Abstract
Epigenetic histone modifications are thought to underlie the rapid memory immune response to recall antigen that develops after vaccination. However, histone-modification patterns in genes encoding transcription factors regulating cytokine production have not been investigated in either memory and naive T cells or as the immune system matures to understand the differences in cytokine response patterns. In the present study, we analyzed histone modifications in promoter regions of T-bet, GATA-3, PU.1, IRF4, and RORC in neonatal naive T cells and in adult naive and memory CD4 T cells, and found a unique and dynamic histone-modification pattern in the PU.1 promoter that was related to age and the naive/memory status of a T cell. Naive T cells required more intense stimulation to switch the chromatin pattern in the PU.1 promoter from a repressive to permissive state, and therefore to produce IL-9 than did memory T cells. Inhibition of repressive histone methylation by the specific inhibitor 3-deazaneplanocin induced Th9-specific PU.1 expression, even in conditions that would normally yield only Th0 cytokines. Conversely, prevention of histone acetylation by the histone acetyltransferase inhibitor curcumin diminished PU.1 expression after IL-9–inducing stimulation. Our findings identify age- and differentiation-status–related epigenetic modifications of PU.1 as a unique regulator of Th9 memory acquisition and Th9 immunity.
Introduction
Upon encountering a specific antigen, memory CD4 Th cells, unlike their naive counterparts, are capable of rapid proliferation and cytokine production. They are also characterized by a considerable degree of phenotypic and functional heterogeneity.1,2 There is compelling evidence that both the maturation of CD4 naive T cells into memory T cells and Th-cell differentiation are underpinned by alterations in histone modifications.3,4 Posttranslational histone modifications control the chromatin structure and thus the accessibility of promoter regions involved in Th-cell differentiation and cytokine production.5,6 Of special interest are permissive and repressive modifications of histone H3, such as histone H3 lysine 4 trimethylation (H3K4me3), histone H3 acetylation (H3ac), and histone H3 lysine 27 trimethylation (H3K27me3). Such histone modifications orchestrate chromatin accessibility in a bivalent manner, either permitting or preventing gene transcription. In fact, their presence or absence in regulatory regions of genes involved in Th-cell differentiation, such as the IFN-G locus, the IL4 locus, the IL17/IL17F locus, the IL12RB2 and TBX loci, or NFATC1-C4 loci determines the transcriptional status of these genes7-11 and thus the outcome of an immune response.
Epigenetic modifications were thought originally to occur in the germline, where they imprint the parental-origin–specific expression of several genes.12 Recently, it has been suggested that epigenetic modifications, in addition to yielding stable imprints governing fetal growth, metabolism, or tissue differentiation, may also occur dynamically over time at specific sites in the genome, for example with increasing age.13 Their presence in particular sites, but not in others seemingly related may, therefore translate into functional differences between subjects with regard to specific gene expression. As a consequence, differences in the epigenetic program between subjects have been related to the development of diseases such as malignancies or autoimmune diseases. For example, the expression of tumor-suppressor genes is inhibited during carcinogenesis in addition to H3K27 methylation by di- and trimethylation of H3K9.14 In systemic lupus erythematosus, a prototype autoimmune disease, changes in the pattern of DNA methylation have been associated with twin discordance.15 The evidence to date indicates that understanding the epigenetic modifications that occur over time might provide a platform for understanding the molecular pathogenesis of diseases to ultimately provide practical approaches to treatment.
In the present study, we show that dynamic changes in epigenetic modifications in a particular and specific site of a master regulator of immune responses do occur, and that these alterations translate into functional differences with regard to specific gene expression and thus lymphocyte functions. We found that Th9-cell differentiation, but not that of Th1, Th2, or Th17 cells, is controlled by unique and dynamic epigenetic modifications in the promoter region of its master transcription factor, PU.1. We showed that naive CD4 T cells are more restricted in their expression of PU.1 than memory CD4 T cells and that this restriction was characterized by a dominant-repressive chromatin configuration. Associated with this was a gradual decrease in cytokine requirements for PU.1 expression and subsequent Th9-cell differentiation from neonatal naive through adult naive to memory CD4 T cells. Our results indicate that epigenetic modifications in the PU.1 promoter uniquely and dynamically control the threshold for Th9-cell development in naive and memory CD4 T cells and therefore functional maturation of the immune system.
Methods
Reagents and Abs
The following Abs were used for T-cell purification, stimulation, and staining: anti-CD45RO (UCHL-1), anti-CD3 (OKT3; ATCC); FITC-labeled mAbs to CD28 (28.2), CD3, CD19, CD27, CD45RA, IL-4, IFN-γ, FITC-labeled mouse IgG1, rat IgG1; PE-labeled mAbs to CD4, CD8, CD20, CD27, CD45RO, PE-labeled mouse IgG1, mouse IgG2b, rat IgG2a, rat IgG2b, allophycocyanin (APC)–labeled mAb to IFN-γ, APC-labeled mouse IgG1, PE/cyanin7 (Cy7)–labeled mouse IgG1, and recombinant human IL-12 (BD Biosciences); PE/Cy7–labeled anti-CD31, APC-labeled anti–IL-9, APC-labeled mouse IgG2b, PE-labeled mAbs to IL-5, and IL-9; PE/Cy7–labeled anti–IL-17 (BioLegend); PE-labeled anti-GATA–binding protein 3 (GATA-3), PE-labeled anti–Th1-specific T box transcription factor (T-bet; eBiosciences); anti-H3K4me3 Ig, anti-H3ac Ig, and rabbit IgG (Millipore); anti-H3K27me3 Ig (Epigentek). PE-labeled anti-PU.1, PE-labeled sheep IgG, recombinant human IL-1β, IL-6, IL-10, IL-23, and TGF-β (R&D Systems); recombinant human IL-2 (Proleukin; Chiron); recombinant human IL-4, neutralizing mAbs to IL-4 and IFN-γ (Endogen); recombinant human IL-21, FCS, PBS (Gibco); ionomycin (Calbiochem); 3-deazaneplanocin A (DZNep) (Cayman Chemical); paraformaldehyde, phorbol myristate acetate, monensin, mouse serum, rat serum, saponin, and curcumin (Sigma-Aldrich).
Blood samples
Peripheral blood samples were obtained from adult healthy donors. All donors provided written informed consent. Umbilical cord blood samples were obtained directly after birth (Klinik für Frauenheilkunde, University of Munich, Germany). The study was approved by the ethics committee of the University of Munich.
T-cell isolation
Mononuclear cells were isolated from peripheral blood and umbilical cord blood by centrifugation over a Ficoll-Hypaque gradient (Lymphoflot; Biotest AG). After rosetting,16 cells were purified by negative selection using the Naive CD4 T-cell Isolation Kit II, the Memory CD4 T-cell Isolation Kit, or the CD4 Recent Thymic Emigrant Isolation Kit (all Miltenyi Biotec). Homogeneity and purity of each cell population was controlled routinely by flow cytometry.
Cell culture
Cell cultures were performed in RPMI 1640 medium supplemented with 50 U/mL of penicillin G, 50 μg/mL of streptomycin, 2nM l-glutamine (Gibco Invitrogen), 10 U/mL of recombinant human IL-2, and 10% normal human serum. Cell cultures were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Flow cytometry
After staining of T cells with saturating amounts of fluorochrome-labeled mAbs, T-cell surface molecules were analyzed by flow cytometry (Cytomics FC500; Beckman Coulter). For intracellular staining, cells were restimulated with 20 ng/mL of phorbol myristate acetate and 1nM ionomycin in the presence of 2μM monensin for 5 hours, fixed with 3% paraformaldehyde, permeabilized with 0.1% saponin in 2% FCS/PBS, incubated with 4% rat and mouse serum, stained with saturating amounts of fluorochrome-labeled Abs, and analyzed by FACS. For costaining of cytokines together with transcription factors, cells were treated according to the manufacturer's instructions with the Human FoxP3 Buffer Set (BD Biosciences) and stained with fluorochrome-labeled Abs against IL-9, GATA-3, or T-bet.
Transfection of siRNA
Naive CD4 T cells from cord blood were transfected with control or SPI1-specific siRNAs (2 μg/5 × 106 cells; Thermo Scientific) using the Human T-cell Nucleofector Kit and the U-14 transfection program (Amaxa Biosystems).
Gene-expression analysis
Total RNA was extracted using the RNeasy Mini Kit (QIAGEN). One microgram of RNA was used to transcribe mRNA to cDNA following standard protocols. Real-time PCR was performed in duplicate using the Universal PCR Master Mix and TaqMan Assays for IFN-γ, IL-4, IL-9, IL-17, PU.1, RAR-related orphan receptor C (RORC), and cyclophilin in the ABI PRISM 7000 Sequence Detection System (Applied Biosystems). Expression of target genes was calculated by a comparative method for relative quantification after normalization to cyclophilin expression.
Stimulation and differentiation of T cells
For the generation of CD4 effector T cells, purified naive and memory CD4 T cells were stimulated in flat-bottomed cell-culture plates (Costar; Corning) coated with 1 μg/mL of OKT3 at a concentration of 0.5 × 106 cells/mL with 1 μg/mL of anti-CD28. Th1 cells were generated by the addition of 40 ng/mL of recombinant human 40 ng/mL of IL-12 and 10 μg/mL of anti–IL-4. Th2 effector cells were generated in the presence of 31.25 ng/mL of recombinant human IL-4. Polarization of Th17 cells was induced by the addition of 10 ng/mL of IL-1β, 100 ng/mL of IL-21, 10 μg/mL of anti–IFN-γ, and 10 μg/mL of anti–IL-4. Th9 cells were generated by the addition of 31.25 ng/mL of recombinant human IL-4 and/or 5 ng/mL of TGF-β. Where indicated, 10 ng/mL of recombinant human IL-1β, 20 ng/mL of IL-6, 20 ng/mL of IL-10, 10 ng/mL of IL-12, 100 ng/mL of IL-21, 50 ng/mL of IL-23, and 5μM DZNep were added at the start of the culture. Treatment with 20μM curcumin was performed for 2 hours before differentiation.
ChIP and DNA quantification
ChIP assays were carried out using the EpiQuik Chromatin Immunoprecipitation Kit (Epigentek). Freshly isolated cells or cells primed for 5 days (3 × 106 cells) were cross-linked with ice-cold 1% formaldehyde for 1 minute. Fixation was stopped by 1.25M glycine for 5 minutes. Cells were resuspended in nuclear lysis buffer and sonicated by a Bioruptor Next Generation (Diagenode). ChIP reactions were performed with lysate and 2-4 μg of anti-H3K4me3, anti-H3ac, anti-H3K27me3, and rabbit IgG as a negative control. Eluted DNA was analyzed by quantitative PCR using the Power SYBR Green PCR Master Mix and 200nM primer mix for SYBR Green detection in the ABI PRISM 7000 Sequence Detection System. Primers for SYBR Green detection were designed using MacVector Version 12.0.2 software (Accelrys). Primer pairs were used as follows: 5′-CGCTCTCCCAAACACCCTG-3 and 5′-AGCCAGCCTGCCCCATTC-3′ for the GATA-3 promoter region; 5′-GGCAACCCGAAAGGTCACTTAG-3′ and 5′-TTCTCCTGGCACTCAGAGGCTC-3′ for the T-bet promoter region; 5′-TCTCCCCTATGCCTGTCACCTG-3′ and 5′-TGATTTTGCCCAAGGACTCACAC-3′ for the RORC promoter region; 5′-GAAGCAGGCATTTGTTGGGTTAG-3′ and 5′-TGGGACTGAGAGGGATGACTTTG-3′ for the PU.1 promoter region; 5′-CCCACCTCGCACTCTCAGTTTC-3′ and 5′-TATCAGCCTCACACCCCTCCTC-3′ for the IRF4 promoter region; 5′-TCTCTTTTCCTTGCTGTCCCTG-3′ and 5′-GTCACTCACCTCTTGCTTCTGGTC-3′ for the PU.1 upstream enhancer region. Data are presented as a log to the base 2 of the ratio of the immunoprecipitated amounts of DNA of H3K4me3 to H3K27me3 and H3ac to H3K27me3 pull-downs or as a ratio of immunoprecipitated DNA to that of input DNA. Binding specificity was verified by amplification of unspecific precipitated DNA with rabbit IgG.
Statistical analysis
Results were analyzed with the Student t test or 1-way ANOVA, followed by the Tukey-Kramer posthoc analysis where indicated. P < .05 was considered statistically significant.
Results
The PU.1 promoter is selectively repressed in neonatal CD4 T cells by H3K27 trimethylation
To dissect epigenetic signatures between naive and memory T cells, we assessed histone modifications in promoter regions of the key transcription factors in Th-cell differentiation, T-bet, GATA-3, PU.1, IRF4, and RORC, in human naive CD4 T cells purified from cord blood and in naive and in memory CD4 T cells from the peripheral blood of adult subjects (Figure 1). The PU.1 promoter region of neonatal CD4 T cells was dominated by the presence of the repressive H3K27me3 histone modification in relation to the permissive H3K4me3, as well as H3ac modifications (Figure 1A-B). The overrepresentation of the repressive H3K27me3 histone modification diminished gradually from neonatal naive through adult naive to memory CD4 T cells (Figure 1A). In contrast, T-bet, GATA-3, IRF4, and RORC promoters showed permissive and repressive histone modifications to a comparable extent in all cell populations analyzed (Figure 1A-B and supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The PU.1 promoter is therefore the only promoter with a strong overrepresentation of repressive modifications in neonatal naive CD4 T cells, suggesting a selective inactivity of this promoter in naive CD4 T cells. Interestingly, the overrepresentation of repressive H3K27me3 histone modifications in the PU.1 promoter was even more strongly represented in neonatal CD4 T cells than in PU.1-nonexpressing CD8 T cells (supplemental Figure 2). Conversely, the extent of permissive and repressive histone modifications in memory CD4 T cells was comparable to that in B cells expressing PU.1 constitutively (supplemental Figure 2).
Histone modifications in the promoter regions of T-bet, GATA-3, PU.1, IRF4, and RORC. Freshly isolated naive CD4 T cells from cord blood or adult naive and memory CD4 T cells from the peripheral blood were processed for ChIP analysis. Relative amounts of precipitated DNA from the T-bet, GATA-3, PU.1, IRF4, and RORC promoter regions were assessed. (A) Log2 of the ratio of relative amounts of DNA precipitated with anti-H3K4me3 to anti-H3K27me3 (top panel) and with anti-H3ac to anti-H3K27me3 (bottom panel). (B) Fraction of the PU.1 promoter DNA precipitated with Abs against H3K4me3, H3ac, or H3K27me3 in relation to the input DNA. The mean ± SEM of at least 3 independent experiments with cells from different donors is demonstrated. Statistical analyses were performed using ANOVA. ***P < .001; *P < .05; n.s. indicates not significant.
Histone modifications in the promoter regions of T-bet, GATA-3, PU.1, IRF4, and RORC. Freshly isolated naive CD4 T cells from cord blood or adult naive and memory CD4 T cells from the peripheral blood were processed for ChIP analysis. Relative amounts of precipitated DNA from the T-bet, GATA-3, PU.1, IRF4, and RORC promoter regions were assessed. (A) Log2 of the ratio of relative amounts of DNA precipitated with anti-H3K4me3 to anti-H3K27me3 (top panel) and with anti-H3ac to anti-H3K27me3 (bottom panel). (B) Fraction of the PU.1 promoter DNA precipitated with Abs against H3K4me3, H3ac, or H3K27me3 in relation to the input DNA. The mean ± SEM of at least 3 independent experiments with cells from different donors is demonstrated. Statistical analyses were performed using ANOVA. ***P < .001; *P < .05; n.s. indicates not significant.
Neonatal CD4 T cells require collaborative cytokine signaling to differentiate into IL-9–producing cells
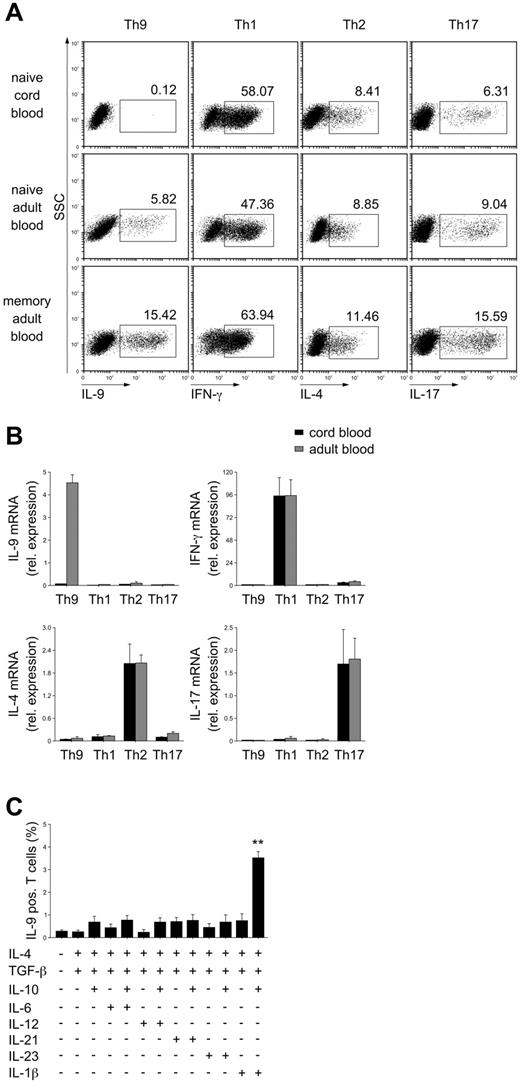
PU.1 has been reported to determine Th9-cell development.17 Therefore, the different histone-modification patterns of its promoter are suggestive of different requirements for PU.1 expression and for Th9-cell differentiation. We analyzed Th9-cell differentiation in response to the “conventional” Th9-inducing conditions, TGF-β and IL-4,18,19 from cord blood CD4 naive T cells and from adult CD4 naive and memory T cells. Whereas TGF-β and IL-4 were sufficient for Th9-cell differentiation from adult naive and memory CD4 T cells, they completely failed to induce IL-9–producing cells from cord blood CD4 T cells (Figure 2A). In contrast, Th1, Th2, and Th17 cell differentiation from neonatal CD4 T cells appeared to be functional and was comparable to that of peripheral blood naive CD4 T cells. A gradual increase in the frequency of Th9 cells after differentiation was observed from neonatal naive through adult naive to memory CD4 T cells. Consistent with this, no IL-9 mRNA could be detected in naive cord blood CD4 T cells after stimulation in the presence of TGF-β and IL-4 (Figure 2B). Conversely, the expression of mRNA for signature cytokines of Th1, Th2, and Th17 cells (IFN-γ, IL-4, and IL-17, respectively) was comparable in neonatal and adult naive T cells after appropriate priming (Figure 2B).
Neonatal CD4 naive T cells require combined IL-10 and IL-1β signaling in addition to TGF-β and IL-4 to differentiate into IL-9–producing cells. Naive or memory CD4 T cells were purified from cord blood or from the peripheral blood of adult subjects and stimulated under Th9, Th1, Th2, and Th17 cell-inducing conditions or in the presence of different cytokine combinations as indicated for 5 days. (A) Cytoplasmic cytokines were analyzed by intracellular flow cytometry after restimulation of the cells with phorbol myristate acetate and ionomycin. Results from 1 of 6 independent experiments with cells from different donors are shown. (B) Cytokine mRNA was assessed by real-time PCR; mRNA levels were normalized for the expression of cyclophilin. Mean ± SEM of 5 independent experiments with cells from different donors is shown. (C) Frequencies of IL-9–producing cells were assessed by intracellular flow cytometry. The mean ± SEM of 4 independent experiments with cells from different donors is demonstrated. **P < .01 by Student t test compared with medium control without any cytokine.
Neonatal CD4 naive T cells require combined IL-10 and IL-1β signaling in addition to TGF-β and IL-4 to differentiate into IL-9–producing cells. Naive or memory CD4 T cells were purified from cord blood or from the peripheral blood of adult subjects and stimulated under Th9, Th1, Th2, and Th17 cell-inducing conditions or in the presence of different cytokine combinations as indicated for 5 days. (A) Cytoplasmic cytokines were analyzed by intracellular flow cytometry after restimulation of the cells with phorbol myristate acetate and ionomycin. Results from 1 of 6 independent experiments with cells from different donors are shown. (B) Cytokine mRNA was assessed by real-time PCR; mRNA levels were normalized for the expression of cyclophilin. Mean ± SEM of 5 independent experiments with cells from different donors is shown. (C) Frequencies of IL-9–producing cells were assessed by intracellular flow cytometry. The mean ± SEM of 4 independent experiments with cells from different donors is demonstrated. **P < .01 by Student t test compared with medium control without any cytokine.
To determine the cytokine requirements for Th9-cell differentiation from neonatal CD4 T cells, these cells were activated in the presence of TGF-β and IL-4 and in the presence of additional cytokines that have been implicated in enhancing Th9-cell differentiation, such as IL-10, IL-1β, IL-6, IL-12, IL-21, and IL-23.20-22 Only the addition of IL-1β in combination with IL-10 potently induced Th9-producing cells from naive cord blood CD4 T cells (Figure 2C). The addition of IL-6, IL-12, IL-21, or IL-23, either alone or in combination with IL-10, did not affect the development of neonatal IL-9–producing T cells significantly.
Different responsiveness of the PU.1 gene to cytokines in naive and memory CD4 T cells determines the stringency of IL-9 expression requirements
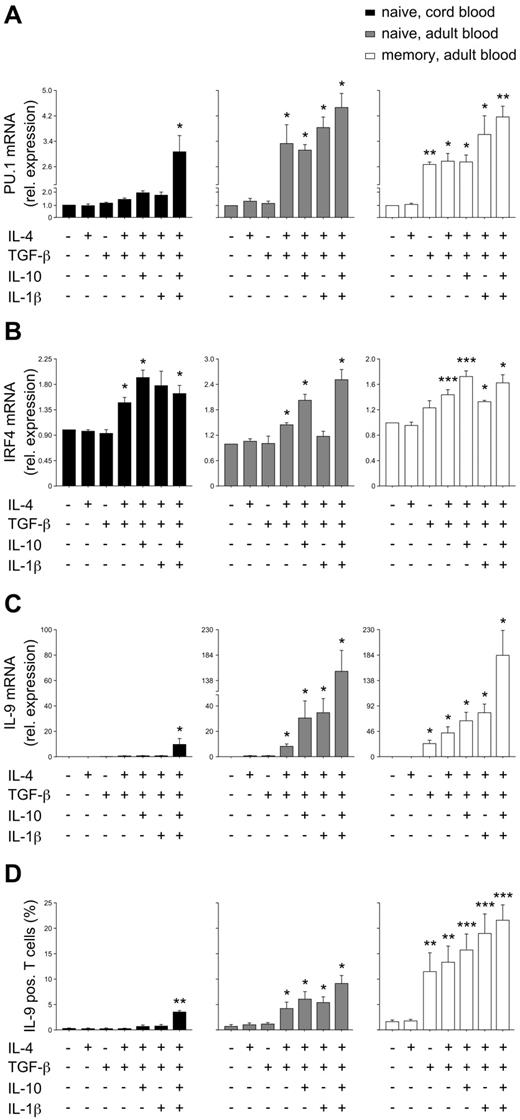
When PU.1 expression was analyzed in response to different cytokines, the collective effect of TGF-β, IL-4, IL-10, and IL-1β (Figure 2C) was required to induce the expression of PU.1 mRNA in cord blood CD4 T cells. In adult naive CD4 T cells, PU.1 mRNA expression was dependent on TGF-β and IL-4, and in memory CD4 T cells, significant PU.1 expression was detectable if only TGF-β was added (Figure 3A). Conversely, the expression of IRF4, another transcription factor essential for Th9-cell development,23 was less fastidious and could be induced in neonatal CD4 T cells with a combination of TGF-β and IL-4 (Figure 3B). The different cytokine requirements for PU.1 expression was well correlated with that for IL-9 mRNA up-regulation and with the increase in Th9-cell frequency in the respective T-cell populations (Figure 3C-D). In control experiments with FACS-sorted naive T cells, we confirmed the observed differences between neonatal and adult naive cells (supplemental Figure 3). Therefore, the data indicate that the decrease in cytokine requirements for Th9-cell development as cells mature from naive neonatal through adult naive to memory CD4 T cells is determined by the cytokines required for PU.1 expression.
Th9 development and expression of the Th9 transcription factors PU.1 and IRF4. Naive cord blood CD4 T cells or adult naive and memory CD4 T cells were stimulated with anti-CD3/28 in the presence of the indicated cytokines. (A-B) Expression of PU.1 and IRF4 mRNA was assessed after the additional stimulation of cells with anti-CD3 for 24 hours; mRNA levels were normalized to cyclophilin and are presented relative to expression in cells differentiated in the absence of any cytokines. (C) IL-9 mRNA was assessed by real-time PCR and normalized for the expression of cyclophilin. (D) Frequencies of IL-9–producing cells were assessed by flow-cytometric analysis after 5 days of culture and restimulation with phorbol myristate acetate and ionomycin. The mean ± SEM of at least 3 independent experiments with cells from different donors is shown. *P < .05; **P < .01; ***P < .001 by ANOVA compared with medium control without any cytokine.
Th9 development and expression of the Th9 transcription factors PU.1 and IRF4. Naive cord blood CD4 T cells or adult naive and memory CD4 T cells were stimulated with anti-CD3/28 in the presence of the indicated cytokines. (A-B) Expression of PU.1 and IRF4 mRNA was assessed after the additional stimulation of cells with anti-CD3 for 24 hours; mRNA levels were normalized to cyclophilin and are presented relative to expression in cells differentiated in the absence of any cytokines. (C) IL-9 mRNA was assessed by real-time PCR and normalized for the expression of cyclophilin. (D) Frequencies of IL-9–producing cells were assessed by flow-cytometric analysis after 5 days of culture and restimulation with phorbol myristate acetate and ionomycin. The mean ± SEM of at least 3 independent experiments with cells from different donors is shown. *P < .05; **P < .01; ***P < .001 by ANOVA compared with medium control without any cytokine.
Cytokines control histone-modification patterns in the PU.1 promoter
Because epigenetic modifications are dynamic and can change after T-cell activation, we next investigated histone modifications in response to different Th9-inducing cytokine combinations (Figure 4). As shown in Figure 4A, an upstream enhancer has been identified in the PU.1 gene locus in both mice and humans.24 Therefore, in addition to the PU.1 promoter region, we also analyzed changes in histone modifications in response to different stimulation conditions in this enhancer. Neonatal CD4 T cells activated in the presence of TGF-β or TGF-β/IL-4 retained their inaccessible chromatin configuration with a predominance of H3K27me3 in the PU.1 promoter (Figure 4B and supplemental Figure 4A). However, after stimulation by TGF-β, IL-4, IL-10, and IL-1β, their chromatin pattern in the PU.1 promoter region switched to an open structure dominated by H3K4me3 and H3ac. Further, adult naive CD4 T cells showed enriched H3K4me3 and H3ac histone modifications in the PU.1 promoter in response to TGF-β and IL-4, but not to TGF-β only, whereas memory CD4 T cells demonstrated a predominance of permissive histone modifications in the PU.1 promoter after stimulation by TGF-β alone (Figure 4B and supplemental Figure 4A). No pronounced changes in the histone modifications in response to intensified stimulation were observed in the enhancer region (Figure 4C and supplemental Figure 4B). Therefore, the acquisition of the active chromatin configuration of the PU.1 promoter but not in the enhancer region requires additional cytokine signals in neonatal naive CD4 T cells compared with adult naive T cells, and both require more cytokine signals than memory CD4 cells. When the IRF4 promoter region was analyzed, concordant with the IRF4 expression shown in Figure 3B, there was no dependency between the different cytokine combinations and the histone-modification pattern (Figure 4D and supplemental Figure 4C). The IL9 promoter was also not affected by alterations of histone modifications during differentiation, further emphasizing that alterations in the PU.1 promoter were functionally relevant for the differences in Th9-cell responses observed (supplemental Figure 5).
Changes in histone modifications in the PU.1 gene locus and IRF4 promoter in response to Th9-inducing conditions. (A) Schematic diagram of the enhancer region of the PU.1 gene located 17 kb upstream of the PU.1 transcription start site. (B-D) Naive cord blood CD4 T cells or adult naive and memory CD4 T cells were stimulated with anti-CD3/28 in the presence of the indicated cytokines. After fixation and DNA shedding, DNA fragments were immunoprecipitated with anti-H3K4me3, anti-H3ac, or anti-H3K27me3 Abs. Eluted DNA was analyzed by quantitative PCR using primers spanning the promoter region of PU.1 (B), the PU.1 upstream enhancer region (C), and the promoter region of IRF4 (D). The mean ± SEM of at least 3 independent experiments with cells from different donors is shown. *P < .05; **P < .01; ***P < .001 by Student t test.
Changes in histone modifications in the PU.1 gene locus and IRF4 promoter in response to Th9-inducing conditions. (A) Schematic diagram of the enhancer region of the PU.1 gene located 17 kb upstream of the PU.1 transcription start site. (B-D) Naive cord blood CD4 T cells or adult naive and memory CD4 T cells were stimulated with anti-CD3/28 in the presence of the indicated cytokines. After fixation and DNA shedding, DNA fragments were immunoprecipitated with anti-H3K4me3, anti-H3ac, or anti-H3K27me3 Abs. Eluted DNA was analyzed by quantitative PCR using primers spanning the promoter region of PU.1 (B), the PU.1 upstream enhancer region (C), and the promoter region of IRF4 (D). The mean ± SEM of at least 3 independent experiments with cells from different donors is shown. *P < .05; **P < .01; ***P < .001 by Student t test.
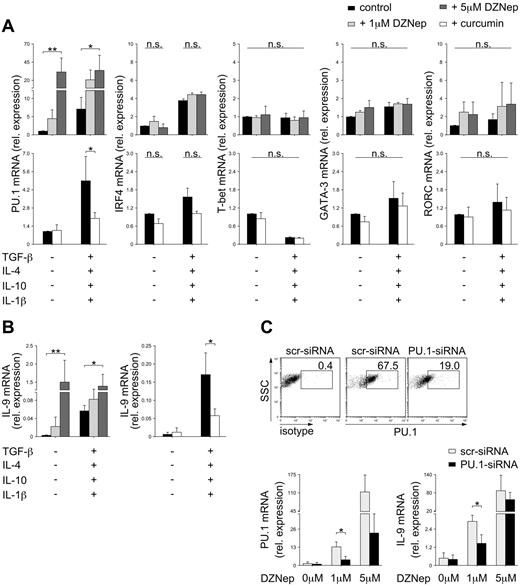
To confirm the control of PU.1 expression by dynamic histone modifications, we inhibited key enzymes governing histone modifications during T-cell differentiation. We activated neonatal CD4 T cells in the presence of DZNep, a specific inhibitor of H3K27 methyltransferases,25 or curcumin, the specific inhibitor of histone acetyltransferases.26 Inhibition of H3K27me3 markedly elevated the levels of PU.1 in a dose-dependent manner even in the absence of Th9-inducing cytokines, whereas inhibition of histone acetyltransferases resulted in significantly decreased expression of PU.1 (Figure 5A). In agreement with the selective control of the PU.1 promoter by histone modifications (Figure 1), the expression of other key transcription factors in Th-cell differentiation, IRF4, T-bet, GATA-3, and RORC, was unaffected by DZNep and curcumin (Figure 5A). Consistent with their effect on PU.1 expression, DZNep and curcumin also modulated the ability of stimulated cells to differentiate and to express IL-9 (Figure 5B). Whereas DZNep facilitated IL-9 production by CD4 T cells, even under Th0-inducing conditions, in a dose-dependent manner, curcumin inhibited IL-9 production despite the presence of cytokines that normally enabled maximum IL-9 secretion. When PU.1 expression was inhibited by siRNAs in DZNep-treated cultures, a significant reduction in IL-9 mRNA production was observed (Figure 5C), supporting a decisive role of PU.1 and not IRF4 in the epigenetic regulation of Th9 development.
Modulation of histone modifications by specific inhibitors. Naive cord blood CD4 T cells were stimulated with anti-CD3/28 in the presence of the indicated cytokines and the presence or absence of increasing concentrations of DZNep or curcumin. (A) Expression of PU.1, IRF4, T-bet, GATA-3, and RORC mRNA were normalized for the expression of cyclophilin. Results are shown in relation to mRNA expression in untreated controls. (B) Expression of IL-9 mRNA was normalized for the expression of cyclophilin. (C) In addition to DZNep treatment, cells were transfected with control, scrambled (scr), or PU.1-specific siRNAs. PU.1 protein expression was assessed by flow cytometry 5 days after anti-CD3/CD28 stimulation. PU.1 and IL-9 mRNA expression were assessed by PCR. mRNA levels were normalized to cyclophilin. The mean ± SEM of at least 3 independent experiments with cells from different donors is shown. *P < .05; **P < .01; ***P < .001; n.s. indicates not significant by Student t test.
Modulation of histone modifications by specific inhibitors. Naive cord blood CD4 T cells were stimulated with anti-CD3/28 in the presence of the indicated cytokines and the presence or absence of increasing concentrations of DZNep or curcumin. (A) Expression of PU.1, IRF4, T-bet, GATA-3, and RORC mRNA were normalized for the expression of cyclophilin. Results are shown in relation to mRNA expression in untreated controls. (B) Expression of IL-9 mRNA was normalized for the expression of cyclophilin. (C) In addition to DZNep treatment, cells were transfected with control, scrambled (scr), or PU.1-specific siRNAs. PU.1 protein expression was assessed by flow cytometry 5 days after anti-CD3/CD28 stimulation. PU.1 and IL-9 mRNA expression were assessed by PCR. mRNA levels were normalized to cyclophilin. The mean ± SEM of at least 3 independent experiments with cells from different donors is shown. *P < .05; **P < .01; ***P < .001; n.s. indicates not significant by Student t test.
Homeostatic proliferation of CD4 T cells in the periphery determines the selective regulation of the PU.1 promoter
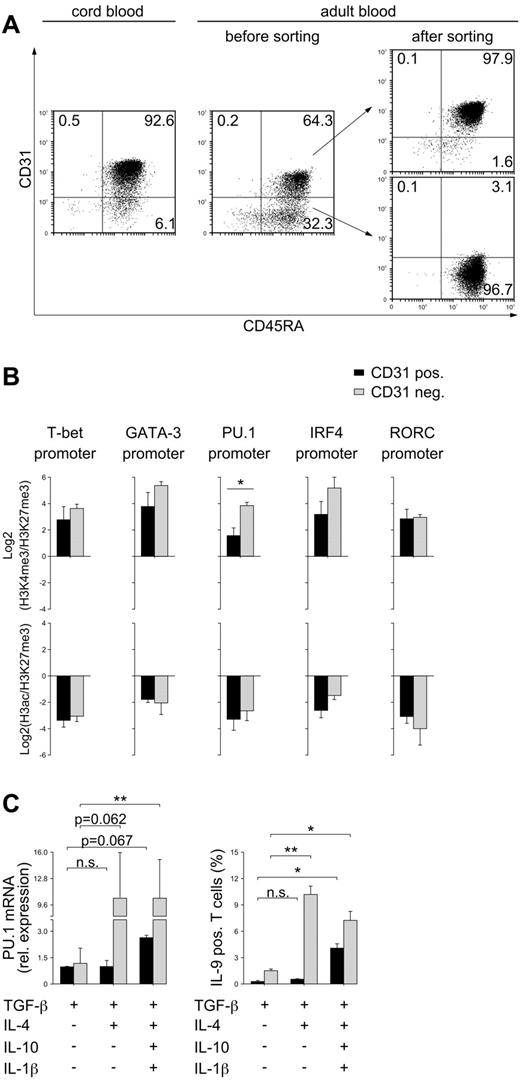
To further clarify whether the histone-modification pattern of the PU.1 promoter is a matter of T-cell aging, we analyzed histone-modification pattern, PU.1 expression, and Th9-cell differentiation in CD31+ and CD31− CD4 T-cell subsets. CD31+ CD4 T cells represent a T-cell population that has recently emigrated from the thymus, recent thymic emigrants (RTEs).27,28 Neonatal naive CD4 T cells contain high numbers of RTEs and are predominantly CD31+ (Figure 6A). Adult naive CD4 T cells represent a mixed population consisting of CD31+ RTEs and peripherally homeostatic proliferating CD31− cells (Figure 6A). Of the T-bet, GATA-3, PU.1, IRF4, and RORC promoters, only the PU.1 promoter demonstrated a significantly different histone-modification pattern between CD31+ and CD31− cells (Figure 6B and supplemental Figure 6). When PU.1 expression and Th9-cell differentiation in response to intensified stimulation were analyzed, profound differences between these T-cell subsets were found (Figure 6C). Whereas, similar to neonatal naive CD4 T cells, CD31+ CD4 T cells initiated PU.1 expression and Th9-cell differentiation first if 4 cytokines were present, CD31− CD4 T cells required just TGF-β and IL-4 for successful PU.1 mRNA up-regulation and Th9-cell development. Therefore, the observed tight epigenetic regulation of PU.1 expression and Th9-cell development from neonatal naive through adult naive and memory CD4 T cells seems to be attributable to the alterations in the PU.1 promoter acquired during homeostatic proliferation.
PU.1 expression and Th9-cell development in CD31+ RTEs. (A) Naive CD4 T cells were purified from cord blood or from peripheral blood of adult subjects and stained for CD31. Adult naive CD4 T cells were sorted into CD31+ and CD31− subpopulations. (B) CD31+ and CD31− cells were processed for ChIP analysis with anti-H3K4me3, anti-H3ac, or anti-H3K27me3 Abs. Eluted DNA was analyzed by quantitative PCR using primers spanning the promoter region of PU.1, T-bet, GATA-3, IRF4, and RORC. The Log2 of the ratio of relative amounts of DNA precipitated with anti-H3K4me3 to anti-H3K27me3 (top panel) and with anti-H3ac to anti-H3K27me3 (bottom panel) are shown. (C) Cells were stimulated with anti-CD3/28 in the presence of the indicated cytokines. Expression of PU.1 mRNA was assessed after additional stimulation of cells with anti-CD3 for 24 hours. mRNA levels were normalized to cyclophilin and are presented relative to expression in CD31+ cells differentiated in the presence of TGF-β only. Frequencies of IL-9–producing cells were assessed by flow cytometry after 5 days of culture and restimulation with phorbol myristate acetate and ionomycin. The mean ± SEM of at least 3 independent experiments with cells from different donors is demonstrated. *P < .05; **P < .01; n.s. indicates not significant by Student t test.
PU.1 expression and Th9-cell development in CD31+ RTEs. (A) Naive CD4 T cells were purified from cord blood or from peripheral blood of adult subjects and stained for CD31. Adult naive CD4 T cells were sorted into CD31+ and CD31− subpopulations. (B) CD31+ and CD31− cells were processed for ChIP analysis with anti-H3K4me3, anti-H3ac, or anti-H3K27me3 Abs. Eluted DNA was analyzed by quantitative PCR using primers spanning the promoter region of PU.1, T-bet, GATA-3, IRF4, and RORC. The Log2 of the ratio of relative amounts of DNA precipitated with anti-H3K4me3 to anti-H3K27me3 (top panel) and with anti-H3ac to anti-H3K27me3 (bottom panel) are shown. (C) Cells were stimulated with anti-CD3/28 in the presence of the indicated cytokines. Expression of PU.1 mRNA was assessed after additional stimulation of cells with anti-CD3 for 24 hours. mRNA levels were normalized to cyclophilin and are presented relative to expression in CD31+ cells differentiated in the presence of TGF-β only. Frequencies of IL-9–producing cells were assessed by flow cytometry after 5 days of culture and restimulation with phorbol myristate acetate and ionomycin. The mean ± SEM of at least 3 independent experiments with cells from different donors is demonstrated. *P < .05; **P < .01; n.s. indicates not significant by Student t test.
Th9 cells from cord blood do not coproduce other cytokines
The aforementioned findings identified the concerted action of TGF-β, IL-4, IL-10, and IL-1β as a prerequisite for Th9-cell development from neonatal naive CD4 T cells. We sought to characterize the phenotype of CD4 effector cells induced in response to this cytokine combination. Th9 cells generated from naive cord blood CD4 T cells produced IL-9 almost exclusively (Figure 7A). Only a minor frequency of cells producing IFN-γ or IL-17 but no IL-9 was detected in neonatal naive CD4 T cells primed under Th9-inducing conditions. In contrast, naive CD4 T cells from the peripheral blood of adult subjects cultured under the same conditions contained substantial numbers of IFN-γ– and IL-17–producing Th9 cells and cells producing only IL-4, IL-5, IFN-γ, or IL-17. The numbers of cells producing cytokines other than IL-9 after “Th9-skewing” further increased in memory CD4 T cells. In memory T cells, 40.0% and 13.4% of Th9 cells were able to coproduce IFN-γ and IL-17, respectively, and 1.0%, 1.1%, 19.9%, and 9.7% of the total population produced only IL-4, IL-5, IFN-γ, or IL-17, respectively (Figure 7A). Consistently, a gradual increase in the expression of the Th1, Th2, and Th17 lineage-specific transcription factors, T-bet, GATA-3, and RORC from neonatal naive through adult naive and memory CD4 T cells in response to Th9-differentiating conditions could be detected (Figure 7B-C).
Analysis of cytokine profiles of Th9 cells developed in response to TGF-β, IL-4, IL-10, and IL-1β. Neonatal naive CD4 T cells from cord blood or adult peripheral blood naive and memory CD4 T cells were stimulated with anti-CD3/28 in the presence of TGF-β, IL-4, IL-10, and IL-1β for 5 days. (A) Cytoplasmic IL-4, IL-5, IFN-γ, and IL-17 were assessed by intracellular flow cytometry. Numbers indicate the percentage of cells in the respective quadrants within viable lymphocytes. Findings from 1 of 4 independent experiments with cells from different donors are shown. (B) Flow cytometric analysis of cytoplasmic IL-9 and nuclear T-bet or GATA-3 expression. Findings from 1 of 3 independent experiments with cells from different donors are shown. (C) Quantitative PCR analysis of RORC gene expression; results were normalized to cyclophilin. The mean ± SEM of 3 independent experiments with cells from different donors is demonstrated.
Analysis of cytokine profiles of Th9 cells developed in response to TGF-β, IL-4, IL-10, and IL-1β. Neonatal naive CD4 T cells from cord blood or adult peripheral blood naive and memory CD4 T cells were stimulated with anti-CD3/28 in the presence of TGF-β, IL-4, IL-10, and IL-1β for 5 days. (A) Cytoplasmic IL-4, IL-5, IFN-γ, and IL-17 were assessed by intracellular flow cytometry. Numbers indicate the percentage of cells in the respective quadrants within viable lymphocytes. Findings from 1 of 4 independent experiments with cells from different donors are shown. (B) Flow cytometric analysis of cytoplasmic IL-9 and nuclear T-bet or GATA-3 expression. Findings from 1 of 3 independent experiments with cells from different donors are shown. (C) Quantitative PCR analysis of RORC gene expression; results were normalized to cyclophilin. The mean ± SEM of 3 independent experiments with cells from different donors is demonstrated.
Discussion
In the present study, we analyzed histone modifications in CD4 T-cell fate-controlling genes in T cells of different ages and/or differentiation status. Of the promoter regions for master transcription factors of T-cell differentiation, the PU.1 promoter was uniquely characterized in naive cells by a striking predominance of the repressive H3K27me3 over the permissive H3K4me3 and H3ac histone modifications. These histone modifications resulted in restrictive PU.1 expression and an associated reduced Th9-cell differentiation from naive T cells. The inaccessibility of the PU.1 promoter gradually decreases from neonatal through adult naive to memory T cells, paralleled by reduced requirements for Th9-cell differentiation. Therefore, the PU.1 promoter represents a gene expression is controlled during aging-related T-cell maturation and differentiation by distinct epigenetic modifications.
Memory T cells are distinct from antigen-inexperienced naive T cells. Whereas naive T cells respond to antigenic stimulation only after a lag period, memory T cells display an enhanced signaling sensitivity and are capable of rapid cytokine production and more robust effector functions. The maturation process from naive to memory T cells is supported by alterations in histone modifications such as acetylation and methylation.4 Histone modifications observed in the PU.1 promoter in the present study reflect the concept of a developmentally related acquisition of epigenetic memory. Both permissive and repressive marks were found in the PU.1 promoter in naive and memory CD4 T cells. However, in memory T cells, the number of repressive H3K27me3 modifications was considerably lower compared with adult naive T cells and even lower in neonatal naive CD4 T cells. This observation is consistent with a reported profound presence of H3K27me3 in several chromosomal regions in naive CD8 T cells.29 Furthermore, the ratio of the permissive to repressive H3K27me3 modifications in the PU.1 promoter was higher in memory than in either naive CD4 T-cell population. The extent of the H3 histone acetylation that has been linked to transcriptional promoter activity3,4 was almost equal in all analyzed resting CD4 T-cell populations. Because gene expression is dependent not only on the presence or absence of the permissive and repressive modifications, but also on their relative levels,11 it is reasonable to assume that transcription of PU.1 can be initiated much faster in memory than in naive CD4 T cells. Consistent with this assumption, PU.1 mRNA expression in CD4 memory T cells was detected after costimulation with TGF-β alone, whereas additional cytokine signals were required in naive CD4 T cells. The initiation of PU.1 expression was associated with further chromatin modifications capable of fostering the opening of the PU.1 promoter. The level of H3 acetylation was at least 10-fold increased in activated cells compared with their resting state. A similarly increased acetylation status has been observed for IL-4 and IFN-γ promoters in effector memory compared with central memory CD4 T cells.8 Finally, in all 3 analyzed CD4 T-cell populations, neonatal naive, adult naive and adult memory, the negative ratio between the permissive and the repressive histone modifications was reversed under Th-cell–polarizing conditions, resulting in the induction of PU.1 expression. The reversion of the H3ac/H3K27me3 and H3K4me3/H3K27me3 ratios appeared in neonatal naive CD4 T cells only in response to stimulation by all 4 cytokines, in adult naive CD4 T cells in response to 2 cytokines, and in memory CD4 T cells in response to TGF-β alone. Remarkably however, the dynamic epigenetic control from naive to memory CD4 T cells was a unique feature of the PU.1 promoter. Whereas promoter regions of the other key transcription factors in Th-cell differentiation, T-bet, GATA-3, IRF4, and RORC, showed both permissive and repressive modifications, these patterns did not differ between the naive and memory cell populations. Consistent with this, Th1, Th2, and Th17 cell differentiation was comparable between both naive cell populations with a further enhancement in memory T cells.
Bivalent histone modifications in GATA-3 and T-bet promoters have been described previously in mouse naive T cells and are maintained in later stages during T-effector cell maturation, where they are thought to be associated with the rapid switch of the phenotype.30 Our data do not exclude that epigenetic histone modifications in regions or genes distinct from the promoter regions analyzed herein or alternative molecular mechanisms such as DNA methylation determine the Th1, Th2, and Th17 epigenetic memory. The unique control of the PU.1 promoter however was confirmed by the analysis of pharmacologic agents targeting histone-modifying enzymes, because these inhibitors affected PU.1 expression exclusively.
Interesting in this context is the observation that of the 2 transcription factors reported to be necessary for successful Th9-cell development, only PU.1 and not IRF4 undergoes strong epigenetic control during different maturation stages of CD4 T cells. PU.1 and IRF4 have been shown to form ternary complexes together with DNA to initiate gene transcription.31 However, it seems that different signals are required for their recruitment to the target gene promoters. Whereas in Th9-cell development, PU.1 results from TGF-β signaling, IRF4 appears in response to IL-4.32 In light of this, different epigenetic control mechanisms of these transcription factors are conceivable. Moreover, in Th-cell differentiation, in addition to Th9-cell development, IRF4 is also involved in the differentiation of Th2 and Th17 cells.33,34 In Th2-cell differentiation, IRF4 has been shown to interact directly with NFATc2 and modulate IL-4 gene expression.35 In Th17-cell development, IRF4 is essential for the stabilization of the Th17 phenotype.34 The important role of IRF4 in Th-cell fate decisions makes it unlikely that IRF4 is controlled by selective epigenetic mechanisms specific for one Th subset (eg, Th9 cells) and thus argues for different epigenetic control of PU.1 and IRF4.
The selective epigenetic inactivation of the PU.1 promoter associated with diminished Th9-cell differentiation and IL-9 production by naive CD4 T cells suggests a special physiologic role for IL-9. Initially, IL-9 was described as a T-cell growth factor,36,37 whereas later studies have shown that mast cells are the main target of IL-9.38 IL-9 has been reported primarily to promote the expansion of mast-cell populations and to be involved in parasite expulsion.39,40 However, once deregulated, IL-9 might be involved in the development of tissue inflammation and autoimmune disorders, as shown in several animal models of asthma, allergy, colitis, diabetes, and experimental autoimmune encephalomyelitis.19,41-44 IL-9 has been suggested to enhance the proinflammatory, Th17-cell–driven immune response by augmenting Th17-cell development42,44,45 or by recruiting mast cells to the sites of inflammation and triggering their effector functions (eg, release of cytokines, histamine, and proteases).46,47 Remarkably, however, IL-9 has also been implicated in preventing allograft rejection by modulating T-cell tolerance via the same effector mechanisms, which involve mast-cell recruitment and activation.48 Under specific conditions, IL-9 seems to promote the proliferation and expansion of T lymphocytes. IL-9 also stimulates the in vitro growth of mouse thymic lymphomas,49 and IL-9–transgenic mice spontaneously develop thymic lymphomas.45 Interestingly, injection of a carcinogen with a thymic tropism into IL-9–transgenic animals increases the tumor incidence. Moreover, overexpression of NPM-ALK, an oncogenic translocation present in approximately 12.5% of non-Hodgkin lymphomas, increases the incidence of the lymphomas in IL-9–transgenic mice even further.50
The rather complex role of IL-9 and Th9 cells in the outcome of specific immune responses may necessitate a tight physiologic control of its expression during aging and the maturation of the immune system. Therefore, it is conceivable that IL-9 promotes tumorigenesis in situations of lymphocyte hyperproliferation, such as the developing thymus in neonates and infants, if presented strongly. Conversely, an eased IL-9 expression in adults might explain its proposed role in autoimmune disorders. It might therefore be beneficial for the development of the immune system to restrict IL-9 expression. Although further studies are necessary to delineate the precise biologic role for different requirements of IL-9 induction, our present data provide a compelling epigenetic mechanism by which Th9-cell development is controlled uniquely during the life of a CD4 T cell.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank C. Schnabel (Division of Rheumatology, University of Munich) for expert technical assistance; Dr P. E. Lipsky (formerly at NIAMS, NIH) for critically reading the manuscript and helpful advice; Dr P. Becker, Dr S. Dulev, and A. Mitterweger (Division of Molecular Biology, Adolf Butenandt Institute, Uni-versity of Munich) for help in performing the ChIP assays; Dr J. Jückstock, Dr L. Rabe, and Dr S. Wagner (Klinik für Frauenheilkunde, University of Munich) for providing cord blood; and Dr M. H. Kaplan (Indiana University School of Medicine) for helpful advice on PU.1 analysis.
This work was supported by the Deutsche Forschungsgemeinschaft (grants SK59/4-1, Schu 786/2-5, and Schu 786/8-1 and training grant GK1202 [oligonucleotides project E2]); the Sonderforschungsbereich 571 (autoimmunity project D9); and the Verbundanträge ArthroMark (projects 1 and 7, OIEC100913) and Impam (project 10, OIEC1008H), Federal Ministry of Education and Research.
Authorship
Contribution: A.R. designed and performed the research, analyzed the data, and wrote the manuscript; D.D. performed the research and analyzed the data; J.L. analyzed the data; and H.S.-K. and A.S. designed the research, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alla Skapenko, Division of Rheumatology, University of Munich, Pettenkoferstr 8a, 80336 Munich, Germany; e-mail: alla.skapenko@med.uni-muenchen.de.