Abstract
Gaucher disease (GD), an inherited macrophage glycosphingolipidosis, manifests with an extraordinary variety of phenotypes that show imperfect correlation with mutations in the GBA gene. In addition to the classic manifestations, patients suffer from increased susceptibility to hematologic and nonhematologic malignancies. The mechanism(s) underlying malignancy in GD is not known, but is postulated to be secondary to macrophage dysfunction and immune dysregulation arising from lysosomal accumulation of glucocerebroside. However, there is weak correlation between GD/cancer phenotype and the systemic burden of glucocerebroside-laden macrophages. Therefore, we hypothesized that genetic modifier(s) may underlie the GD/cancer phenotype. In the present study, the genetic basis of GD/T-cell acute lymphoblastic lymphoma in 2 affected siblings was deciphered through genomic analysis. GBA gene sequencing revealed homozygosity for a novel mutation, D137N. Whole-exome capture and massively parallel sequencing combined with homozygosity mapping identified a homozygous novel mutation in the MSH6 gene that leads to constitutional mismatch repair deficiency syndrome and increased cancer risk. Enzyme studies demonstrated that the D137N mutation in GBA is a pathogenic mutation, and immunohistochemistry confirmed the absence of the MSH6 protein. Therefore, precise phenotype annotation followed by individual genome analysis has the potential to identify genetic modifiers of GD, facilitate personalized management, and provide novel insights into disease pathophysiology.
Introduction
Gaucher disease (GD), an autosomal recessive inborn error of metabolism, is the most prevalent lysosomal storage disorder. It is caused by mutations in the GBA gene, which results in defective acid β-glucosidase and progressive accumulation of glucocerebroside in the lysosomes of mononuclear phagocytes. The result is systemic accumulation of tissue macrophages engorged with lysosomal glucocerebroside (the eponymous Gaucher cells).1 The phenotypes of GD resulting from biallelic mutations in GBA and defective acid β-glucosidase include variable combinations of hepatosplenomegaly, skeletal disease, cytopenias, and a complex pattern of bone marrow (BM) involvement. In the rare neuronopathic types 2 and 3 forms of the disease, patients also develop chronic neurodegenerative disease.2 Nonneuronopathic type 1 GD (GD1) is the most common form, accounting for approximately 90% of currently known GD patients. GD1 is treated effectively with macrophage-targeted enzyme replacement therapy (ERT) with recombinant glucocerebrosidase.3
GD1 manifests with extraordinarily diverse phenotypes among patients harboring identical GBA mutations and even among affected sibling pairs.4,5 Phenotype diversity manifests as variations in the overall severity of the disease, as well as in the pattern of organ involvement (eg, visceral/hematologic vs skeletal vs pulmonary involvement).2,6-8 Another category of phenotype variation in GD1 is exemplified by the occurrence of unusual manifestations such as Parkinson disease,9 pulmonary hypertension,10 and cancers,11-16 which, despite their potentially life-threatening nature, appear to show imperfect correlation with the overall severity of the classic manifestations.11,12 These observations are consistent with the notion that modifier genes may underlie these phenotypes.
There is an increased risk of hematologic and nonhematologic malignancies and recurrent malignancies in individual patients in GD1.11-16 However, the pathophysiologic basis of increased cancer risk is not known. Several mechanisms have been proposed to be involved in the pathogenesis of malignancy in GD1, including alternatively activated macrophages,17 immune dysregulation,18-20 splenectomy,12 hyperferritinemia,21 lysosomal dysfunction, and endoplasmic reticulum stress.22 Moreover, in vitro studies have suggested that the cellular accumulation of glucocerebroside itself is conducive to cancer progression. Conversely, reducing the cellular burden of glucocerebroside via chemical inhibition of its synthesis has been proposed as cancer adjuvant therapy.23 Clarification of the pathways underlying predisposition to cancer in GD1 may provide novel insights into disease mechanisms and enhance personalized management of individual patients.
To investigate the hypothesis that genetic modifiers underlie phenotypic diversity in GD, we focused on a pair of siblings with a remarkable concordance of GD1 and T-cell acute lymphoblastic lymphoma (T-LBL). Genome analysis through exome capture and massively parallel sequencing revealed a novel GBA mutation and a genetic modifier underlying the GD/cancer phenotype represented by biallelic mutations in the MSH6 gene.
Methods
Patients
This research was approved by the Yale University School of Medicine Human Investigation Committee. All participants gave written informed consent to take part in the study in accordance with the Declaration of Helsinki.
Genotype/phenotype analysis of GD
GD was confirmed via demonstration of > 95% reduction of acid β-glucosidase activity in peripheral blood leukocytes. The GBA gene was analyzed by meta-PCR and Sanger sequencing of the coding regions. The clinical phenotype was characterized by volumetric measurements of the liver and spleen, by assessment of skeletal/BM involvement, and by serum biomarkers to assess the total body burden of Gaucher cells, as described previously.11
DNA preparation, whole-exome capture, and massively parallel sequencing
Genomic DNA was extracted from peripheral blood using the Puregene genomic DNA purification kit (Gentra Systems) according to the manufacturer's protocol.
Targeted capture of human whole exome using the solution-based 2.1M NimbleGen Exome array followed by Illumina Genome Analyzer IIx sequencing was performed as described previously.24 One lane of paired-end sequence reads 74 bp in length was generated per sample following the manufacturer's protocol. Subsequent image analysis and base calling was done with Illumina Pipeline Version 1.5 with default settings at the Yale Center for Genome Analysis. Genomic sequences were mapped to the human genome (hg18) using MAC Version 0.7.1 and BWA Version 0.5.0 software.24,25 Reads aligned to the targeted sequences were retrieved and subjected to further analyses using Perl scripts. Variant calling was performed using SAMtools Version 0.1.7 software25 and variants were annotated using an in-house Perl script. dbSNP (build 132) and 1000 genomes database (release May 11, 2011) were used to test the novelty of variations.
Whole-genome genotyping and identification of loss-of-heterozygosity intervals
Single-nucleotide polymorphism (SNP) genotyping of genomic DNAs was performed on the Illumina Human Omni-1M quad DNA Analysis BeadChip. The image data were analyzed and SNP genotypes were called using Beadstudio Version 3.2 software (Illumina). Sample processing and labeling were performed following the manufacturer's protocols. Plink Version 1.07 software was used to identify homozygous intervals. A sliding window of 50 SNPs was used on the tag SNPs and included no more than 1 possible heterozygous genotype. Resulting intervals had to have met the limit of at least 100 SNPs and 1 Mb in size.
Orthologs of GBA
Full-length orthologous protein sequences from vertebrate and invertebrate species were extracted from GenBank. GBA orthologs were confirmed based on database identity of annotation. If an ortholog could not be identified, the closest paralog (ie, top “hit” of a BLAST search of the respective proteome) was studied. Protein sequences were aligned using the ClustalW algorithm. GenBank accession numbers are as follows: human GBA, NP_000148.2; mouse acid β-glucosidase, EDL15229; frog MGC84284 protein, NP_001087746.1; zebrafish glucosylceramidase-like, XP_687471.3; Ciona intestinalis acid β-glucosidase, XP_002129322.1; and Caenorhabditis elegans F11E6.1d, CCD31056.1.
MMR investigations
Immunohistochemistry for mismatch repair (MMR) proteins encoded by 4 MMR genes (MLH1, MSH2, PMS2, and MSH6) was performed on the mediastinal mass from the proband. Gene sequencing of the MSH6 gene was performed on BM from the proband and peripheral blood from the sibling and both parents (Associated Regional and University Pathologists Laboratories, University of Utah, Salt Lake City, UT).
NF1 investigation
To investigate the prior clinical diagnosis of NF1 in the index case and her sibling, NF1 gene sequencing was performed on DNA isolated from the BM and peripheral blood, respectively (Medical Genomics Laboratory, University of Alabama, Birmingham, AL). In addition, NF1 gene analysis was performed on DNA isolated from melanocytes derived from a skin biopsy taken from a café-au-lait macule (CALM) in patient 2 (Medical Genomics Laboratory, University of Alabama). Finally, confirmatory NF1 gene sequencing (of DNA extracted from skin fibroblasts) was repeated in patient 2 at a second outside laboratory (Boston University School of Medicine Center for Human Genetics, Boston, MA).
Mutant D137N GBA characterization
To elucidate the precise intracellular distribution of the active GBA molecules, Inhibody probes were used to allow visualization of the active GBA molecules in situ in living cells.26 Fibroblast cultures derived from 2-mm punch-skin biopsies from family members were grown to confluence, labeled with inhibody, and the outcomes analyzed on gel and by FACS, as described previously.26 In situ GBA activity was determined using the artificial substrates 4MU-β-D-glucopyranoside and fluorescein di-β-D-galactopyranoside as described previously.26
Mutant D137N GBA-docking experiments
To determine its functional consequence, D137N mutation was modeled on the crystal structure (PDB 2V3E) of wild-type GBA1 enzyme. GBA1 crystal structures were cleaned, water and ions removed (PyMOL), structure energy minimized (Charmming), and flexibility set for side chains and protonation of nonpolar residues (AutoDock Tools). Structures of wild-type GBA1 and D137N mutant enzyme were obtained by in silico folding with algorithms of PHYRE2, CPH models, and SWISS-MODEL, and treated equally to obtain crystal structures. The ligand molecule 4MU-β-D-glucopyranoside (4MU-Glc) was created and saved as energy minimized PDB files (MarvinDraw Version 5.5.1.0), with the flexibility and protonation status set (AutoDock Tools Version 1.54). Docking of ligand was performed 9 times within a 100 Å-box encompassing the entire protein structure, with exhaustiveness set to 12 (AutoDock Vina). Results were analyzed and rendered in PyMOL.
Results
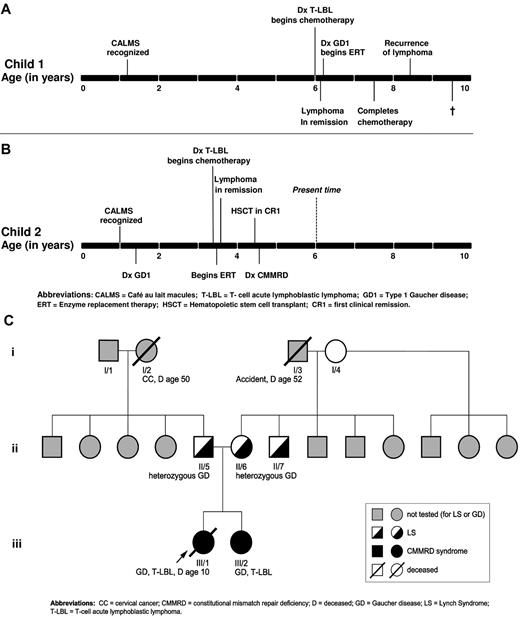
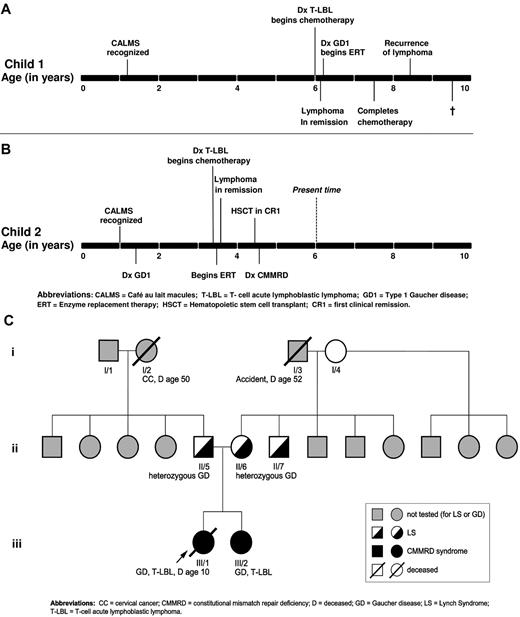
The clinical and genetic characteristics of the proband (patient 1) and her sibling (patient 2) are summarized in Table 1. The timeline of the natural history in each patient is depicted in Figure 1A-B.
Gross phenotype of T-LBL and GD in the proband and her affected sister. (A) Biopsy of the mediastinal mass showing a T-LBL with diffuse infiltration of the soft tissues by a population of monomorphic lymphoid cells with hyperchromatic nuclei and scant cytoplasm. Immunophenotyping revealed the tumor cells to be positive for CD34 and negative for CD4 and CD8. (B) Chest radiograph of patient 1 demonstrating mediastinal mass at time of initial diagnosis of T-LBL. (C) BM biopsy performed several months later showing the presence of many large macrophages with pale eosinophilic cytoplasm and typical wrinkled tissue paper appearance typical of GD. (D) Electron microscopy performed on the BM shows many Gaucher cells with cytoplasmic striations that represent abundant tubular bodies. (E) Biopsy of the mediastinal mass in patient 2 showing features of T-LBL. (F) BM biopsy from patient 2 showing the presence of many Gaucher cells similar to her sibling.
Gross phenotype of T-LBL and GD in the proband and her affected sister. (A) Biopsy of the mediastinal mass showing a T-LBL with diffuse infiltration of the soft tissues by a population of monomorphic lymphoid cells with hyperchromatic nuclei and scant cytoplasm. Immunophenotyping revealed the tumor cells to be positive for CD34 and negative for CD4 and CD8. (B) Chest radiograph of patient 1 demonstrating mediastinal mass at time of initial diagnosis of T-LBL. (C) BM biopsy performed several months later showing the presence of many large macrophages with pale eosinophilic cytoplasm and typical wrinkled tissue paper appearance typical of GD. (D) Electron microscopy performed on the BM shows many Gaucher cells with cytoplasmic striations that represent abundant tubular bodies. (E) Biopsy of the mediastinal mass in patient 2 showing features of T-LBL. (F) BM biopsy from patient 2 showing the presence of many Gaucher cells similar to her sibling.
Patient 1 was first evaluated at age 15 months for multiple (> 6) CALMs and axillary freckling, consistent with the diagnosis of NF1. She remained in good health until 6 years of age, when she presented with easy fatigability, right cervical lymphadenopathy, and dyspnea. The physical examination was significant for right supraclavicular lymphadenopathy and moderate hepatosplenomegaly. She had multiple small CALMs and axillary freckling. Her WBC count was 4.0 × 109/L, hemoglobin was 10.2 g/dL, and platelets were 157 × 109/L. Radiology revealed a solid homogenous anterior mediastinal mass (4.8 × 6.6 cm) with areas of necrosis, cervical lymphadenopathy, and splenomegaly (Figure 1B). An open biopsy of the anterior mediastinal mass was performed. Flow cytometry revealed an immunophenotype of CD34+CD4−CD8−. The histological and immunohistochemical findings were consistent with T-LBL (Figure 1B). Bilateral BM biopsies of her iliac crests did not show any evidence of infiltration by lymphoma, but revealed abundant sheets of CD68+ cells with eosinophilic cytoplasm with a “wrinkled paper” appearance, which is considered to represent pseudo-Gaucher cells associated with lymphoma7 (Figure 1C-D).
Patient 1 achieved remission on Children's Oncology Group protocol A5971 for T-LBL. However, the patient had persistent hepatosplenomegaly that prompted further evaluation (Table 1). Acid β-glucosidase activity in peripheral blood leukocytes on 2 separate occasions was almost undetectable: 0.01 units/1010 cells and 0 units/1010 cells (normal range, 0.080-0.235 units/1010), which was diagnostic of GD. Serum biomarkers of GD27 were elevated: chitotriosidase activity was elevated 104-fold (6147 nmol/h/mL) and CCL18 approximately 20-fold (749 ng/mL). Serum ferritin was elevated 9-fold at 2710 ng/mL, a common finding in untreated GD.21 Allele-specific PCR for the common mutations in the GBA gene was negative. MRI revealed a liver volume of 1159 cc (1.9 × normal) and a spleen volume of 308 cc (6.3 × normal), bilateral Erlenmeyer-flask deformity, and cellular BM but no avascular osteonecrosis. BM biopsies were reexamined, and in the context of additional clinical and laboratory data, were determined to represent authentic Gaucher cells (Figure 1C-D). Sequencing of the entire coding region of the GBA gene revealed homozygous D137N mutation in the patient. The patient was started on imiglucerase enzyme replacement therapy (ERT), which resulted in reversal of organomegaly, amelioration of cytopenia, a growth spurt, and resolution of bone pain. One year after the diagnosis of GD, she completed chemotherapy for lymphoma and was in complete remission. However, 8 months later, she suffered relapse with a large PET-avid mediastinal mass compressing the trachea. The patient's T-LBL did not respond to several courses of chemotherapy/mediastinal radiation and the patient died from disseminated disease. The timeline of the natural history in patient 1 is depicted in Figure 2A.
Evolution of phenotype in the two siblings and family history. (A) Timeline of the natural history in the proband. (B) Timeline of the natural history in the sibling. (C) Pedigree with respect to GD, CMMRD/LS, and malignancy.
Evolution of phenotype in the two siblings and family history. (A) Timeline of the natural history in the proband. (B) Timeline of the natural history in the sibling. (C) Pedigree with respect to GD, CMMRD/LS, and malignancy.
At 12 months of age, patient 2, the younger sister of patient 1, was diagnosed with NF1 according to National Institutes of Health clinical criteria of the presence of CALMs and axillary freckling. After diagnosis of GD1 in her sister, the patient underwent screening at age 18 months (Table 1). GBA gene analysis revealed a D137N homozygous mutation; acid β-glucosidase activity in peripheral blood leukocytes was extremely low at 0.01 units/1010 (normal range, 0.08-0.35 units/1010). The serum biomarkers of GD were elevated: chitotriosidase activity was 1408 nmol/h/mL (28.2 × upper normal limit of normal) and CCL18 was 240 ng/mL (6.9 × normal). However, the patient was asymptomatic with no evidence of bone pain, bone crisis, bruising, cytopenias, or organomegaly. Given the lack of overt clinical manifestations of GD, the patient was followed expectantly for optimal timing to start ERT. The timeline of the natural history in patient 2 is shown in Figure 2B.
At the age of 3.5 years, patient 2 presented with a 2-week history of cough and orthopnea. On physical examination, she had mild splenomegaly and lymphadenopathy, but no hepatomegaly. Blood work revealed mild microcytic anemia, but other cell lines were normal: (hemoglobin, 9.8 g/dL; WBC count, 13.3 × 109/L; platelets, 481 × 109/L, and a normal differential). Chest CT revealed a heterogeneous anterior mediastinal mass with predominantly soft tissue attenuation without calcifications or fatty components and measuring 5 × 10 × 10 cm with tracheal compression. Ultrasound-guided biopsy of the mass revealed monoclonal T cells that were morphologically and immunophenotypically consistent with T-LBL (Figure 1E). Bilateral BM biopsies demonstrated no evidence of lymphoma, but extensive infiltration by Gaucher cells (Figure 1F). Chemotherapy according to Children's Oncology Group protocol A5971 was commenced, and the patient simultaneously began imiglucerase ERT and went into remission after induction. However, because of the unusually aggressive nature of her sister's disease, patient 2 underwent a cord blood transplantation in first clinical remission. There has been no recurrence of lymphoma or GD.
The parents of the affected siblings were of Mexican-American ancestry and originated from the same village; they denied consanguinity and reported a minimal history of cancer in the parents and extended family. The pedigree is depicted in Figure 2C.
Initial genetic analysis
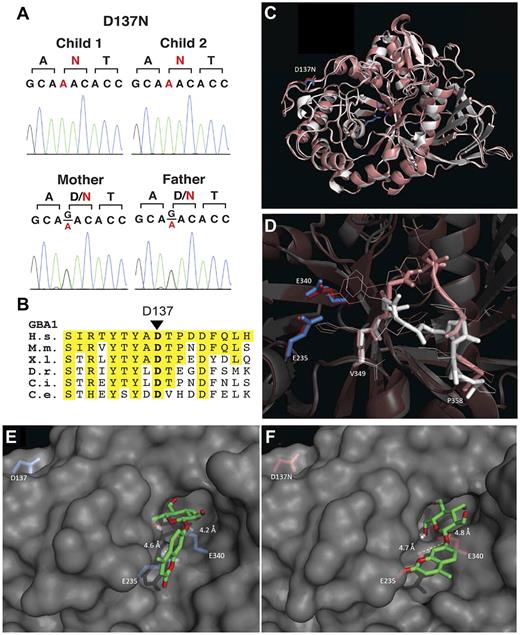
GBA gene analysis revealed a novel mutation, D137N, which was present in homozygous form in both siblings and in heterozygous form in both parents (Figure 3A). Clinical phenotype suggestive of NF1 prompted NF1 gene analysis in the proband and sibling; the result was wild-type sequence (reliability > 97%). Moreover, ophthalmologic evaluation revealed no Lisch nodules or other NF1-associated findings. Therefore, we performed genomic analysis to delineate the genetic basis of the striking concordance of GD and T-LBL in these 2 siblings.
Studies delineating the novel GBA1 D137N mutation. (A) Sanger traces showing heterozygous G > A changes from the parents and homozygous changes from the affected children. (B) Conservation of D137 among orthologs of GBA1. The amino acid segment 129-145 of human GBA1 is shown. Positions identical to human amino acids are highlighted in yellow. H.s. indicates Homo sapiens; M.m., Mus musculus; X.l., Xenopus laevis; D.r., Danio rerio; C.i., Ciona intestinalis; and C.e., C elegans. (C). Overlay of wild-type GBA (2V3E, white) and in silico folded D137N GBA (SWISS-MODEL algorithm, pink) and detailed view of D137N-induced loop change (D). Docking analysis of artificial substrate 4MUGlc on wild-type GBA1 (E) and D137N GBA1 (SWISS-MODEL, F), with distances depicted in Angstroms. Associated energies are depicted in supplemental Figure 3.
Studies delineating the novel GBA1 D137N mutation. (A) Sanger traces showing heterozygous G > A changes from the parents and homozygous changes from the affected children. (B) Conservation of D137 among orthologs of GBA1. The amino acid segment 129-145 of human GBA1 is shown. Positions identical to human amino acids are highlighted in yellow. H.s. indicates Homo sapiens; M.m., Mus musculus; X.l., Xenopus laevis; D.r., Danio rerio; C.i., Ciona intestinalis; and C.e., C elegans. (C). Overlay of wild-type GBA (2V3E, white) and in silico folded D137N GBA (SWISS-MODEL algorithm, pink) and detailed view of D137N-induced loop change (D). Docking analysis of artificial substrate 4MUGlc on wild-type GBA1 (E) and D137N GBA1 (SWISS-MODEL, F), with distances depicted in Angstroms. Associated energies are depicted in supplemental Figure 3.
Molecular genetic analysis and whole-exome capture sequence
The major findings from whole-exome capture sequencing are presented in Table 2. We achieved a mean coverage of 82×, and 96.3% of all targeted bases were read more than 4 times, sufficient to identify novel homozygous and heterozygous variants with high specificity. Comparison of called variants with SNP genotyping data demonstrated 98% of sensitivity and 99.9% of specificity of variant detection (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). We anticipated finding disease-causing mutation(s) that were homozygous by descent, so novel and rare mutations were sought that altered the encoded protein. Several thousand homozygous and heterozygous variations were identified, including nonsynonymous substitutions, synonymous coding variants, canonical splice site variants, and coding region indels. Substitutions at positions that are completely conserved from invertebrates to humans are highly likely to disrupt normal protein function and, therefore, we ranked the novel missense variants by conservation scores to identify the most likely functional mutations. Single-nucleotide variants were annotated for effect on the encoded protein and for conservation by comparison against sequences of 43 vertebrate species and orthologs in the fly and the worm, as described previously.24 We prioritized mutations that introduce truncations of the encoded protein and therefore would be predicted to have functional effects. Finally, databases with exome sequences of large numbers of subjects have become available, providing the ability to distinguish low-frequency alleles descendent from ancient ancestors from de novo or extremely rare mutations introduced recently into the population.
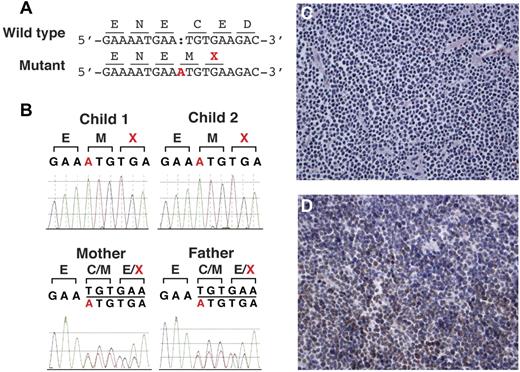
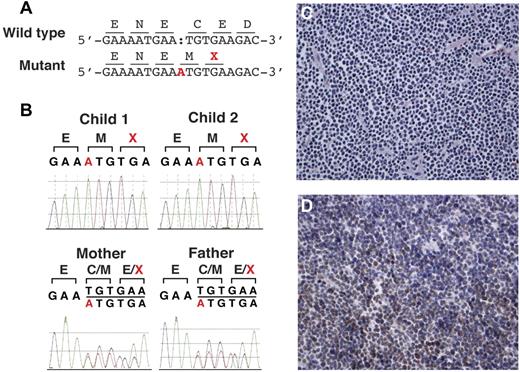
Among the variants that were shared between the 2 affected children and not found in the dbSNP and 1000 genomes, 6 mutations were in homozygous form in the 2 affected patients (Table 2). Two of these 6 mutations were immediately relevant to the phenotypes displayed by the affected patients: a single-base substitution that introduced a missense variant in GBA and a single-base insertion that introduced a frame-shift mutation and premature termination in MSH6 (Table 2). These 2 mutations were not found from 5000 control chromosomes and were confirmed by Sanger sequencing of PCR-amplified segments to be homozygous in both affected children and to be present in heterozygous form in both parents. The GBA gene mutation D176N is depicted in Figure 3A. Alignment of GBA1 protein sequences to orthologs demonstrated that the D176 residue is highly conserved to the worms (Figure 3B). Novel mutation c.3822dupA in the MSH6 gene is depicted in Figure 4A-B. Short-read alignments for GBA and MSH6 mutations are displayed in supplemental Figures 1 and 2. No compound heterozygous mutations were detected.
Studies delineating the homozygous insertion mutation in MSH6. (A) Insertion of single base in mutant allele (A, highlighted in red) causes frame-shift and a premature termination codon (X, highlighted in red). (B) Sanger traces showing homozygous insertion of base A from the affected children and heterozygous insertion from the parents. (C) Immunostaining for MSH6 showing loss of normal nuclear staining in the lymphoma cells. (D). Immunostaining for MLH1 showing normal nuclear staining in tumor cells. The results were similar with MSH2 and PMS2 immunostaining (data not shown).
Studies delineating the homozygous insertion mutation in MSH6. (A) Insertion of single base in mutant allele (A, highlighted in red) causes frame-shift and a premature termination codon (X, highlighted in red). (B) Sanger traces showing homozygous insertion of base A from the affected children and heterozygous insertion from the parents. (C) Immunostaining for MSH6 showing loss of normal nuclear staining in the lymphoma cells. (D). Immunostaining for MLH1 showing normal nuclear staining in tumor cells. The results were similar with MSH2 and PMS2 immunostaining (data not shown).
Table 2 depicts 4 additional homozygous mutations. The variant in MUC1 is in very low PhyloP, implying that the variant is dispensable during evolution. Moreover, the MUC1 gene is expressed specifically in cell types not involved in GD, such as the lung and gastric epithelial cells. The MUC1 homozygous variant is therefore not relevant to the phenotype we were investigating. For the variant in C2orf63, the father is homozygous and it is in very low PhyloP, both of which make this variant very unlikely to be relevant to the phenotypes under investigation. C2orf63 encodes a hypothetical protein with estimated 586 amino acids. No functional information on the protein has been reported. For the 2 variants in MAGEC1, because the parents carry the variants in homozygous status and both variants are seen at approximately 50% frequency in the control population of 5000 chromosomes, we did not consider these variants to be relevant to the pathology of our patients.
Immunohistochemistry and genetic analysis of MSH6
Immunohistochemistry on the mediastinal mass from the proband demonstrated an absence of staining of MSH6 in both the tumor and background nonneoplastic cells (Figure 4C); all other MMR proteins were normal (Figure 4D). Analysis of the MSH6 gene by PCR and sequencing in the proband and the sibling confirmed homozygous pathogenic mutations, c.3822dupA (Figure 4A-B). Heterozygous MSH6 mutations were confirmed in both parents. Loss-of-heterozygosity analysis using SNP genotype data revealed shared loss-of-heterozygosity intervals between the 2 patients, including MSH6 (chr2:47,740k-56,109k), which was not observed in the parents (data not shown). Mutation C.3822dupA in MSH6 represents an insertion frame-shift mutation resulting in premature protein truncation. The insertion variant is located in exon 9 and the frame-shift causes premature termination at position 1276 in a protein of normal length of 1360 amino acids. This variant has not been reported previously. However, several truncating mutations have been reported in cancer patients in close proximity to the mutation identified in our patients (summarized in supplemental Table 2). These considerations are consistent with the pathogenic nature of the C.3822dupA mutation in the MSH6 gene.
Functional analysis of D137N mutant GBA
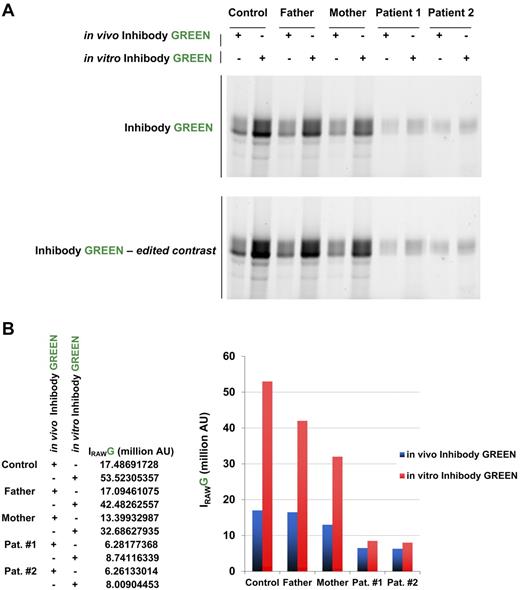
To define the functional consequence of the D137N GBA1 mutation, in situ acid β-glucosidase enzyme activity was determined in vivo (in intact skin fibroblasts) and in vitro (in cell lysates) using the novel inhibody probes.26,27 In contrast to the strikingly low acid β-glucosidase activity measured in peripheral blood leukocytes using the 4MU-Glc substrate (Table 1), functional enzyme activity measured by inhibody probes and fluorescein di-β-D-galactopyranoside substrate in intact fibroblasts was less severely impaired (Figure 5A). In vitro inhibody labeling using skin fibroblasts revealed a marked reduction of active GBA1 in parent and patient fibroblasts compared with control: approximately 70% and 15%, respectively (Figure 5B).
Characteristics of mutant D137N acid β-glucosidase. (A) In vivo labeling of active acid β-glucosidase in intact skin fibroblasts (10nM inhibody, green) compared with total labeling in vitro (cell lysates of untreated cells). (B) Quantification of gels shown in panel A.
Characteristics of mutant D137N acid β-glucosidase. (A) In vivo labeling of active acid β-glucosidase in intact skin fibroblasts (10nM inhibody, green) compared with total labeling in vitro (cell lysates of untreated cells). (B) Quantification of gels shown in panel A.
To assess the possible influence of the D137N substitution on the structure of GBA1, it was modeled into the crystal structure. Although located distant from the active site (Figure 3C), in silico folding predicts an attenuation of the size of the active site because of a shift in a loop directly in front of it (V349-P358, VHWYLDFLAP; Figure 3D). To determine whether the D137N substitution influences the affinity of the enzyme for the artificial substrate 4MU-Glc, we performed molecular docking analyses. In wild-type GBA1, 4MU-Glc can be positioned in close proximity to both the nucleophile (E340) and the acid/base (E235). Calculated energies associated with this interaction are in the range of −8.5 kcal/mol (Figure 3E-F; for energies, see supplemental Figure 5). All proposed models of D137N GBA have lower associated energies: approximately −7 kcal/mol or higher (supplemental Figure 3). These data suggest that the D137N substitution alters the affinity for the substrate, and therefore could have a profound effect on the enzymatic characteristics of D137N GBA1.
Discussion
In the present study, we investigated the molecular genetic basis of a highly concordant phenotype of GD1 and T-LBL in 2 affected siblings. The patients harbored novel homozygous mutations in the GBA1 and MSH6 genes that were responsible for T-LBL and progressive manifestations of GD. Our findings suggest that the MSH6 gene mutation resulted in defective MMR, leading to a vulnerability of tumor suppressor genes and oncogenes to develop mutations and ultimately lymphomagenesis in a cancer-promoting background of systemic macrophage dysfunction and immune dysregulation that exists in GD. These results validate the concept that genetic modifier(s) underlie increased cancer risk in GD and that individual genome analysis is an effective approach for the identification of modifier genes with large effect size in GD. The findings also suggest that the full potential of genomic analysis through exome capture can be harnessed maximally by scrupulous attention to phenotype annotation. For example, because of our patients' CALMs and axillary freckling, initially NF1 was suspected to underlie cancer susceptibility. However, whole-exome and targeted neurofibromin gene sequencing ruled out NF1. In the search for a gene defect underlying familial cancer in this pedigree, among the variants revealed by exome sequencing, the homozygous mutation in MSH6 came into sharp focus because of the presence of CALMs. The central role of the MSH6 gene mutation was further underscored by homozygosity mapping data and its correlation with the clinical phenotype.
Our patients developed T-LBL associated with constitutional MMR deficiency (CMMRD) syndrome due to biallelic germ-line mutations in the MSH6 gene, a member of the MMR family of genes. The MMR system is involved in maintaining genome integrity through the correction of base-base mismatches and insertion/deletion loops that arise during DNA replication.28 The contribution of defective MMR to the development of human cancer is well recognized. Heterozygous inactivating germline mutations in any of the 4 MMR genes (MLH1, MSH2, MSH6, and PMS2) cause the autosomal-dominant adult cancer predisposition syndrome Lynch syndrome (LS) or hereditary nonpolyposis colorectal cancer.29,30 In LS, somatic loss of MMR gene heterozygosity leads to impaired MMR and accumulation of somatic mutations in cancer genes and microsatellites, thereby leading to tumorigenesis.30 There is a paucity of information regarding the phenotype of those rare individuals who harbor biallelic (homozygous or compound heterozygous) MMR mutations, such as the siblings reported herein.31 Biallelic mutations cause CMMRD syndrome, a childhood cancer syndrome associated with onset of primarily hematologic, CNS, and LS-associated tumors in the first decade of life.32-34 Children with CMMRD syndrome are known to manifest CALMs and axillary freckling.35 Few cases have been reported in the literature and therefore the full phenotypic spectrum or the expressivity of CMMRD syndrome is not known.
The approach described in the present study to discover the bases of distinct phenotype variants of GD represents an attractive opportunity to enhance personalized management of patients and families affected by GD. Timely genomic analysis would have influenced the management of the patients and the parents in 3 important ways. First, the finding of Gaucher cells in the patients was initially attributed to pseudo-Gaucher cells; earlier diagnosis would have permitted timely ERT to prevent disease progression and ameliorate the tumor-promoting environment created by GD. The latter is supported by the observation that in the new generation of patients in whom ERT is commenced in a timely fashion and in whom splenectomy rates have declined dramatically, the cancer rates appear to declining concomitantly.36 Second, because our proband suffered from treatment-resistant, fatal lymphoma, her sibling was treated for her lymphoma with a stem cell transplantation during the first clinical remission (before the diagnosis of CMMRD syndrome). Stem cell transplantation cured her lymphoma (as well as her GD); nevertheless, on a background of DNA instability, she is now at greater risk for second cancers, including CNS- and LS-associated malignancies, because of alkylating agents and total body irradiation administered during her treatment and conditioning regimens. It should be noted that GD patients are at higher risk of developing multiple malignancies12 and therefore the above considerations may be of general applicability to patients exhibiting a GD/cancer phenotype. Finally, the parents in this pedigree are obligate carriers of heterozygous MMR mutations and therefore have the genotype for LS. Therefore, the parents were referred for appropriate genetic counseling and cancer screening.
Of the various cancers, GD1 patients harbor the highest risk of multiple myeloma, with a lifetime relative risk of > 30-fold compared with the general population.11,37 Microsatellite instability (MSI) has been described in multiple myeloma,38 and it would be of interest to investigate whether the GD/multiple myeloma phenotype is associated with mutations in MMR genes. MSI occurs frequently in tumor cells harboring MMR mutations. Failure to correct base changes in DNA leads to the accumulation of randomly occurring mutations in genes, including tumor suppressor genes and oncogenes, causing an increased risk of malignancy. Interestingly, high-level MSI in LS has been associated with increased macrophage and dendritic cell infiltration, as well as increased tumor-infiltrating T-regulatory cells.39 The immune dysregulation described in GD likely impairs immune responses to neoplastic cells, further accentuating cancer risk.18,20,40
Characterization of the D137N mutant GBA1 enzyme suggests an interesting dichotomy. Although acid β-glucosidase activity in peripheral blood leukocytes using standard fluorescent substrate was extremely low, inhibody probing and measurement of in situ enzyme activity in patient fibroblasts revealed that the actual impairment of enzyme activity was less severe (residual activity < 1% vs approximately 15%, respectively, compared with wild-type enzyme activity). Consistent with these findings, the clinical phenotype of GD in our pedigree was initially mild enough to attribute the finding of Gaucher cells to pseudo-Gaucher cells.7 It was only after the onset of T-LBL that overt manifestations of GD developed. This observation raises the intriguing possibility that the pseudo-Gaucher cells described in several hematologic malignancies may in fact represent a mild form of GD that is unmasked in the setting of high cell turnover triggered by the onset of malignancy. A similar situation has been described in the occurrence of multiple myeloma in older adults homozygous for the N370S mutation, which tends to occur in the setting of mild overall GD severity.11
The mechanisms of lymphomagenesis are beginning to be elucidated at the molecular level. Both T-LBL and T-cell lymphoblastic leukemias are neoplasms of lymphoblasts committed to the T-cell lineage. The malignant clones in patients with T-LBL and T-cell lymphoblastic leukemia are thought to originate from normal lymphoid progenitor cells arrested at the early stages of T-cell maturation.41,42 Recent demonstration of impaired T-cell maturation in a mouse model of GD suggests that GD patients might be especially vulnerable to T-cell neoplasms, by tipping the delicate balance between normal differentiation and malignant transformation.18 The thymus is a site for Gaucher cell accumulation, highlighting the need to study the effects of lipid (glucocerebroside, glucosylsphingosine, and secondary metabolites such as ceramide and sphingosine) accumulation in GD on cell differentiation and proliferation.43
Conclusions
Defective lysosomal GBA due to mutations in the GBA1 gene in GD leads to an accumulation of glucosylceramide in tissue macrophages, systemic macrophage activation, immune dysregulation, and a complex multisystemic phenotype. There is increased incidence for hematologic malignancies in GD, but the mechanisms are incompletely understood. We hypothesized a prominent role of genetic modifier(s) in the development of the GD1/cancer phenotype. In the present study, we used whole-exome capture and massively parallel sequencing in a pedigree with a complete concordance of GD1 and childhood-onset T-LBL. The patients harbored a novel homozygous mutation in the GBA gene and a novel homozygous mutation in the MSH6 gene. Our results underscore the concept that a modifier gene(s) may underlie the extreme diversity of phenotypes in GD1. The delineation of modifier genes for GD using the approach described herein should facilitate personalized management of individual patients and illuminate disease mechanisms.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the family for their participation in the study; Robert Brown, a pediatric oncologist, and James McGrath, a medical geneticist, for assistance; Pei Hui for assistance with the MMR immunohistochemistry of the tumor specimen; the Medical Genomics Laboratory of the University of Alabama and the Boston University School of Medicine Center for Human Genetics for performing the mutation analysis of the NF1 gene; and the Associated Regional and University Pathologists (ARUP) Laboratories (University of Utah, Salt Lake City, UT) for performing the gene sequencing of the MSH6 gene. P.K.M. thanks Dr Jeffrey Gruen and Mary Nathan for advice. This work is dedicated to the late Diana Beardsley, MD, PhD, Associate Professor of Pediatrics and Program Director of the Post-doctoral Training Program in Pediatric Hematology/Oncology, Yale University School of Medicine, New Haven, CT.
This research was supported by the Mendelian Disorders Genome Center at Yale University School of Medicine (principal investigators, R.L. Lifton, M. Gunel, and S. Mane). S.M.L. was supported by the National Institutes of Health T32 postdoctoral training program in investigative hematology (principal investigator, M. Dhodapkar). S.M.L. is the 2011 Lysosomal Disease Network fellowship recipient. P.K.M. was supported by a National Institute of Diabetes and Digestive and Kidney Diseases midcareer clinical investigator award (K24DK066306) and by the National Gaucher Foundation.
National Institutes of Health
Authorship
Contribution: S.M.L., G.M.K., R.P.L., and P.K.M., developed the hypothesis; S.M.L., F.P., and P.K.M. recruited the patients and provided medical care; S.M.L., M.C., J.L., R.G.B., J.M.F.G.A., and S.M. performed the laboratory analyses; S.M.L. and M.C. compiled the database and performed the statistical analyses; D.J. performed the pathology evaluation; R.G.B., W.W.K., and J.M.F.G.A. performed the GBA enzyme analysis and the molecular modeling; S.M.L., M.C., R.P.L., and P.K.M. wrote the manuscript; and all authors edited multiple versions of the manuscript.
Conflict-of-interest disclosure: P.K.M. receives research support from Genzyme Corporation for participation in the International Gaucher Registry. The remaining authors declare no competing financial interests.
Correspondence: Pramod K. Mistry, Professor of Pediatrics and Medicine, Section of Pediatric Gastroenterology and Hepatology, Section of Digestive Diseases, Yale School of Medicine, 333 Cedar St, PO Box 208064, New Haven, CT 06520-8064; e-mail: pramod.mistry@yale.edu.
References
Author notes
S.M.L. and M.C. contributed equally to this work.