In this issue of Blood, Zardo et al describe an emerging mechanism by which microRNAs can regulate gene expression in blood cells via transcriptional gene silencing (TGS).1 The authors provide evidence that miR-223 binds to specific sites within the promoter of its target gene Nfia and represses transcription by influencing epigenetic events. These observations support the notion that multiple mechanisms of miRNA-mediated gene repression exist and should be considered when investigating gene targets of a given miRNA of interest.
MicroRNAs (miRNAs) clearly regulate gene expression at the posttranscriptional level by recruiting the RNA-induced silencing complex to the 3′ untranslated regions (UTRs) of target mRNAs, and this has been shown to be a major mechanism underlying their function in mammalian cells.2 However, the current study demonstrates a role for miRNAs in the direct regulation of gene transcription in mammalian cells, and is among only a few reports having made this connection to date.3,4
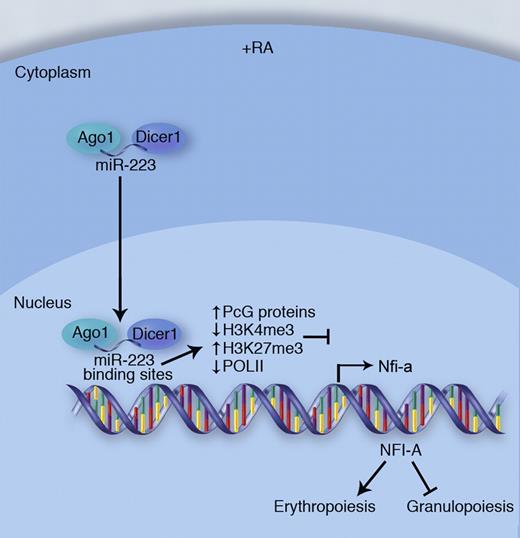
In response to all-trans-retinoic acid (RA), a signal that promotes granulocyte development, miR-223 translocates into the nucleus along with Dicer1 and Ago1. Once in the nucleus, miR-223-RNAi complexes bind to miR-223 targets sites located in the 5′ regulatory region of the Nfia gene. This triggers a number of epigenetic changes leading to transcriptional silencing of Nfia. Reduced expression of NFI-A redirects cellular development away from erythropoiesis and toward granulopoiesis. Professional illustration by Marie Dauenheimer.
In response to all-trans-retinoic acid (RA), a signal that promotes granulocyte development, miR-223 translocates into the nucleus along with Dicer1 and Ago1. Once in the nucleus, miR-223-RNAi complexes bind to miR-223 targets sites located in the 5′ regulatory region of the Nfia gene. This triggers a number of epigenetic changes leading to transcriptional silencing of Nfia. Reduced expression of NFI-A redirects cellular development away from erythropoiesis and toward granulopoiesis. Professional illustration by Marie Dauenheimer.
The article focuses on the epigenetic silencing of the gene encoding the transcription factor NFI-A by miR-223 (see figure). NFI-A is an established repressor of granulopoiesis and a previously identified posttranscriptional target of miR-223.5 Experiments found that miR-223, which localized to the nucleus after the induction of granulopoiesis by all-trans-retinoic acid, directly interacted with complementary binding sites in the promoter of Nfia and served as a guide for the localization of specific protein complexes that mediated epigenetic silencing of the Nfia gene. Chromatin immunoprecipitation (ChIP) experiments identified increased H3K27me3 and decreased H3K4me3 and Pol-II promoter binding after induction of granulopoiesis. Polycomb group proteins (PcGs) and components of the RNAi machinery, including Dicer1 and Ago1, also interacted with the Nfia promoter region under these conditions. Importantly, these events were shown to occur through a mechanism involving miR-223. While the experimental system used here demonstrates that this regulatory mechanism influences granulocyte development, these findings imply that other blood cell types might use a similar miRNA-dependent approach to accomplishing TGS during lineage fate decisions that occur during hematopoiesis.
Along with these new and exciting observations that miRNAs can directly regulate transcription in mammalian blood cells come many questions about the process. For instance, how widespread is this mechanism? miRNA transcriptional regulation could be specific to certain types of genes, like Nfia, or might be a common regulatory mechanism used to modulate a majority of genes. It would be interesting to know if miRNAs commonly target the same gene at both the transcriptional and posttranscriptional levels to ensure repression as appears to be the case for Nfia. In addition, while the authors indicate some degree of evolutionary conservation of the miR-223 binding site sequences in the promoter of Nfia, it remains to be determined whether sequence conservation of miRNA promoter binding sites are common like they are in 3′ UTRs. Other questions include how Ago1 and Dicer1 function to impact histone marks and other epigenetic events that control gene transcription after their recruitment by miRNAs, and whether or not miRNAs can activate gene transcription in certain contexts. This is clearly a new frontier in miRNA biology that warrants further investigation.
A variety of different types of noncoding RNAs (ncRNAs) are now known to modulate target gene transcription. Examples include long intergenic ncRNAs (lincRNAs),6 promoter associated RNAs7 and exogenously delivered small interfering RNAs (siRNAs) designed to target gene promoters.8 Endo-genously expressed miRNAs can now be added to this group of transcription-regulating ncRNAs, which can take advantage of their Watson-Crick base pairing with promoter DNA sequences to recruit regulatory protein complexes to specific gene promoters to repress transcription.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■