To the editor:
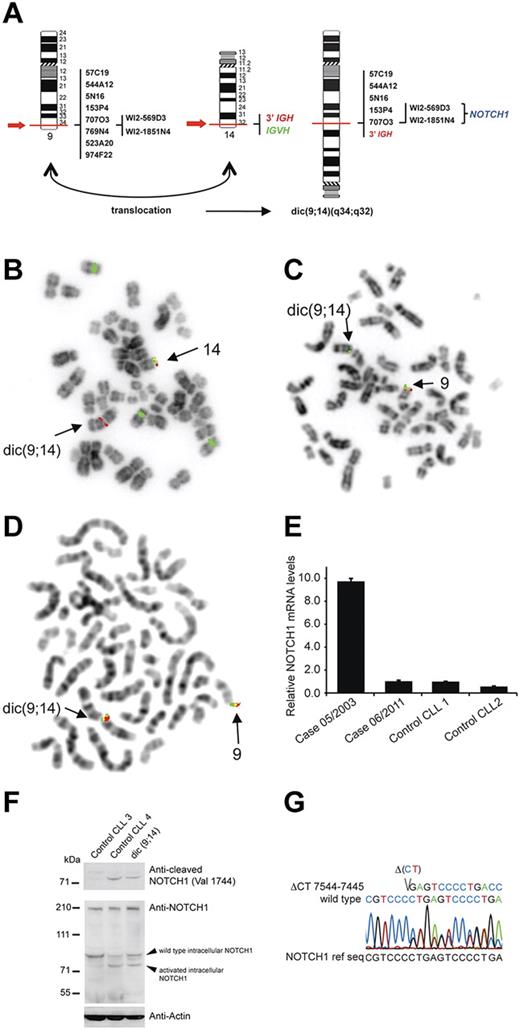
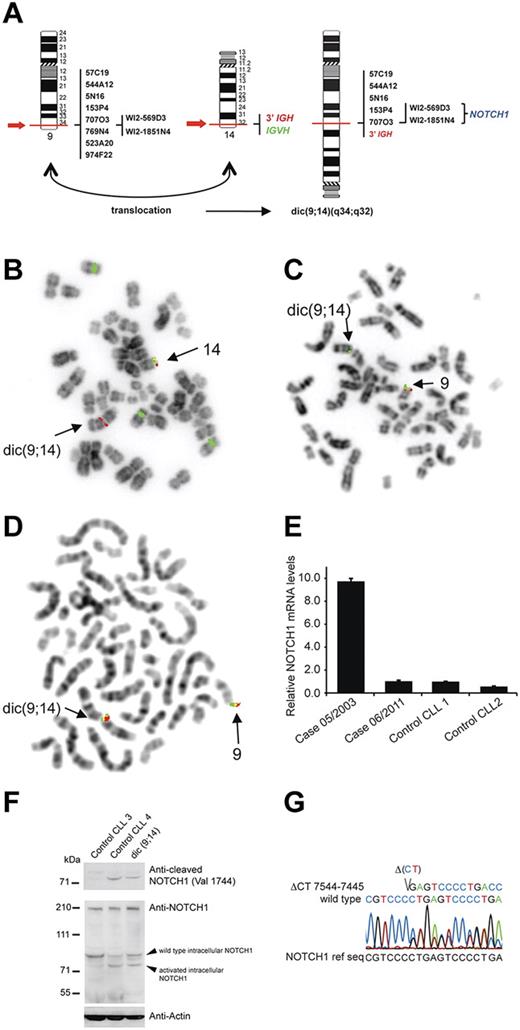
Recent data indicate that NOTCH1 mutations significantly increase the risk of CLL progression toward Richter syndrome (RS) and chemoresistance,1,2 and that activation of NOTCH1 at time of CLL diagnosis is an independent prognostic factor of poor survival.1,3 We report here a case of CLL with a novel rearrangement of NOTCH1 identified at the time of RS. The patient, a 58-year-old male, was diagnosed with CLL (unmutated VH) in RS in June 2003. Cytogenetic analysis and FISH on peripheral blood (PBL), bone marrow (BM), and lymph node (LN) cells showed 2 related clones: one with an isolated +12 and a second with +12 and dic(9;14)(q34;q32) (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). FISH analysis of dic(9;14)(q34;q32) indicated that this aberration resulted in juxtaposition of 3′IGH and 5′NOTCH1, as evidenced by the loss of sequences telomeric to the breakpoints (Figure 1A-D). These imbalances were confirmed by array CGH (data not shown). The targeting of NOTCH1 by dic(9;14) was evidenced by qRT-PCR analysis, which showed a 10-fold up-regulation of NOTCH1 mRNA (Figure 1E) and a low expression of the neighboring genes (GPSM11, CARD9, DNL2). Immunoblotting of a cell lysate from LN with a NOTCH1 antibody recognizing active, cleaved NOTCH1 (Val1744) identified a band corresponding to activated intracellular NOTCH1 (Figure 1F), suggesting an additional truncating mutation. Indeed, sequence analysis identified a 2 basepair deletion, ΔCT7544–7545/P2515fs, in the nucleotide sequence encoding the PEST domain (Figure 1G). This mutation resulting in expression of a truncated intracellular NOTCH1 allele is recurrent in T-ALL4 and CLL.1-3,5
Genetic and molecular analysis of NOTCH1 aberrations. (A) Graphic representation of dic(9;14)(q34;q32) with indicated probes applied for FISH mapping of both breakpoints and their hybridization pattern. (B-D) Examples of FISH analysis performed on a diagnostic LN sample with (B) LSI IGH dual color break apart probe and CEP12 (green), (C) RP11-707O3 (green) and RT11-769N4 (red), (D) WI2-569D3 (G248P80019F4, green) and WI2-1851N4 (G248P8957B2, red). BAC and fosmid clones were selected using the UCSC Genome Browser on Human May 2004 (NCBI35/hg17) Assembly. Note in panel B the presence of the 3′IGH/red signal on dic(9;14) and loss of the distal IGVH/green signal in cell with trisomy 12, in panel C hybridization of RP11-707O3 (green) with dic(9;14) and loss of the distal RT11-769N4 (red) sequences, and in panel D hybridization of both fosmids covering NOTCH1 with dic.(9;14). (E) qRT-PCR analysis of NOTCH1 mRNA expression levels. The patient was analyzed at 2 different timepoints, at diagnosis (05/2003; PBL; 57% of cells with dic(9;14)) and during disease evolution (06/2011; PBL; dic(9;14)-negative), and relative NOTCH1 expression levels were compared with 2 control CLL samples (CLL1 and CLL2) with unmutated VH and trisomy12. (F) Western blot analysis of protein extract of diagnostic LN cells of the index case (15% of cells with dic(9;14)) and 2 control CLLs (unmutated VH, trisomy12); CLL3 with unmutated NOTCH1 and CLL4 positive for ΔCT7544-7545/P2515fs. The top panel shows detection of active, cleaved NOTCH1 (Val 1744 antibody; Cell Signaling Technology) in CLL4 and the index case, but not in CLL3 cells. The bottom panel shows expression of NOTCH1 with a general anti-NOTCH1 antibody (c-20 Santa Cruz Biotechnology). Because of a low content of cells with dic(9;14) in the only available LN sample, overexpression of an activated form of NOTCH1 could not be evidenced. (G) Sanger chromatogram illustrating a heterozygous ΔCT7544-7545/P2515fs of the NOTCH1 cDNA in the diagnostic BM sample. FISH images acquired with a 63×/1.4 oil immersion objective in an Axioplan 2 fluorescence microscope equiped with an Axiophot 2 camera (Carl Zeiss) and an Isis imaging system (MetaSystems). Images were imported directly into PowerPoint (Microsoft).
Genetic and molecular analysis of NOTCH1 aberrations. (A) Graphic representation of dic(9;14)(q34;q32) with indicated probes applied for FISH mapping of both breakpoints and their hybridization pattern. (B-D) Examples of FISH analysis performed on a diagnostic LN sample with (B) LSI IGH dual color break apart probe and CEP12 (green), (C) RP11-707O3 (green) and RT11-769N4 (red), (D) WI2-569D3 (G248P80019F4, green) and WI2-1851N4 (G248P8957B2, red). BAC and fosmid clones were selected using the UCSC Genome Browser on Human May 2004 (NCBI35/hg17) Assembly. Note in panel B the presence of the 3′IGH/red signal on dic(9;14) and loss of the distal IGVH/green signal in cell with trisomy 12, in panel C hybridization of RP11-707O3 (green) with dic(9;14) and loss of the distal RT11-769N4 (red) sequences, and in panel D hybridization of both fosmids covering NOTCH1 with dic.(9;14). (E) qRT-PCR analysis of NOTCH1 mRNA expression levels. The patient was analyzed at 2 different timepoints, at diagnosis (05/2003; PBL; 57% of cells with dic(9;14)) and during disease evolution (06/2011; PBL; dic(9;14)-negative), and relative NOTCH1 expression levels were compared with 2 control CLL samples (CLL1 and CLL2) with unmutated VH and trisomy12. (F) Western blot analysis of protein extract of diagnostic LN cells of the index case (15% of cells with dic(9;14)) and 2 control CLLs (unmutated VH, trisomy12); CLL3 with unmutated NOTCH1 and CLL4 positive for ΔCT7544-7545/P2515fs. The top panel shows detection of active, cleaved NOTCH1 (Val 1744 antibody; Cell Signaling Technology) in CLL4 and the index case, but not in CLL3 cells. The bottom panel shows expression of NOTCH1 with a general anti-NOTCH1 antibody (c-20 Santa Cruz Biotechnology). Because of a low content of cells with dic(9;14) in the only available LN sample, overexpression of an activated form of NOTCH1 could not be evidenced. (G) Sanger chromatogram illustrating a heterozygous ΔCT7544-7545/P2515fs of the NOTCH1 cDNA in the diagnostic BM sample. FISH images acquired with a 63×/1.4 oil immersion objective in an Axioplan 2 fluorescence microscope equiped with an Axiophot 2 camera (Carl Zeiss) and an Isis imaging system (MetaSystems). Images were imported directly into PowerPoint (Microsoft).
The patient was treated and achieved complete remission (supplemental Table 1). In 2007, however, CLL relapsed and an examination of BM identified a clone with a sole +12 negative for ΔCT7544–7545/P2515fs. Two years later, CLL progressed and BM revealed an evolved clone with complex aberrations including +12 and t(14;19)(q32;q13)/IGH-BCL3 but lacking dic(9;14)(q34;q32). Of note, BM was again positive for ΔCT7544–7545/P2515fs. Despite treatment, 2 clones harboring +12, one with t(14;19) and a second with new additional karyotypic changes, were seen in the analyzed BM (06/2011) positive for ΔCT7544–7545/P2515fs. TP53 disruption frequently associated with RS6 was not observed in the analyzed samples (supplemental Table 1). Four months later, the patient developed peripheral T-cell lymphoma, a rare recurrent event in CLL.7 Altogether, the present case allows us to deduce the sequence of multiple genetic defects driving development and progression of CLL. An initial clone with +12, likely present at a presymptomatic CLL phase, later acquired an activating mutation of NOTCH1. Subsequent acquisition of dic(9;14)/IGH-NOTCH1 triggered Richter transformation. After a few years, a persistent, chemorefractory NOTCH1-mutated clone underwent another hit, t(14;19)/IGH-BCL3, which initiated the second progression that was followed by a fatal CLL-unrelated T-cell lymphoma. Our findings confirm the risk of activating mutations of NOTCH1 in Richter transformation of CLL,1,2 particularly CLL with +12,3 and highlight that a residual chemotherapy-resistant NOTCH1-mutated clone is at risk of acquiring further progression-associated hits. Besides the known t(14;19)(q32;q13)/IGH-BCL3,8-10 we also identified dic(9;14)(q34;q32)/IGH-NOTCH1, which so far has not been reported in B-cell leukemia/lymphoma, as a novel genomic aberration capable of triggering RS.
Authorship
The online version of this article contains a data supplement.
Acknowledgments: This work was supported by the European Research Council (ERC-starting grant to J.C.), KU Leuven (concerted action grant to J.C., P.V., I.W.), and the Stichting Tegen Kanker (grant to P.V.). K.D.K. is a postdoctoral researcher of the Research Foundation-Flanders (FWO) and P.V. is a senior clinical investigator of FWO. The authors thank Ursula Pluys for technical assistance and Rita Logist for editorial help.
Contribution: K.D.K. and I.W. designed and performed the research, analyzed data, and wrote and approved the manuscript; L.M. analyzed data and wrote and approved the manuscript; A.B. and C.G. treated the patient, contributed vital patient information, and approved the manuscript; J.F.F. performed array CGH, analyzed data, and approved the manuscript; P.V. analyzed data and wrote and approved the manuscript; and J.C. designed the research, analyzed data, and wrote and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Iwona Wlodarska, Center for Human Genetics, KU Leuven, Gasthuisberg, Herestraat 49, Box 602, B-3000 Leuven, Belgium; e-mail: iwona.wlodarska@uzleuven.be.