish cpa5 was performed at 30 and 48 hpf. Bar graphs represent the percent of embryos categorized by WISH expression levels. (A) Treatment with 50μM CpdE results in a decrease in cpa5 expression at 30 hpf (n = 35), whereas treatment with 75μM CpdE shows absent cpa5 expression (n = 10) compared with untreated controls (n = 44). Similarly, at 48 hpf, treatment with CpdE shows a dose-dependent decrease in cpa5 expression (n = 25 for 50μM CpdE, n = 30 for 75μM CpdE, n = 24 for wild-type embryos). DMSO 0.5% was used as a control, and from this group 24 and 23 embryos were analyzed at 30 and 48 hpf, respectively. (B) Treatment with 25 or 50μM CpdE results in a dose-dependent decrease of cpa5 expression in mibta52b mutants at 48 hpf, both anteriorly and posteriorly (n = 12 for 25μM, n = 11 for 50μM CpdE). Insets: Magnified view of the area of interest (5× objective, MZ6 microscope; Leica).
Zebrafish notch1b regulates mast cell development through gata2
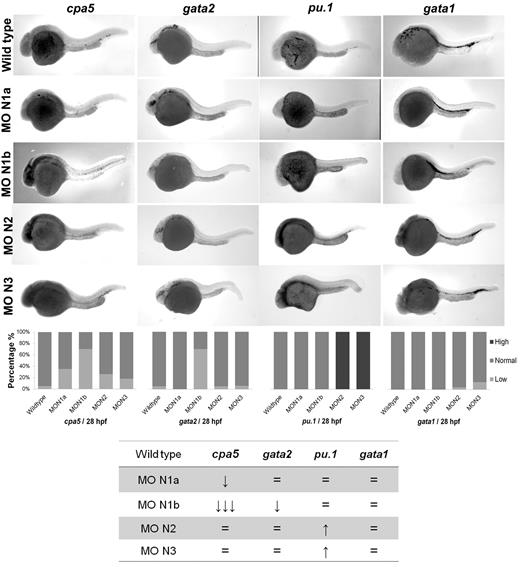
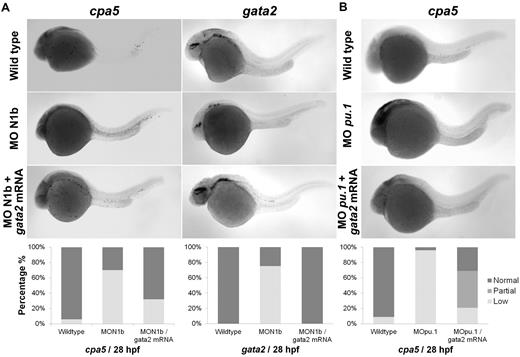
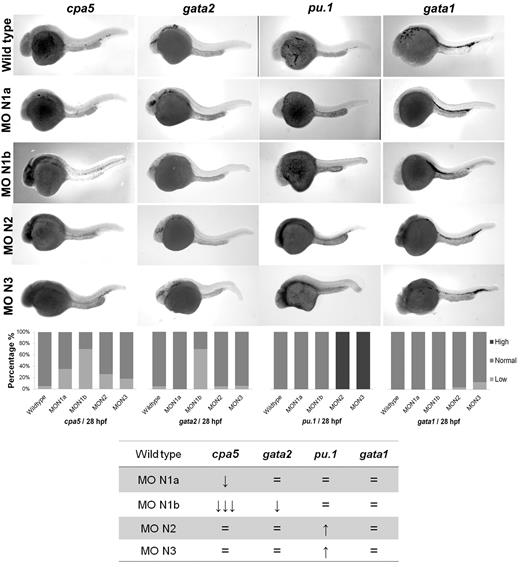
Morpholino oligonucleotides effectively permit transient knockdown of zebrafish protein translation to identify the relative requirement of a particular gene in developmental programming in vivo. We next used a panel of morpholinos targeting each of the zebrafish notch gene homologs, using whole-mount in situ hybridization to cpa5 and upstream hematopoietic transcription factors implicated in mast cell development as readouts. At 28 hpf, cpa5 expression was almost completely absent in notch1b morphants, in contrast to a moderate decrease noted in notch1a morphants and wild-type levels found in notch2 and notch3 morphants. Zebrafish notch1b morphants similarly demonstrated loss of gata2 expression, which was not observed in the other notch morphants. Levels of gata1 expression were not affected by knockdown of any of the notch genes, whereas pu.1 levels were maintained in notch1a and notch1b morphants and, indeed, were somewhat increased in notch2 and notch3 morphants (Figure 3). Similarly, mpx expression was found at wild-type levels in notch1b morphants but was elevated in all other notch morphants (supplemental Figure 4). Taken together, these data implicate the Notch pathway and, in particular, notch1b in mast cell fate determination, with gata2 highlighted as the key downstream transcription factor. To further support this hypothesis, notch1b morphants were injected with gata2 mRNA, which completely rescued both gata2 and cpa5 expression to wild-type levels (Figure 4A). We previously showed that, like notch1b morphants, pu.1 morphant embryos showed an absence of cpa5 expression.9 Because gata2, but not pu.1, was affected in notch morphants, we hypothesized that rescue of gata2 could restore mast cell development. However, when gata2 mRNA was injected into pu.1 morphants, it only partially rescued cpa5 expression, confirming a requirement for pu.1 in mast cell fate determination that may be independent of gata2 and Notch signaling (Figure 4B).
Morpholino knockdown reveals a specific requirement of notch1b for embryonic mast cell development in zebrafish. Morpholinos to zebrafish notch gene homologs were injected in 1 to 4 cell stage wild-type embryos followed by WISH for cpa5, gata2, pu.1, and gata1. notch1b morphants demonstrate a dramatic decrease in the level of cpa5 expression and reduced gata2 expression at 28 hpf, implicating a key role for notch1b in mast cell development mediated through gata2. Bar graphs represent the percent of embryos categorized by WISH expression levels. The table summarizes the predominant phenotype observed for each probe with each morpholino. The numbers of wild-type embryos, notch1a, notch1b, notch2, and notch3 morphants analyzed for each probe were n = 37, 17, 33, 19, and 11 for cpa5; n = 13, 9, 5, 10, and 9 for pu.1; n = 36, 9, 8, 23, and 24 for gata1; and n = 19, 12, 10, 22, and 16 for gata2. MO indicates morpholino; N1a, notch1a; N1b, notch1b; N2, notch2; and N3, notch3. 5× objective, MZ6 microscope (Leica).
Morpholino knockdown reveals a specific requirement of notch1b for embryonic mast cell development in zebrafish. Morpholinos to zebrafish notch gene homologs were injected in 1 to 4 cell stage wild-type embryos followed by WISH for cpa5, gata2, pu.1, and gata1. notch1b morphants demonstrate a dramatic decrease in the level of cpa5 expression and reduced gata2 expression at 28 hpf, implicating a key role for notch1b in mast cell development mediated through gata2. Bar graphs represent the percent of embryos categorized by WISH expression levels. The table summarizes the predominant phenotype observed for each probe with each morpholino. The numbers of wild-type embryos, notch1a, notch1b, notch2, and notch3 morphants analyzed for each probe were n = 37, 17, 33, 19, and 11 for cpa5; n = 13, 9, 5, 10, and 9 for pu.1; n = 36, 9, 8, 23, and 24 for gata1; and n = 19, 12, 10, 22, and 16 for gata2. MO indicates morpholino; N1a, notch1a; N1b, notch1b; N2, notch2; and N3, notch3. 5× objective, MZ6 microscope (Leica).
Rescue of embryonic mast cells completely in notch1b morphants and partially in pu.1 morphants by injecting gata2 mRNA. Zebrafish notch1b or pu.1 morpholino and gata2 mRNA were injected in 1 to 4 cell stage wild-type embryos followed by WISH for cpa5 expression at 28 hpf. (A) notch1b morphants demonstrate decreased cpa5 expression (n = 33) and gata2 expression (n = 8) compared with wild-type embryos (n = 34 and 16, respectively) that is completely rescued by injecting gata2 mRNA (n = 25). (B) pu.1 morphants demonstrate a dramatic decrease in cpa5-positive mast cells (n = 24) compared with wild-type embryos (n = 34) that is partially rescued by injecting gata2 mRNA (n = 29; Leica MZ6 microscope, 5× objective). Bar graphs represent the percent of embryos categorized by WISH expression levels.
Rescue of embryonic mast cells completely in notch1b morphants and partially in pu.1 morphants by injecting gata2 mRNA. Zebrafish notch1b or pu.1 morpholino and gata2 mRNA were injected in 1 to 4 cell stage wild-type embryos followed by WISH for cpa5 expression at 28 hpf. (A) notch1b morphants demonstrate decreased cpa5 expression (n = 33) and gata2 expression (n = 8) compared with wild-type embryos (n = 34 and 16, respectively) that is completely rescued by injecting gata2 mRNA (n = 25). (B) pu.1 morphants demonstrate a dramatic decrease in cpa5-positive mast cells (n = 24) compared with wild-type embryos (n = 34) that is partially rescued by injecting gata2 mRNA (n = 29; Leica MZ6 microscope, 5× objective). Bar graphs represent the percent of embryos categorized by WISH expression levels.
Constitutive Notch activation increases numbers of embryonic mast cells, which can be abrogated by pharmacologic inhibition of Notch signaling
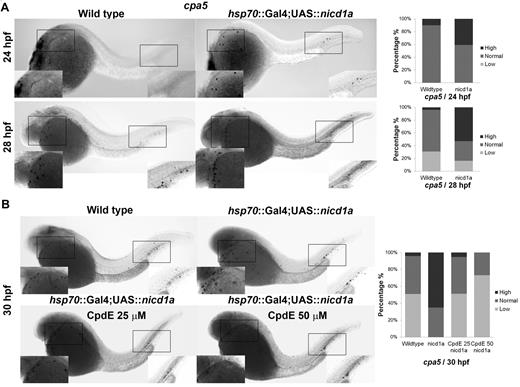
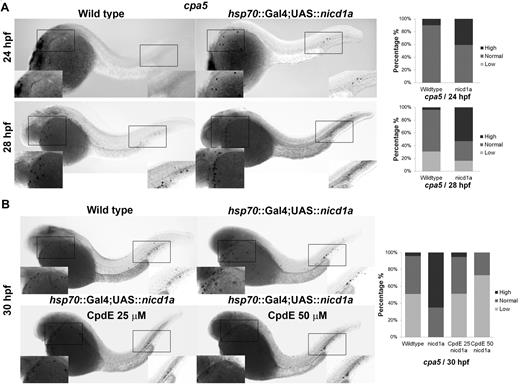
To complement Notch pathway knockdown assays, we examined the mast cell phenotype in the context of Notch pathway hyperactivation using a transgenic zebrafish line overexpressing nicd1a. Crossing of hsp70::GAL4 and UAS::nicd1a lines followed by heat shock at 13 to 15 hpf produced constitutively active expression of the NICD of notch1a. Double-transgenic hsp70::GAL4;UAS::nicd1a (= nicd1a transgenic) embryos displayed increased cpa5 expression at 24 and 28 hpf (Figure 5A) with similarly increased levels of gata2 expression, whereas gata1 and mpx expression levels remained unchanged (supplemental Figure 5). Expression of pu.1 was increased at 18 to 24 hpf, but not at later time points (supplemental Figure 6). Moreover, BrdU incorporation studies performed at 28 hpf revealed similar numbers of cells in S-phase of the cell cycle in hsp70::GAL4;UAS::nicd1a and wild-type embryos, indicating that overall proliferation was not increased in nicd1a transgenic embryos (data not shown). Taken together, these results suggest that the increased cpa5 expression observed in nicd1a transgenic embryos is a result of Notch preferentially driving mast cell development. Specifically, pu.1 functions to promote increased myeloid differentiation, of which a subset of myeloid cells subsequently acted on by Notch signaling through gata2, evolves into the mast cell lineage.
Zebrafish nicd1a overexpression results in an increase of cpa5-positive embryonic mast cells, which can be reversed by Notch inhibition using CpdE. WISH using a digoxigenin-labeled RNA antisense probe to zebrafish cpa5 was performed at different time points in hsp70::GAL4;UAS::nicd1a embryos heat shocked at 13 to 15 hpf. Bar graphs represent the percent of embryos categorized by WISH expression levels. (A) Overexpression of nicd1a results in an increase in the number of cpa5-positive mast cells at 24 hpf (n = 22) and 28 hpf (n = 49) compared with wild-type embryos (n = 20 and 52, respectively). (B) Treatment of hsp70::GAL4;UAS::nicd1a transgenic embryos (n = 43) with the Notch inhibitor CpdE for 8 hours (22-30 hpf) restores wild-type cpa5 expression levels at 25μM (n = 39) with further reduced expression at 50μM (n = 41) compared with wild-type embryos (n = 47). Insets: Magnified view of the area of interest (5× objective, MZ6 microscope, Leica).
Zebrafish nicd1a overexpression results in an increase of cpa5-positive embryonic mast cells, which can be reversed by Notch inhibition using CpdE. WISH using a digoxigenin-labeled RNA antisense probe to zebrafish cpa5 was performed at different time points in hsp70::GAL4;UAS::nicd1a embryos heat shocked at 13 to 15 hpf. Bar graphs represent the percent of embryos categorized by WISH expression levels. (A) Overexpression of nicd1a results in an increase in the number of cpa5-positive mast cells at 24 hpf (n = 22) and 28 hpf (n = 49) compared with wild-type embryos (n = 20 and 52, respectively). (B) Treatment of hsp70::GAL4;UAS::nicd1a transgenic embryos (n = 43) with the Notch inhibitor CpdE for 8 hours (22-30 hpf) restores wild-type cpa5 expression levels at 25μM (n = 39) with further reduced expression at 50μM (n = 41) compared with wild-type embryos (n = 47). Insets: Magnified view of the area of interest (5× objective, MZ6 microscope, Leica).
nicd1a.
nicd1a transgenic embryos were subsequently treated with CpdE to determine whether the increase in cpa5 expression could be reversed by targeting the Notch pathway. A 25μM dose of CpdE reduced cpa5 expression in nicd1a transgenic embryos to wild-type levels. cpa5 expression was further reduced below baseline levels with 50μM of CpdE (Figure 5B), suggesting a phenotype amenable to treatment with Notch inhibitors.
Definitive mast cells arise first from EMPs, before the emergence of HSCs
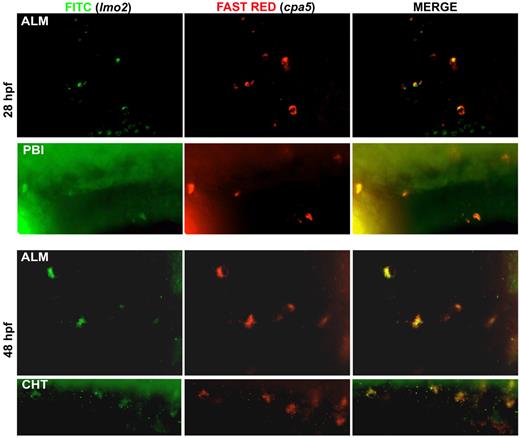
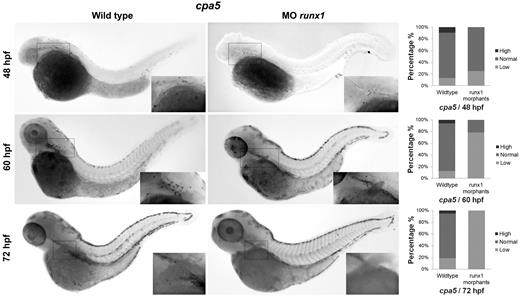
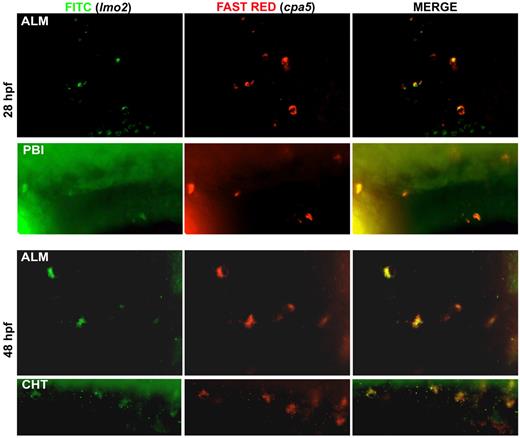
Recent progress in identifying specific markers of zebrafish HSCs35,36 and the more transient EMPs that specify the first wave of definitive hematopoiesis37 have enabled studies detailing the origins of cell lineages. We were interested in examining the origin of the zebrafish mast cell lineage throughout early development. EMPs are labeled by the early stem cell-like marker lmo2 as well as gata1. Using lmo2 as a marker of EMPs, we observed colocalization with cpa5 at 28 and 48 hpf, suggesting that mast cells arise from the EMP population during these early developmental time points (Figure 6). In support of these findings, when we eliminated the HSC population (measured by loss of c-myb) using runx1 double morpholinos,16 cpa5 expression at 48 hpf was only slightly decreased compared with wild-type embryos. This suggests that the majority of mast cells seen in early hematopoiesis delineate from EMPs. At later developmental time points, 60 hpf and 72 hpf, runx1 double morphant embryos showed a progressive reduction in cpa5 expression (Figure 7), indicative of the HSC population assuming production of the mast cell lineage between 48 and 60 hpf. These temporal studies suggest that the first wave of definitive mast cells originates from EMPs and that the emergence of HSCs marks a transition in mast cell development. HSCs subsequently become the source of mast cell generation for the duration of the life of the animal, in keeping with the established model of zebrafish definitive hematopoiesis.37
Mast cells originate from EMP at 28 to 48 hpf. Double WISH using digoxigenin-labeled RNA antisense probe to zebrafish cpa5 stained with Fast Red and fluorescein-labeled RNA antisense probe to zebrafish lmo2 demonstrate coexpression of the zebrafish EMP marker lmo2 with the mast cell specific marker cpa5 at the ALM and posterior blood island (PBI) at 28 hpf, and at both the ALM and the CHT at 48 hpf (40× objective, Z1 inverted stereo-microscope; Carl Zeiss).
Mast cells originate from EMP at 28 to 48 hpf. Double WISH using digoxigenin-labeled RNA antisense probe to zebrafish cpa5 stained with Fast Red and fluorescein-labeled RNA antisense probe to zebrafish lmo2 demonstrate coexpression of the zebrafish EMP marker lmo2 with the mast cell specific marker cpa5 at the ALM and posterior blood island (PBI) at 28 hpf, and at both the ALM and the CHT at 48 hpf (40× objective, Z1 inverted stereo-microscope; Carl Zeiss).
Mast cells arise from HSCs from 48 to 72 hpf. Two morpholinos targeting the zebrafish runx1 gene were injected in 1 to 4 cell stage wild-type embryos followed by WISH for cpa5. runx1 morphants demonstrate a mild decrease in the number of cpa5-positive mast cells at 48 hpf (n = 28) and a progressive decrease at later time points, 60 hpf (n = 14) and 72 hpf (n = 41), compared with wild-type embryos (n = 31, 16, and 17, respectively), implicating that HSCs contribute to the production of the definitive mast cell lineage (5× objective, MZ6 microscope; Leica). Insets: Higher magnification views of the anterior of the embryos. Bar graphs represent the percent of embryos categorized by WISH expression levels. MO indicates morpholino.
Mast cells arise from HSCs from 48 to 72 hpf. Two morpholinos targeting the zebrafish runx1 gene were injected in 1 to 4 cell stage wild-type embryos followed by WISH for cpa5. runx1 morphants demonstrate a mild decrease in the number of cpa5-positive mast cells at 48 hpf (n = 28) and a progressive decrease at later time points, 60 hpf (n = 14) and 72 hpf (n = 41), compared with wild-type embryos (n = 31, 16, and 17, respectively), implicating that HSCs contribute to the production of the definitive mast cell lineage (5× objective, MZ6 microscope; Leica). Insets: Higher magnification views of the anterior of the embryos. Bar graphs represent the percent of embryos categorized by WISH expression levels. MO indicates morpholino.
Discussion
The controversy regarding the origin and signals underlying mast cell development is reflected in the different published models that exist to explain mast cell lineage commitment. Gata2, a transcription factor typically expressed early in multipotent progenitor cells,38 is strongly expressed in mast cells,39 lending credence to a model postulating that mast cell lineage branches off directly from the HSCs.40 However, coexpression of Gata1 and Pu.1 in mast cells in a unique cooperative interaction, not present in other mature cell types, suggests that the mast cell lineage may derived from a more differentiated common myeloid-erythroid progenitor cell and cell fate determination is dependent on the ratio of Gata1 to Pu.1 expression levels.9,39,41 Although both Gata2 and Gata1 appear necessary for mast cell development, they perform different and non-overlapping roles. Gata2 is required for early mast cell development,39 whereas Gata1 is needed for mast cell maturation42 and, indeed, appears itself to be directly regulated by Gata2 and Pu.1.43
The zebrafish model affords the opportunity for elucidating the transcriptional regulation of many vertebrate hematopoietic lineages in vivo, including mast cells.9,10 We previously identified cpa5 as a unique marker of zebrafish mast cells, and we characterized both pu.1 and gata2 as key transcription factors for primitive zebrafish mast cell development.9 Our present study confirms the requirement for gata2 in early mast cell development with new insight into its upstream regulation being directed through Notch signaling. However, gata2 alone is not sufficient to specify mast cell fate as evidenced by morpholino rescue studies. Absent cpa5 expression seen in notch1b morphants could be rescued by gata2 mRNA, but only in the presence of endogenous levels of pu.1 expression. By contrast, in pu.1 morphants, gata2 mRNA only partially restored cpa5 expression, demonstrating the need for both of these transcription factors in mast cell development (Figure 4).
In the case of notch1b morphant embryos, pu.1 appears to function independently while gata2 appears dependent on Notch signaling. However, we found that forced notch1a signaling achieved in the nicd1a transgenic line revealed increased cpa5 (Figure 5), gata2 (supplemental Figure 5), and pu.1 (supplemental Figure 6) expression levels. Thus, although both gata2 and pu.1 are necessary to specify mast cell lineage fate, gata2 appears to be more broadly controlled by the Notch signaling pathway and pu.1 may be more selectively regulated by notch1a. These findings have enabled us to further expand on our previous model of zebrafish mast cell development9 to incorporate the role of Notch signaling (supplemental Figure 7). These results are consistent with recent mammalian in vitro studies.25,26 Sakata-Yanagimoto et al elegantly demonstrated that both common myeloid progenitors and granulocyte-monocyte progenitors cultured with Dll1 ligand in the presence of Notch2 preferentially matured into mast cells at the expense of other myeloid lineages, with either Gata3 or Gata2 together with Hes1 identified as necessary intermediaries.25
Notch signaling has been previously investigated in zebrafish hematopoiesis.16,19 Initial studies by Burns et al using mibta52b mutants demonstrated no difference in myeloid or erythroid cell gene expression at 28 hpf, leading the authors to conclude that Notch signaling was not required for primitive hematopoiesis in the zebrafish, although mast cells were not evaluated.16 Our results similarly show conserved expression of pan-myeloid markers in mibta52b mutants at this developmental time point, with the notable exception of cpa5. Given the colocalization of cpa5-positive cells with lmo2, we hypothesize that these mast cells at 28 hpf probably arise from EMPs, indicating a unique dependence of pre-HSC-derived mast cells on Notch signaling in contrast to other myeloid lineages (see later paragraphs in the “Discussion”). Expression of gata2 was also maintained in mibta52b mutants, in contrast to the decrease observed after CpdE treatment. However, the dose-dependent reduction in gata2 expression after increasing CpdE concentrations suggests that gata2 expression may be sustained with even minimal Notch signaling. The mibta52b mutants may still permit Notch signaling, potentially via Jag ligands or via maternally provided mindbomb19 and thus may not serve as robust a model for Notch inhibition as γ-secretase inhibition. This supposition is demonstrated by the dose-dependent decrease in cpa5 expression observed in mibta52b mutants after treatment with CpdE (Figure 2B).
Our studies suggest that early mast cells in the zebrafish embryo arise from the EMP population. EMPs represent the first definitive wave of hematopoiesis and was initially described by Bertrand et al as originating from the posterior blood island beginning at 24 to 30 hpf,37 which parallels a similar transient period of definitive hematopoiesis in mammals.44 The emergence of EMPs overlaps with the latter half of primitive hematopoiesis and the emergence of HSCs from the dorsal aorta at 36 to 48 hpf.37,45 EMPs are labeled by lmo2 and gata1. This group showed that Notch signaling was necessary for HSCs, but not for lineages derived from EMPs, although mast cells were not examined.19 By contrast, our studies demonstrate the novel involvement of Notch signaling in an early hematopoietic lineage, before the emergence of HSCs. Previously, we determined that gata1 is not required for early mast cell progenitor development in the zebrafish.9 However, we found colocalization of lmo2 and cpa5 at both 28 and 48 hpf in the region of the posterior blood island/CHT, suggesting mast cells arise from EMPs (Figure 6). These findings support our original observations of minimal cpa5 expression observed anteriorly at 24 hpf (which may have been derived from primitive myelopoiesis), with more robust cpa5 expression both anteriorly and posteriorly beginning at 28 hpf once EMPs are present.9 EMPs are implicated as the origin for mast cells both for their anatomic location and the fact that HSCs do not yet significantly contribute to hematopoiesis at this developmental stage. In support of this hypothesis, runx1 double morphants, which effectively have no c-myb–positive HSCs, show the presence of cpa5 positive mast cells between 48 and 60 hpf (Figure 7). Taken together, these results imply that Notch signaling plays a critical role in mast cell development before the emergence of HSCs (supplemental Figure 7). By 72 hpf, mast cell production is assumed by HSCs, as evidenced by the gradual loss of cpa5-positive mast cells in runx1 morphants after 48 hpf. In agreement with other studies, colocalization studies of cpa5 and various zebrafish notch gene homologs at 7 dpf (supplemental Figure 1) suggest that these definitive mast cells arising from HSCs may similarly have a requirement for Notch signaling.19
Perturbed mast cell development and proliferation can result in a myeloproliferative neoplasm called systemic mastocytosis, which can progress to mast cell leukemia, a variant of acute myeloid leukemia.46 Currently, systemic mastocytosis and mast cell leukemia are without curative therapies given that tyrosine kinase inhibitors, such as imatinib mesylate (Gleevec), are ineffective in the presence of the common c-KIT D816V mutation.46 Our studies suggest, for the first time, that Notch pathway inhibition may serve as an attractive novel therapeutic strategy in the treatment of mast cell diseases. Here we report that a transgenic line overexpressing nicd1a, which demonstrates increased embryonic mast cell numbers, can indeed be effectively “treated” using a γ-secretase inhibitor (Figure 5B). In addition, a transgenic zebrafish line that harbors the human c-KIT D816V mutation that has been developed in our laboratory (T.B.B., manuscript in preparation) may provide a further in vivo tool that can be exploited to rapidly screen for prospective Notch pathway inhibitors, as well as synergistic drug combinations for mast cell diseases. Parallel and supportive studies in human cell lines and mammalian model systems can be used to validate these findings and identify potentially clinically valuable compounds and/or drug combinations.
Using the elegance and simplicity afforded by the zebrafish model, we have confirmed an essential role for the Notch pathway in vertebrate mast cell fate determination for the first time in vivo and determined a developmental time course for the origin of the mast cell lineage during definitive hematopoiesis. In addition to establishing normal mast cell developmental pathways, our studies have revealed new potential therapeutic targets that can be explored in mast cell-related diseases. This work further highlights the rapidly expanding literature demonstrating the utility of the zebrafish model for elucidating the regulatory networks underlying blood cell development and its potential as an innovative tool for drug discovery.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Nathan Lawson for provision of the notch1a, notch1b, notch2, and notch3 constructs; Dr Leonard Zon for the mibta52b mutants, hsp70::GAL4 and UAS::nicd1a zebrafish lines; Dr David Traver and A. Michael Forrester for thoughtful review of the manuscript; Angela Young for zebrafish care and maintenance; and Jocelyn Jaques for administrative support.
This work was supported by Canadian Institutes of Health Research (grant 211071, J.N.B.), National Institutes of Health (grant R01CA120196, A.A.F.), and the Leukemia & Lymphoma Society Scholar Award (A.A.F.).
National Institutes of Health
Authorship
Contribution: S.I.D. designed and performed experiments, analyzed data, and wrote the manuscript; A.J.C. and C.A.G. performed experiments and analyzed data; T.B.B. oversaw experiments, analyzed data, and wrote the manuscript; A.A.F. provided expertise and guidance, helped design experiments, and edited the manuscript; and J.N.B. conceived of the study concept, designed experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jason N. Berman, Division of Pediatric Hematology/Oncology, Departments of Pediatrics, Microbiology & Immunology, and Pathology, Dalhousie University, Aquatics Laboratory, IWK Health Centre, 5850/5980 University Ave, Halifax, NS, Canada, B3K 6R8; e-mail: jason.berman@iwk.nshealth.ca.