Cells remove unstable polypeptides through protein quality-control (PQC) pathways such as ubiquitin-mediated proteolysis and autophagy. In the present study, we investigated how these pathways are used in β-thalassemia, a common hemoglobinopathy in which β-globin gene mutations cause the accumulation and precipitation of cytotoxic α-globin subunits. In β-thalassemic erythrocyte precursors, free α-globin was polyubiquitinated and degraded by the proteasome. These cells exhibited enhanced proteasome activity, and transcriptional profiling revealed coordinated induction of most proteasome subunits that was mediated by the stress-response transcription factor Nrf1. In isolated thalassemic cells, short-term proteasome inhibition blocked the degradation of free α-globin. In contrast, prolonged in vivo treatment of β-thalassemic mice with the proteasome inhibitor bortezomib did not enhance the accumulation of free α-globin. Rather, systemic proteasome inhibition activated compensatory proteotoxic stress-response mechanisms, including autophagy, which cooperated with ubiquitin-mediated proteolysis to degrade free α-globin in erythroid cells. Our findings show that multiple interregulated PQC responses degrade excess α-globin. Therefore, β-thalassemia fits into the broader framework of protein-aggregation disorders that use PQC pathways as cell-protective mechanisms.
Introduction
The production of functional hemoglobin A (HbA) tetramers (α2β2) requires the coordinated synthesis and assembly of α- and β-globin protein chains and iron-containing heme groups. Individually, all HbA components are toxic to RBCs and their precursors, as illustrated by β-thalassemias, a common hemoglobinopathy in which β-globin gene (HBB) mutations cause the buildup of free α-globin.1 These unpaired α chains initiate an oxidative damage cascade and form damaging precipitates that contribute largely to the clinical problems associated with β-thalassemia.
The pathophysiology of β-thalassemia bears similarities to a diverse group of protein-aggregation diseases affecting multiple organs (for review, see Khandros and Weiss2 ). These disorders, which include Parkinson disease, Alzheimer disease, Huntington disease, amyotrophic lateral sclerosis, and α1-antitrypsin deficiency, are caused by the accumulation of unstable, relatively insoluble proteins. It is believed that the affected cells can detoxify and remove these damaging proteins via multiple interacting biochemical pathways called protein quality-control (PQC) pathways, but that disease ensues when such compensatory mechanisms are overwhelmed (for review, see Ciechanover and Brundin,3 Ding and Yin,4 and Jaeger and Wyss-Coray5 ). Cellular PQC systems include molecular chaperones, ubiquitin-mediated proteolysis, and autophagy. Several lines of evidence suggest that β-thalassemic erythroid cells use PQC pathways to detoxify free α-globin (for review, see Khandros and Weiss2 ): (1) the clinical severity of β-thalassemia is proportional to the degree of α-globin excess; (2) there is a threshold below which excess α-globin is less harmful, as illustrated by subjects with the β-thalassemia trait, who experience 50% reduced β-globin synthesis with minimal clinical manifestations or accumulation of α-globin precipitates; and (3) there is direct biochemical evidence that α-globin interacts with cellular PQC components.
Numerous studies have shown that normal and β-thalassemic erythroid precursors can balance globin ratios through selective α-chain proteolysis. Pulse-chase experiments using intact human β-thalassemic erythroid cells6,,,,,,–13 and cell lysates11,13,14 showed that excessive α chains are actively degraded and accumulate mainly in the late stages of erythroid maturation, presumably as the proteolytic capacity becomes exceeded. The ubiquitin proteasome system (UPS; for review, see Ciechanover and Brundin3 ), originally characterized in reticulocyte lysates using denatured Hb as a substrate,15 is responsible for physiologic degradation of native proteins and for removing misfolded proteins as part of the PQC pathway in all cells. Studies by Shaeffer et al showed that normal and β-thalassemic hemolysates can ubiquitinate and degrade exogenous α-globin,12,14 although the associated pathways remain largely uncharacterized. RBC precursors also use autophagy, a group of related processes in which targeted proteins or organelles are fused to lysosomes and degraded.16 For example, autophagy-related genes are up-regulated by the master erythroid transcription factor GATA-1 during terminal erythropoiesis.17 During reticulocyte maturation, mitochondria are eliminated by “macroautophagy” (for review, see Ding and Yin4 and Mizushima et al16 ), a process in which cells form double-membrane vesicles (autophagosomes) around cytoplasmic contents for delivery to lysosomes.18,–20 Interestingly, electron micrographs of β-thalassemic erythroblasts identify a subset of α-globin precipitates within lysosomes.2,21,22 More recent work indicates that autophagic processes are increased in HbE/β-thalassemia.23
Most studies linking PQC to detoxification of free α-globin in β-thalassemia were performed before the development of newer genetic and pharmacologic approaches to interrogating the relevant mechanisms. Moreover, recent proof-of-principle studies have shown that the induction of PQC can improve phenotypes of various aggregation disorders in murine models. For example, pharmacologic activation of autophagy can attenuate liver damage in α1-antitrypsin deficiency,24 and genetic up-regulation of proteasome activity can alleviate proteotoxic heart disease.25 However, before such principles can be applied to β-thalassemia, the relevance and contributions of PQC pathways involved in neutralizing free α-globin must be better defined. In the present study, we investigated this problem in a mouse model of β-thalassemia and in human patient cells, and found that the processes of UPS, autophagy, and heat shock all likely participate in the detoxification of free α-globin and are coordinately regulated.
Methods
Mice
β-thalassemic (β-globinTh3/+) mice were kindly provided by Oliver Smithies (University of North Carolina, Chapel Hill, NC),26 and were backcrossed onto a C57BL/6J background for 9-10 generations. All animal experiments were done in accordance with protocols approved by the Institutional Animal Care and Use Committee of The Children's Hospital of Philadelphia.
Human samples
Blood samples were collected from β-thalassemia patients with pretransfusion reticulocyte counts of 3%-10%) and healthy controls at the Children's Hospital of Philadelphia per research protocols approved by the local institutional review board. Written informed consent was obtained from all participants.
Isolation and analysis of insoluble globin fractions
Analysis of globin precipitates in circulating erythrocyte membrane skeletons was performed as described previously.27 For fetal liver cultures, cells were lysed in RIPA buffer containing 1mM DTT, 1mM PMSF, and protease inhibitor cocktail (Sigma-Aldrich). Insoluble fractions were collected by centrifugation at 16 000g, washed, and solubilized by boiling in 2× Laemmli buffer (Sigma-Aldrich). See supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) for detailed denaturing immunoprecipitation and Western blotting protocols.
Reticulocyte pulse-chase analysis
Freshly collected mouse erythrocytes and reticulocytes were used for pulse-chase experiments with 35S-labeled methionine and cysteine (PerkinElmer),27 as described in supplemental Methods. Where indicated, chase medium contained 100μM chloroquine (Sigma-Aldrich), 0.5μM epoxomicin, 10μM MG132 (Enzo Life Sciences), or 0.1% DMSO as a control.
Fetal liver cultures
Fetal livers were collected from embryonic day 14.5 embryos from crosses of wild-type or Th3/+ × Th3/+ mice. Embryos were genotyped and erythroid precursors were isolated from individual embryos using the EasySep hematopoietic progenitor enrichment kit (StemCell Technologies) supplemented with biotin-conjugated CD71 Ab (BioLegend). Purified CD71−Lin− cells were cultured in differentiation medium consisting of IMDM with 10% FCS, 10% PDS, 2mM l-glutamine, 10μM 1-thioglycerol, 1% penicillin/streptomycin, 5% PFHM-II medium, and 5 U/mL of erythropoietin (Amgen). For retroviral infections, cells were cultured in expansion medium consisting of StemPro34 medium (Invitrogen) supplemented with 2mM l-glutamine, 1% penicillin/streptomycin, 10μM 1-thioglycerol, 1μM dexamethasone, 0.5 U/mL of erythropoietin, and 1% murine SCF-conditioned medium.28 After 72 hours of expansion, cells were washed and resuspended in differentiation medium. For Nrf1 and Nrf2 induction experiments, cells in differentiation medium were treated for 24 hours with 0.1μM MG132 or 1μM R-sulforaphane (LKT laboratories). For proteasome-inhibition studies, cells were incubated for 12 hours with 1 or 10μM MG132.
Retroviral shRNA delivery
shRNA constructs were purchased from OpenBiosystems (see supplemental Methods) and selected hairpins cloned into the MSCV-PIG (puromycin-IRES-GFP) vector. Cells (5 × 104) were spinfected at 1300g with 50 μL of retroviral supernatant and 8 μg/mL of polybrene for 90 minutes at 30°C.
Flow cytometry
Flow cytometry staining is described in supplemental Methods. For proteasome activity quantification, cells were first stained with 1μM MV151 (Chemical Proteomics Reagents, Leiden Institute for Chemistry, Leiden, The Netherlands) for 4 hours in culture medium.29 Human patient erythrocytes were similarly treated, but were stained with Hoescht33342 and thiazole orange (ReticCOUNT; BD Biosciences) instead of specific Abs. Cells were analyzed on an LSRII or an LSRFortessa instrument (BD Biosciences) maintained by the Flow Cytometry Core Laboratory at The Children's Hospital of Philadelphia Research Institute.
Microarray analysis
CD71+Ter119+FSChigh cells were sorted from E14.5 fetal livers of Th3/+ × Th3/+ mouse crosses (3 embryos per genotype) using a FACSAriaII cell sorter (BD Biosciences). Cells were sorted directly into TRIzol LS reagent (Invitrogen) and RNA was prepared using the RNeasy kit (QIAGEN). Samples were processed for microarray analysis using the Mouse Gene 1.0ST Array (Affymetrix) by the microarray core facility at the University of Pennsylvania. Microarray data reported herein were submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/44) as accession number GSE34125. Further details are provided in supplemental Methods.
Bortezomib treatment and hematologic analysis
Mice were treated by IP injection of 0.25, 0.5, or 1.0 mg/kg of bortezomib (Velcade; Millennium Pharmaceuticals) in normal saline or of saline control every 3 days. Blood was collected by submandibular bleeding, anticoagulated with EDTA, and analyzed on a Hemavet HV950FS analyzer (Drew Scientific). Reticulocyte counts were done using Retic-COUNT reagent (BD Biosciences). BM erythroid precursors were purified from mice using the EasySep PE Selection Kit (StemCell Technologies) with Ter119-PE Ab (BioLegend).
Real-time RT-PCR
Quantitative real-time RT-PCR used the standard curve method with SYBR Green dye on an ABI 7900 real-time machine (PE Applied Biosystems). Target gene expression was normalized to the average of Actb and Hprt values. For proteasome subunit PCR, multiple randomly selected subunits were examined, and the average fold change is presented. Primer sequences are described in supplemental Methods.
Statistical methods
Statistical analysis was performed using GraphPad Prism Version 4.0 software. All multigroup comparisons were done using 1-way ANOVA. Comparisons between 2 groups were done using the Student t test.
Results
Insoluble α-globin accumulates in proteasome-inhibited β-thalassemic reticulocytes.
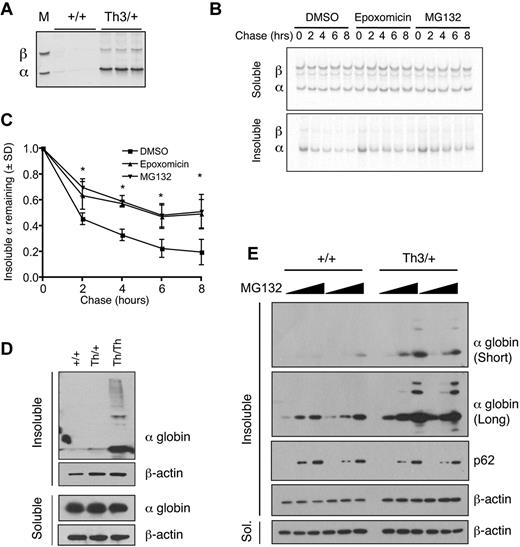
We investigated whether pharmacologic inhibition of the UPS impairs α-globin turnover and accumulation in β-thalassemic erythroid cells. We used Th3 mutant mice in which both the β1 (Hbb1) and β2 (Hbb2) adult globin genes are deleted.26 Homozygous mutants (Th3/Th3) die in utero or perinatally of severe anemia. Heterozygous animals (Th3/+) are viable and exhibit microcytic hypochromic anemia with accumulation of insoluble α-globin chains in erythroid precursors and in the membranes of circulating erythrocytes (Figure 1A). In general, the extent of α-globin precipitation within the latter reflects the severity of β-thalassemia and can be used as a surrogate marker to gauge disease modifiers.27,30,31
Insoluble α-globin accumulates after proteasome inhibition in β-thalassemic erythroid cells. (A) Coomassie-stained triton acetic acid urea gel showing α-globin precipitates in Th3/+ erythrocyte membranes. The marker lane M shows purified α and β-globins. (B) Th3/+ reticulocytes were pulse labeled with 35S-methionine and 35S-cysteine and chased with unlabeled amino acids for the indicated periods of time with or without proteasome inhibitors. Radiolabeled soluble and insoluble globins were isolated from equal numbers of cells, fractionated by triton acetic acid urea gel electrophoresis, and visualized by autoradiography. (C) Quantification of autoradiographs from panel B; n = 3 mice/group. *P < .05 versus DMSO. (D) α-globin Western blots of soluble and insoluble fractions from mouse fetal liver erythroid cultures (48 hours differentiation) of wild-type, Th3/+, and Th3/Th3 erythroblasts. (E) α-globin Western blots of soluble and insoluble fractions from mouse fetal liver erythroid cultures (72 hours of differentiation) treated with vehicle or MG132 (1 or 10μM) for 12 hours.
Insoluble α-globin accumulates after proteasome inhibition in β-thalassemic erythroid cells. (A) Coomassie-stained triton acetic acid urea gel showing α-globin precipitates in Th3/+ erythrocyte membranes. The marker lane M shows purified α and β-globins. (B) Th3/+ reticulocytes were pulse labeled with 35S-methionine and 35S-cysteine and chased with unlabeled amino acids for the indicated periods of time with or without proteasome inhibitors. Radiolabeled soluble and insoluble globins were isolated from equal numbers of cells, fractionated by triton acetic acid urea gel electrophoresis, and visualized by autoradiography. (C) Quantification of autoradiographs from panel B; n = 3 mice/group. *P < .05 versus DMSO. (D) α-globin Western blots of soluble and insoluble fractions from mouse fetal liver erythroid cultures (48 hours differentiation) of wild-type, Th3/+, and Th3/Th3 erythroblasts. (E) α-globin Western blots of soluble and insoluble fractions from mouse fetal liver erythroid cultures (72 hours of differentiation) treated with vehicle or MG132 (1 or 10μM) for 12 hours.
Pulse radiolabeling, followed by chase with unlabeled amino acids, demonstrated that insoluble α-globin was degraded with a half-life of approximately 2 hours in murine Th3/+ reticulocytes (Figure 1B-C). Similar degradation of membrane-associated insoluble α-globin has been observed in human samples.12,13 Treatment with 2 different proteasome inhibitors, MG132 and epoxomicin, during the chase period decreased the turnover of α-globin aggregates, indicating that α-globin is degraded at least in part by ubiquitin-mediated proteolysis (Figure 1B-C).
To study α-globin degradation further, we used a mouse fetal liver erythroid culture system in which purified erythroid progenitors mature synchronously over 72 hours.32 In this system, cultured murine Th3/Th3 cells accumulate insoluble α-globin and begin to die at approximately 36-48 hours of differentiation, whereas the Th3/+ cells mature at a slightly delayed rate relative to wild-type cells. Remarkably, after 48 hours of culture, Th3/+ erythroblasts contained relatively little insoluble α-globin compared with Th3/Th3 cells (Figure 1D), which is consistent with the presence of compensatory proteolytic mechanisms as observed in human thalassemic erythroblasts.7,9,12 Treatment with MG132 caused a dose-dependent accumulation of insoluble α-globin chains in Th3/+ cultured erythroblasts and to a lesser extent in control cells (Figure 1E top 2 panels). The latter finding suggests that the UPS may degrade the free α chains that are synthesized at slight excess relative to β chains during normal erythropoiesis.33 SQSTM1/p62, which serves as a marker of insoluble protein aggregates in structures called aggresomes34 (see “Discussion”), also copurified with this fraction after proteasome inhibition in wild-type and Th3/+ cells.
Free α-globin is polyubiquitinated in β-thalassemic cells
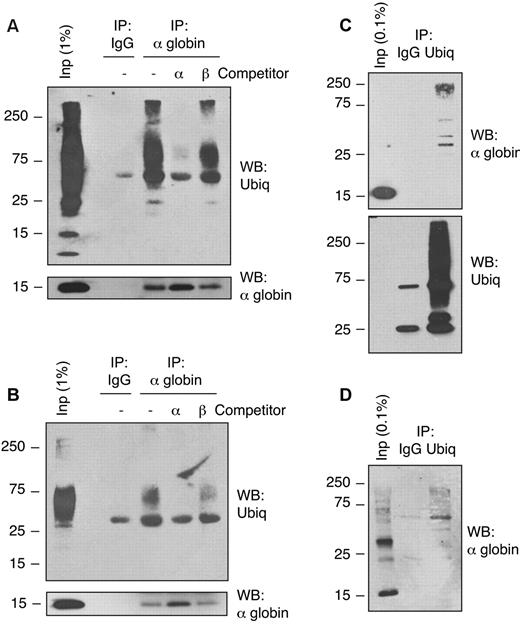
Previous studies of globin chain removal by the UPS have focused primarily on degradation of destabilized globins in vivo or artificially denatured Hbs in reticulocyte lysates. Shaeffer et al showed that exogenous purified α-globin chains are ubiquitinated and degraded by the proteasome in β-thalassemic patient hemolysates,14 but ubiquitination of endogenous α-globin has not been demonstrated previously. We investigated whether α-globin is actually ubiquitinated in vivo in thalassemic cells. We used denaturing immunoprecipitation from samples boiled in 1% SDS to purify α-globin chains from native circulating β-thalassemic erythroid cells under conditions that prevent noncovalent protein-protein associations during immunoprecipitation. Because most soluble α-globin in Th3/+ RBCs exists in stable complexes with β-globin as HbA, we performed the immunoprecipitation experiments using material from α-globin–enriched insoluble fractions. Immunoprecipitation by α-globin–specific Ab, followed by Western blot analysis using an anti-ubiquitin Ab, revealed high-molecular-weight polyubiquitinated α-globin in RBCs from Th3/+ mice (Figure 2A) and a β-thalassemia major human patient (Figure 2B). Inclusion of competitor α-globin, but not β-globin, during the immunoprecipitation blocked the recovery of polyubiquitinated proteins, demonstrating Ab specificity. Denaturing immunoprecipitation using an Ab that recognizes mono- and polyubiquitinated proteins, but not free ubiquitin, followed by α-globin Western blotting, also revealed ubiquitinated α-globin in mouse (Figure 2C) and human (Figure 2D) thalassemic RBCs. Ubiquitinated α-globin was not detected in wild-type RBCs (not shown). Therefore, α-globin is polyubiquitinated in mouse and human β-thalassemic RBCs in vivo.
Free α-globin is ubiquitinated in β-thalassemic erythroid cells. Immunoprecipitation-Western blot analysis of solubilized α-globin aggregates from Th3/+ erythrocytes. Membrane-associated α-globin was isolated from mouse Th3/+ (A,C) or human β-thalassemia major (B,D) erythrocytes and solubilized in SDS. Samples were immunoprecipitated with anti–α-globin (A-B) or anti-ubiquitin (C-D) Abs and analyzed by Western blotting using the indicated Abs. Input fractions and preimmune serum (IgG) immunoprecipitation are included as controls. Immunoprecipitation reactions containing purified α-globin or β-globin competitors demonstrate Ab specificity.
Free α-globin is ubiquitinated in β-thalassemic erythroid cells. Immunoprecipitation-Western blot analysis of solubilized α-globin aggregates from Th3/+ erythrocytes. Membrane-associated α-globin was isolated from mouse Th3/+ (A,C) or human β-thalassemia major (B,D) erythrocytes and solubilized in SDS. Samples were immunoprecipitated with anti–α-globin (A-B) or anti-ubiquitin (C-D) Abs and analyzed by Western blotting using the indicated Abs. Input fractions and preimmune serum (IgG) immunoprecipitation are included as controls. Immunoprecipitation reactions containing purified α-globin or β-globin competitors demonstrate Ab specificity.
Proteasome subunits are up-regulated in β-thalassemic erythroblasts
Total proteolytic activity and α-globin–specific proteolysis are increased in β-thalassemic erythroblasts relative to normal patient samples,10,11 but the identity of the proteases and the associated mechanisms have not been determined. We compared gene-expression profiles of flow cytometry–purified erythroblasts from embryonic day 14.5 fetal livers of wild-type, Th3/+, and Th3/Th3 mice. We examined Ter119+CD71+FSChigh cells to compare developmental stage-matched samples at a point when Hb synthesis becomes active and when Th3/Th3 homozygous erythroblasts remain viable (supplemental Figure 1A). Hierarchical clustering of significantly up-regulated or down-regulated (P < .01 by ANOVA) genes grouped mice on the basis of genotype and showed that wild-type and Th3/+ cells are more similar to each other than to Th3/Th3 cells (supplemental Figure 1B). There were relatively few changes in Th3/+ cells compared with wild-type samples, with an R2 value of 0.9925 for +/+ versus Th3/+, compared with 0.8930 for +/+ versus Th3/Th3, suggesting that heterozygous mutant erythroblasts compensate for α-globin synthetic excess, thereby minimizing its impact on gene expression.
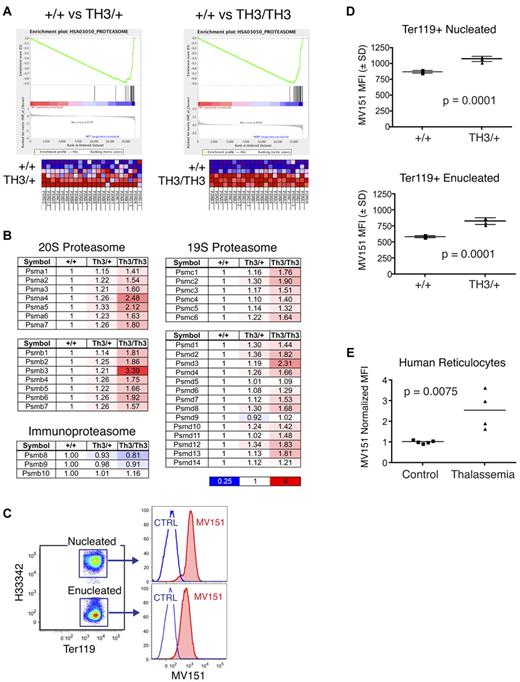
We performed Gene Set Enrichment Analysis (GSEA) to assess whether transcripts that are differentially expressed in β-thalassemia are enriched for specific biologic properties or functions.35 The most highly enriched gene set in both Th3/+ and Th3/Th3 erythroblasts was the proteasome pathway (Figure 3A), which was overrepresented in β-thalassemia at a high statistical significance (normalized enrichment scores of 2.44 for Th3/+ and 2.11 for Th3/Th3, with false discovery rate q-values less than 0.0001). Messenger RNAs encoding all subunits of the catalytically active 20S proteasome complex, and almost all subunits of the regulatory 19S complex, were up-regulated in an α-globin–dosage dependent manner (Figure 3B). The β subunits specific for the immunoproteasome that is involved in the generation of peptides for MHC display were not altered. We used MV151, a fluorescent proteasome inhibitor that binds specifically to active proteasome subunits,29 to determine whether proteasome activity is enhanced in β-thalassemic cells (Figure 3C). Costaining of cultured fetal liver erythroid cells with MV151 and stage-specific markers revealed elevated proteasome activities in Th3/+ cells, in both immature nucleated erythroblasts (Ter119+Hoechst33342+) and enucleated reticulocytes (Ter119+Hoechst33342−). The magnitude of the increase was similar to the transcript changes shown in Figure 3B. We were not able to examine Th3/Th3 cultures because of high toxicity of MV151 in these cells. We also used MV151 to analyze proteasome activity in circulating reticulocytes from human β-thalassemia major patients, whose reticulocytes also had increased proteasome activity relative to reticulocytes from healthy controls (Figure 3E). These data indicate that up-regulation of proteasome subunit transcripts in β-thalassemia leads to the production of new functional proteasomes.
Proteasome components are up-regulated in β-thalassemic erythroblasts. (A) Transcriptome analysis was performed on FACS-purified, developmental stage–matched fetal liver erythroblasts from wild-type, heterozygous (Th3/+), and homozygous (Th3/Th3) thalassemic mouse embryos (supplemental Figure 1). Gene Set Enrichment Analysis (GSEA) reveals the induction of proteasome subunit mRNAs in Th3/+ (left) and Th3/Th3 (right) erythroblasts compared with wild-type controls. (B) Relative expression levels of proteasome subunit mRNAs in β-thalassemic erythroblasts normalized to wild-type controls. (C) Proteasome activities in fetal liver erythroid cultures using the fluorescent proteasome activity indicator MV151. Samples were costained for expression of the erythroid-specific antigen Ter119 and for DNA using Hoescht33342 to distinguish nucleated and enucleated erythroid cells. (D) MV151 mean fluorescence intensity (MFI) for nucleated (top) and enucleated (bottom) erythroid cells (Ter119+) in 48-hour erythroid cultures from wild-type and Th3/+ fetal livers; n = 4 embryos/genotype. (E) MV151 MFI for circulating human reticulocytes (Hoescht33342−, thiazole orange+) from control or β-thalassemia major patients. Values are normalized to controls.
Proteasome components are up-regulated in β-thalassemic erythroblasts. (A) Transcriptome analysis was performed on FACS-purified, developmental stage–matched fetal liver erythroblasts from wild-type, heterozygous (Th3/+), and homozygous (Th3/Th3) thalassemic mouse embryos (supplemental Figure 1). Gene Set Enrichment Analysis (GSEA) reveals the induction of proteasome subunit mRNAs in Th3/+ (left) and Th3/Th3 (right) erythroblasts compared with wild-type controls. (B) Relative expression levels of proteasome subunit mRNAs in β-thalassemic erythroblasts normalized to wild-type controls. (C) Proteasome activities in fetal liver erythroid cultures using the fluorescent proteasome activity indicator MV151. Samples were costained for expression of the erythroid-specific antigen Ter119 and for DNA using Hoescht33342 to distinguish nucleated and enucleated erythroid cells. (D) MV151 mean fluorescence intensity (MFI) for nucleated (top) and enucleated (bottom) erythroid cells (Ter119+) in 48-hour erythroid cultures from wild-type and Th3/+ fetal livers; n = 4 embryos/genotype. (E) MV151 MFI for circulating human reticulocytes (Hoescht33342−, thiazole orange+) from control or β-thalassemia major patients. Values are normalized to controls.
Transcription factor Nrf1 induces proteasome subunit mRNAs in normal and β-thalassemic erythroblasts
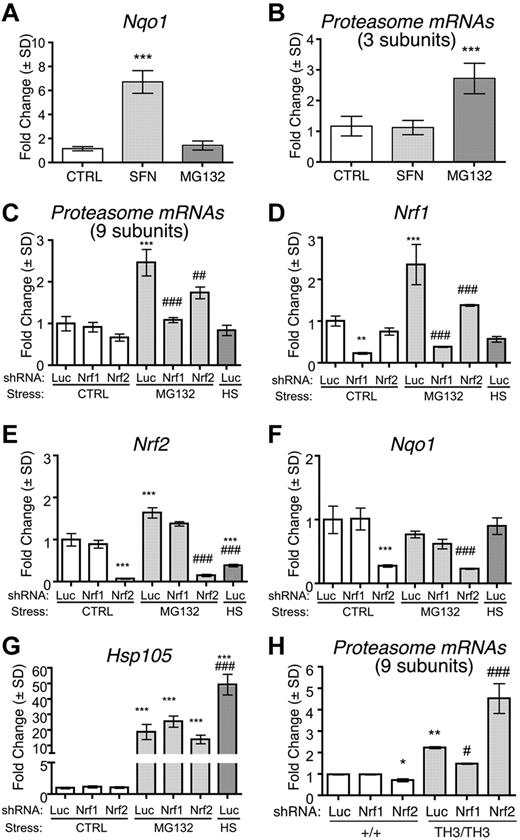
Proteasome subunit genes contain antioxidant response elements (AREs) that are bound by members of the NFE2-like protein family, Nrf1 and Nrf2, which accumulate in cells during specific stresses.36,–38 Typically, oxidative stress stabilizes Nrf2 and proteotoxic stress/proteasome inhibition stabilizes Nrf1.37,38 We examined how these transcription factors regulate erythroid proteasome subunit gene expression. First, we investigated whether activation of Nrf1 or Nrf2 up-regulates proteasome subunits in wild-type fetal liver erythroblasts. Treatment with the Nrf2 activator sulforaphane39 induced the Nrf2-specific target gene Nqo1 (Figure 4A), but had no effect on proteasome subunit mRNAs (Figure 4B). In contrast, treatment with a low dose (0.1μM) of the proteasome inhibitor MG132 induced proteasome subunit mRNAs (Figure 4B) in a manner similar to what occurs in nonerythroid cells through Nrf1-mediated effects.37,38 MG132 did not up-regulate Nqo1 mRNA (Figure 4A), indicating that Nrf2 was not activated. Proteasome subunit mRNA induction by MG132 was blocked entirely by shRNA-mediated knock-down of Nrf1 (Figure 4C-D). These data indicate that activation of Nrf1 by MG132 induces proteasome subunit gene expression, whereas activation of Nrf2 by sulforaphane does not. Nrf2 shRNA partially blocked proteasome subunit mRNA induction by MG132 (Figure 4C,E-F), but this effect may be indirect because Nrf1 expression was also inhibited (Figure 4D). Proteasome inhibition by MG132 also activates heat-shock responses with up-regulation of molecular chaperones (Figure 4G).40 Incubation at 42°C for 1 hour to induce heat shock increased Hsp105 transcripts (Figure 4G), but failed to up-regulate proteasome subunits (Figure 4C). Therefore, the induction of proteasome subunits by proteasome inhibition is independent of heat-shock responses.
Proteasome subunit up-regulation in β-thalassemia is Nrf1 dependent. (A-B) Wild-type murine fetal liver erythroid cultures were treated for 24 hours with 1μM sulforaphane or 0.1μM MG132 to activate Nrf2 or Nrf1, respectively. (A) Expression of Nqo1, an Nrf2 target gene; n = 4 embryos/group. ***P < .001 versus control (CTRL). (B) Proteasome subunit mRNA expression (average fold change of 3 randomly selected subunits) normalized to control. (C-G) Wild-type fetal liver erythroid precursors were infected with retroviruses encoding control (anti-luciferase, Luc), Nrf1, or Nrf2 targeted shRNAs and differentiated for 24 hours with or without 0.1μM MG132. As an additional control, Luc shRNA–infected cells were also heat-shocked at 42°C for 1 hour and allowed to recover for 1 hour at 37°C (HS). Transcript expression was normalized to β-actin and Hprt mRNAs and compared among experimental groups with control-treated Luc shRNA samples assigned an arbitrary value of 1.0. Proteasome subunit expression is shown as the average fold change of an expanded panel of 9 proteasome subunit mRNAs (C), Nrf1 (D), Nrf2 (E), Nqo1 (F), and Hsp105 (G); n = 3 embryos per group. **P < .01 versus Luc Ctrl; ***P < .001 versus Luc Ctrl; ##P < .01 versus Luc MG132; ###P < .001 versus +/+ Luc. (H) Proteasome subunit mRNA expression in wild-type or Th3/Th3 fetal liver erythroblasts infected with retroviruses expressing shRNAs targeting Nrf1, Nrf2, or luciferase (Luc). Analysis was performed after 48 hours of expansion and 44 hours of differentiation. Data shown are the average fold change for 9 subunit transcripts; n = 3 embryos/group. *P < .05 versus +/+ Luc; **P < .01 versus +/+ Luc, #P < .05 versus Th3/Th3 Luc; ## P < .001 versus +/+ Luc.
Proteasome subunit up-regulation in β-thalassemia is Nrf1 dependent. (A-B) Wild-type murine fetal liver erythroid cultures were treated for 24 hours with 1μM sulforaphane or 0.1μM MG132 to activate Nrf2 or Nrf1, respectively. (A) Expression of Nqo1, an Nrf2 target gene; n = 4 embryos/group. ***P < .001 versus control (CTRL). (B) Proteasome subunit mRNA expression (average fold change of 3 randomly selected subunits) normalized to control. (C-G) Wild-type fetal liver erythroid precursors were infected with retroviruses encoding control (anti-luciferase, Luc), Nrf1, or Nrf2 targeted shRNAs and differentiated for 24 hours with or without 0.1μM MG132. As an additional control, Luc shRNA–infected cells were also heat-shocked at 42°C for 1 hour and allowed to recover for 1 hour at 37°C (HS). Transcript expression was normalized to β-actin and Hprt mRNAs and compared among experimental groups with control-treated Luc shRNA samples assigned an arbitrary value of 1.0. Proteasome subunit expression is shown as the average fold change of an expanded panel of 9 proteasome subunit mRNAs (C), Nrf1 (D), Nrf2 (E), Nqo1 (F), and Hsp105 (G); n = 3 embryos per group. **P < .01 versus Luc Ctrl; ***P < .001 versus Luc Ctrl; ##P < .01 versus Luc MG132; ###P < .001 versus +/+ Luc. (H) Proteasome subunit mRNA expression in wild-type or Th3/Th3 fetal liver erythroblasts infected with retroviruses expressing shRNAs targeting Nrf1, Nrf2, or luciferase (Luc). Analysis was performed after 48 hours of expansion and 44 hours of differentiation. Data shown are the average fold change for 9 subunit transcripts; n = 3 embryos/group. *P < .05 versus +/+ Luc; **P < .01 versus +/+ Luc, #P < .05 versus Th3/Th3 Luc; ## P < .001 versus +/+ Luc.
We also investigated whether proteasome subunit mRNA up-regulation in β-thalassemic erythroblasts was attenuated by shRNAs against Nrf1 or Nrf2. Only shRNAs against Nrf1 blocked the up-regulation of proteasome subunits in Th3/Th3 cells (Figure 4H and supplemental Figure 2). Somewhat paradoxically, Nrf2 knock-down increased proteasome subunit gene expression specifically in Th3/Th3 cells (Figure 4H and supplemental Figure 2). This may reflect competition between Nrf1 and Nrf2 for binding to AREs within proteasome subunit genes and/or for obligate heterodimerization partner Maf proteins.36 In this case, suppression of Nrf2 would favor binding of the more potent activator Nrf1 to AREs within proteasome subunit genes to further induce their expression. Overall, our findings indicate that Nrf1 largely mediates the induction of proteasomal subunit mRNAs in normal erythroblasts exposed to proteotoxic stress by MG132 and in β-thalassemic erythroblasts, which experience proteotoxic stress through the accumulation of free α-globin.
In vivo proteasome inhibition impairs erythropoiesis in wild-type and β-thalassemic mice
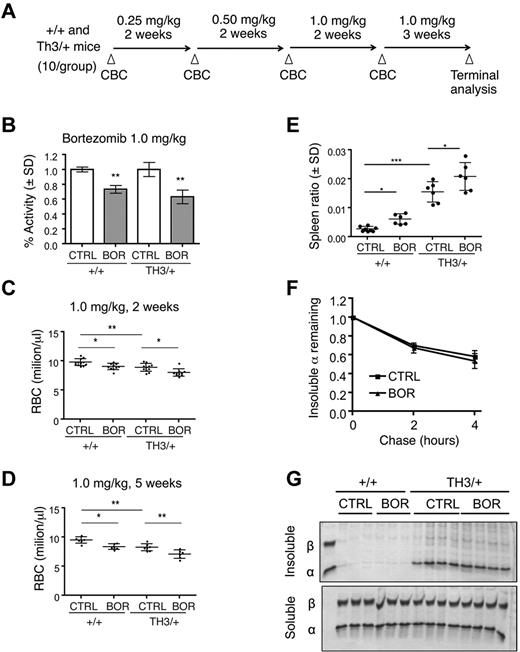
To determine the in vivo importance of the UPS in modulating α-globin toxicity, we treated β-thalassemic mice with the proteasome inhibitor bortezomib.41 Proteasome inhibition is potentially toxic to all cells and has been shown to impair normal erythroid differentiation in vitro.42 However, we reasoned that if β-thalassemic erythroblasts are dependent on the UPS for degradation of excess α-globin, then affected mice may exhibit increased bortezomib-induced toxicities compared with wild-type animals. We first confirmed that bortezomib can impair α-globin turnover ex vivo in Th3/+ mouse reticulocyte pulse-chase experiments (supplemental Figure 3). We then conducted a dose-escalation trial in which wild-type or Th3/+ mice were treated with bortezomib at doses similar to those used in human patients (Figure 5A).43 The timing of treatment at each dose was 2-3 weeks, which represents approximately one half-life for wild-type murine RBCs. Examination of circulating RBCs 24 hours after systemic treatment with bortezomib revealed an approximately 25% inhibition of proteasome activity at 0.25 mg/kg and approximately 30%-40% inhibition at 1.0 mg/kg (Figure 5B), which is consistent with previous findings.44 Lower doses of drug (0.25 and 0.5 mg/kg) produced minimal effects on RBC parameters (supplemental Table 1). At 1.0 mg/kg of bortezomib, RBC counts and Hb decreased by similar extents in both wild-type and Th3/+ mice (Figure 5C-D and supplemental Table 2). Similar results were obtained with a 3-week treatment at 1.0 mg/kg without dose escalation (data not shown). Bortezomib treatment caused an increase in spleen size (Figure 5E) and expansion of early erythroid precursors in the BM and spleens of both wild-type and Th3/+ mice, although the effects were more prominent in wild-type animals (supplemental Figure 4).31,32 Bortezomib did not alter the turnover of insoluble α-globin in (Th3/+) reticulocytes or increase the levels of insoluble globins in wild-type or Th3/+ RBCs (Figure 5F-G). Overall, UPS inhibition by bortezomib generally impaired erythropoiesis with subtle differences in drug effects on wild-type versus Th3/+ mice. However, bortezomib did not cause the accumulation of α-globin precipitates or enhanced toxicity in the β-thalassemic animals.
Systemic proteasome inhibition by bortezomib impairs both thalassemic and normal erythropoiesis in vivo. (A) Scheme for bortezomib dosing in wild-type and Th3/+ mice. (B) Proteasome activity in erythrocytes from mice 24 hours after treatment with 1.0 mg/kg of bortezomib. Activity, normalized to total protein, was measured by fluorescence release from Suc-LLVY-AMC proteasome substrate and normalized to control treated mice; n = 3 mice/group. **P < .01 versus control (CTRL). (C) RBC counts of +/+ or Th3/+ mice treated with vehicle or 1.0 mg/kg of bortezomib for 2 weeks; n = 10 mice/group. (D) RBC counts of +/+ or Th3/+ mice treated with vehicle or 1.0 mg/kg of bortezomib for 5 weeks; n = 6 mice/group. *P < .05; **P < .01; ***P < .001. (E) Spleen weight normalized to total body weight in bortezomib-treated and control mice n = 6 mice/group. *P < .05; ***P < .001. (F) Pulse-chase analysis of insoluble α-globin in reticulocytes from control or bortezomib-treated Th3/+ mice. (G) Coomassie-stained insoluble globin aggregates (top) from equal numbers of circulating erythrocytes from wild-type or Th3/+ mice treated with vehicle or bortezomib at 1.0 mg/kg. Soluble fractions are included as loading controls (bottom).
Systemic proteasome inhibition by bortezomib impairs both thalassemic and normal erythropoiesis in vivo. (A) Scheme for bortezomib dosing in wild-type and Th3/+ mice. (B) Proteasome activity in erythrocytes from mice 24 hours after treatment with 1.0 mg/kg of bortezomib. Activity, normalized to total protein, was measured by fluorescence release from Suc-LLVY-AMC proteasome substrate and normalized to control treated mice; n = 3 mice/group. **P < .01 versus control (CTRL). (C) RBC counts of +/+ or Th3/+ mice treated with vehicle or 1.0 mg/kg of bortezomib for 2 weeks; n = 10 mice/group. (D) RBC counts of +/+ or Th3/+ mice treated with vehicle or 1.0 mg/kg of bortezomib for 5 weeks; n = 6 mice/group. *P < .05; **P < .01; ***P < .001. (E) Spleen weight normalized to total body weight in bortezomib-treated and control mice n = 6 mice/group. *P < .05; ***P < .001. (F) Pulse-chase analysis of insoluble α-globin in reticulocytes from control or bortezomib-treated Th3/+ mice. (G) Coomassie-stained insoluble globin aggregates (top) from equal numbers of circulating erythrocytes from wild-type or Th3/+ mice treated with vehicle or bortezomib at 1.0 mg/kg. Soluble fractions are included as loading controls (bottom).
Systemic proteasome inhibition activates alternate PQC pathways
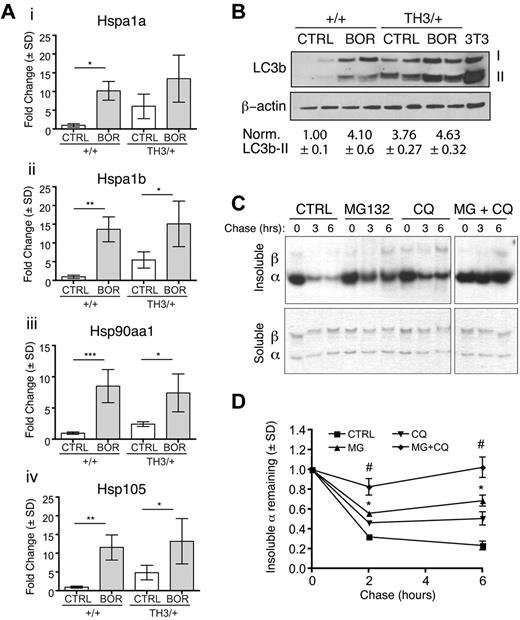
The failure of bortezomib to enhance α-globin accumulation in β-thalassemic mice contrasts with our findings that proteasome inhibition blocks degradation of free α-globin in erythroid cultures and in reticulocytes. In vivo, early-stage erythroid progenitors may activate alternate PQC pathways in response to systemic proteasome inhibition. To investigate this, we analyzed purified Ter119+ erythroid precursors from the BM of bortezomib-treated wild-type and Th3/+ mice. Proteasome inhibition activates heat-shock factor transcription factors that induce the expression of heat-shock proteins (HSPs), molecular chaperones that bind misfolded proteins to stabilize their structures and prevent precipitation.40 In wild-type fetal liver erythroblasts, proteasome inhibition induced mRNAs encoding Hsp105 (Figure 4G) and Hsp90aa1 (not shown). Without bortezomib treatment, numerous HSP mRNAs trended toward up-regulation in Th3/+ erythroblasts, likely reflecting proteotoxic stress and activation of the heat-shock factor pathway (Figure 6A). However, a significantly greater HSP response resulted from systemic treatment with bortezomib in both wild-type and Th3/+ erythroid precursors. Therefore, one compensatory response to proteasome inhibition is the induction of HSPs, which may alleviate some of the toxic effects of free α-globin in β-thalassemia.
Systemic proteasome inhibition activates alternate α-globin detoxification pathways. (A) Real-time RT-PCR quantification of HSP mRNA expression in Ter119+ erythroblasts from wild-type or Th3/+ mice treated with vehicle (CTRL) or bortezomib (BOR). Expression is normalized to β-actin and Hprt mRNA levels. Relative expression between different experimental groups is shown with vehicle-treated wild-type mice assigned an arbitrary value of 1.0. Data are shown for Hspa1a (i), Hspa1b (ii), Hsp90aa1 (iii), and Hsp105 (iv); n = 4 mice/group. *P < .05; *P < .01; ***P < .001. (B) Purified BM Ter119+ cells from mice treated with bortezomib (BOR, 1.0 mg/kg for 14 days) or vehicle (CTRL) were analyzed by Western blotting for the autophagosome marker LC3b. Two forms of LC3b are indicated: the unmodified form (I) and the phosphatidylethanolamine-conjugated form (II), which indicates active autophagosomes. β-actin expression was examined as a loading control. Mean ± SEM LC3b-II signal normalized to β-actin is shown for 4 mice for each group, with wild-type control treated mice set at 1. (C) Reticulocytes from β-thalassemic (Th3/+) mice were labeled with 35S-cysteine and 35S-methionine and chased with unlabeled amino acids in the presence or absence of proteasome (MG132, 10μM) or lysosome (CQ, chloroquine, 100μM) inhibitors. Soluble and insoluble fractions were purified and analyzed for labeled α-globin by triton acetic acid urea gel electrophoresis, followed by autoradiography. (D) Quantification of autoradiographs from panel C; n = 3 mice. *P < .001 versus control; #P < .001 versus MG or CQ. Note that the drugs inhibit the loss of insoluble α-globin additively.
Systemic proteasome inhibition activates alternate α-globin detoxification pathways. (A) Real-time RT-PCR quantification of HSP mRNA expression in Ter119+ erythroblasts from wild-type or Th3/+ mice treated with vehicle (CTRL) or bortezomib (BOR). Expression is normalized to β-actin and Hprt mRNA levels. Relative expression between different experimental groups is shown with vehicle-treated wild-type mice assigned an arbitrary value of 1.0. Data are shown for Hspa1a (i), Hspa1b (ii), Hsp90aa1 (iii), and Hsp105 (iv); n = 4 mice/group. *P < .05; *P < .01; ***P < .001. (B) Purified BM Ter119+ cells from mice treated with bortezomib (BOR, 1.0 mg/kg for 14 days) or vehicle (CTRL) were analyzed by Western blotting for the autophagosome marker LC3b. Two forms of LC3b are indicated: the unmodified form (I) and the phosphatidylethanolamine-conjugated form (II), which indicates active autophagosomes. β-actin expression was examined as a loading control. Mean ± SEM LC3b-II signal normalized to β-actin is shown for 4 mice for each group, with wild-type control treated mice set at 1. (C) Reticulocytes from β-thalassemic (Th3/+) mice were labeled with 35S-cysteine and 35S-methionine and chased with unlabeled amino acids in the presence or absence of proteasome (MG132, 10μM) or lysosome (CQ, chloroquine, 100μM) inhibitors. Soluble and insoluble fractions were purified and analyzed for labeled α-globin by triton acetic acid urea gel electrophoresis, followed by autoradiography. (D) Quantification of autoradiographs from panel C; n = 3 mice. *P < .001 versus control; #P < .001 versus MG or CQ. Note that the drugs inhibit the loss of insoluble α-globin additively.
Autophagy is active during normal erythropoiesis17 and has been shown to degrade unstable proteins in numerous diseases.5 In the present study, we investigated whether autophagy compensates for proteasome inhibition during normal and thalassemic erythropoiesis. We used Western blotting to examine the autophagosome marker LC3b in purified Ter119+ BM cells (Figure 6B). LC3b levels, including the phosphatidylethanolamine-conjugated (type II) form indicative of active autophagy, were increased in Th3/+ erythroblasts compared with wild-type, similar to what was reported for human thalassemic cells.23 Systemic bortezomib treatment increased LC3b in both genotypes. Therefore, the highest levels of autophagy were observed in Th3/+ mice treated with the proteasome inhibitor. We also investigated whether autophagy participates in the turnover of α-globin aggregates in β-thalassemia. In pulse-chase studies of Th3/+ mouse reticulocytes, ATP depletion (which inhibits both the UPS and autophagy) reduced the degradation of α-globin aggregates to a greater extent than proteasome inhibition alone (supplemental Figure 5). Inhibition of lysosomal acidification with chloroquine inhibited α-globin degradation to a similar extent as proteasome inhibition, whereas both inhibitors together produced additive effects (Figure 6C-D). These results indicate that in β-thalassemia, excess α-globin is degraded by autophagy, and that this process is enhanced by inhibition of the UPS. Therefore, both HSP and autophagy pathways are induced by bortezomib in wild-type and Th3/+ mice. The induction of these PQC components likely explains why systemic proteasome inhibition does not enhance the accumulation of precipitated α-globin in Th3/+ mice (Figure 7).
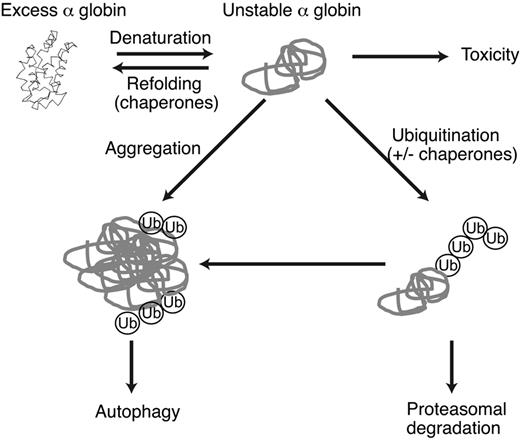
Model of α-globin detoxification pathways in β-thalassemia. Excess free α-globin is unstable and cytotoxic to RBC precursors and mature RBCs. Unstable α-globin is polyubiquitinated and degraded via the proteasome. If ubiquitin-proteasome activity is insufficient, α-globin forms relatively insoluble aggregates that serve as a substrate for macroautophagy. Chaperones may be involved in refolding α-globin or in targeting excess α-globin for degradation. Molecular cross-talk exists between these pathways; for example, inhibition of proteasome activity results in the accumulation of unfolded proteins, activation of stress pathways, and consequent induction of autophagy and heat-shock/molecular chaperone responses.
Model of α-globin detoxification pathways in β-thalassemia. Excess free α-globin is unstable and cytotoxic to RBC precursors and mature RBCs. Unstable α-globin is polyubiquitinated and degraded via the proteasome. If ubiquitin-proteasome activity is insufficient, α-globin forms relatively insoluble aggregates that serve as a substrate for macroautophagy. Chaperones may be involved in refolding α-globin or in targeting excess α-globin for degradation. Molecular cross-talk exists between these pathways; for example, inhibition of proteasome activity results in the accumulation of unfolded proteins, activation of stress pathways, and consequent induction of autophagy and heat-shock/molecular chaperone responses.
Discussion
More than 40 years ago, Fessas, Bank, Nathan, Weatherall, and others demonstrated that β-thalassemia is caused by globin chain imbalance and that the accumulation of cytotoxic free α-globin is a primary determinant of disease pathophysiology.45,,–48 Additional early studies, interpreted in light of more recent work, indicate striking similarities between β-thalassemia and a class of diseases called “protein-aggregation disorders” (for review, see Khandros and Weiss2 ). Common features include the accumulation of unstable, misfolded proteins that can be detoxified to some extent by cellular PQC systems, with disease ensuing when protective mechanisms are overwhelmed. It is likely that lessons learned from recent studies of protein-aggregation disorders, including α1 antitrypsin deficiency,24 cardiomyopathy, myocardial ischemia,25 and some neurodegenerative diseases,5 can be exploited to better understand and treat β-thalassemia. Conversely, defining how α-globin is detoxified in β-thalassemia may further elucidate PQC mechanisms in nonerythroid protein-aggregation disorders. In the present study, we investigated further how β-thalassemia fits into emerging paradigms of protein-aggregation disorders. We show that numerous interregulated PQC pathways, including the UPS, autophagy, and HSP responses, are used to detoxify and remove free α-globin in β-thalassemic erythroid cells. Furthermore, we demonstrate for the first time that the UPS is regulated dynamically at the transcriptional level in β-thalassemic erythroblasts through a Nrf1 stress-response pathway, suggesting a new potential therapeutic target for β-thalassemia.
Previous studies have shown that free α chains can be ubiquitinated in cell-free systems or in reticulocyte lysates in vitro14 and that ATP-dependent degradation of α chains occurs in β-thalassemic patient reticulocytes.12 In the present study, we used a mouse model of β-thalassemia to establish a simple system for following the fate of excess α-globin in live cells and show that newly synthesized excess α-globin chains rapidly associate with an insoluble stromal fraction and are degraded, mirroring earlier findings in human cells.12 Both proteasomal and lysosomal inhibitors alter turnover of insoluble α chains, implicating the UPS and autophagy in this process. Most likely, autophagy mediates direct turnover of insoluble α-globin, whereas the UPS degrades soluble α-globin, which accumulates and shifts to the insoluble fraction after proteasome inhibition in isolated reticulocytes. This interpretation of the current data is consistent with electron microscopy analysis of human β-thalassemic erythroblasts, in which putative electron-dense α-globin inclusions were observed being engulfed by and within lysosomes.21,22 In addition, our denaturing immunoprecipitation experiments detected insoluble polyubiquitinated α-globin chains in β-thalassemic erythrocytes, even without proteasome inhibitor treatment. These findings show for the first time that free α-globin is polyubiquitinated in vivo and suggest that the form of α-globin recognized by ubiquitin ligases is misfolded and/or unstable. Most likely, a proportion of polyubiquitinated α-globin precipitates in β-thalassemic erythroblasts because proteasomal degradation systems are saturated.
Our results indicate that in β-thalassemia, unstable α-globin does not form static precipitates, but rather exists in dynamic subcellular fractions that interact with both UPS and autophagy pathways. These features raise the possibility that free α-globin interacts physically with the aggresome, a recently described intracellular structure in which unstable proteins colocalize with PQC machinery, specialized adapter proteins, and the cytoskeleton.49 Further, electron micrographs of β-thalassemic erythroblasts demonstrate that some precipitated α-globin is perinuclear, ubiquitin-associated, and pericentriolar, all features of aggresomes.21,22,50 It is believed that aggresomes sequester abnormal proteins to minimize cellular damage and provide a staging area for chaperone-mediated refolding, proteasome degradation, or bulk autophagy (macroautophagy) through scaffolding proteins such as SQSTM1/p62 that interact with the autophagosome protein LC3.34,49 Therefore, we speculate that thalassemic erythroblasts degrade precipitated free α-globin via aggresome-mediated macroautophagy.
Previous work suggests that β-thalassemic erythroblasts exhibit increased capacity for protein degradation,10,11 but the identity of the proteases and the mechanisms of their increased activity are not known. We have demonstrated herein that within β-thalassemic cells, most proteasome subunits are coordinately up-regulated according to the dosage of free α-globin. Whereas it has been shown previously that proteasome subunit gene transcription increases in response to pharmacologic proteasome inhibition,37,38 similar effects have not been demonstrated in any disease process, but are predicted to occur in nonerythroid protein-aggregation disorders according to the present findings. Proteasome subunit genes contain AREs that are recognized by a family of cap-n-collar basic leucine zipper (CNC-bZip) transcription factors, including Nrf1 (TCF11) and Nrf2, which activate distinct and overlapping sets of stress-response genes.36,–38 The TCF11 isoform of Nrf1 up-regulates proteasome subunit genes in cell lines treated with low doses of proteasome inhibitor. A role for Nrf2 in proteasome gene regulation has been suggested, but is less well defined.51,52 Mice lacking Nrf1 in neurons exhibit reduced brain proteasome activity and neurodegeneration.53 Global deletion of Nrf1 leads to embryonic lethality from anemia, but this defect is not cell autonomous, so the role of Nrf1 in erythroid development, and in proteasome function therein, remains unknown.54 We used drugs and shRNAs to show that Nrf1 mediates erythroid proteasome up-regulation in β-thalassemia. TCF11, the human isoform of Nrf1 known to activate proteasome subunit gene transcription, is normally constitutively degraded via the proteasome.38 Therefore, proteasome inhibition stabilizes TCF11, which translocates to the nucleus and activates transcription via AREs. Mechanistically, this likely occurs through proteotoxic stress and proteasome “clogging” by aggregated α subunits in β-thalassemia. Pharmacologic up-regulation of Nrf1 activity may be a potential route to further increase proteasome activity in thalassemic mice and patients in future studies.
Systemic bortezomib impaired erythropoiesis in a dose-dependent fashion, which is consistent with in vivo studies of proteasome inhibition42 and reports of anemia as a drug toxicity in patients.55 Contrary to our initial expectations, bortezomib did not produce a disproportionately adverse effect or enhance α-globin accumulation in β-thalassemic mice. This may be explained by our observations that multiple erythroid PQC pathways are activated additively by β-thalassemia and systemic protease inhibition. These induced pathways include an HSP response, which produces molecular chaperone proteins that bind denatured “client” proteins to either promote their refolding or target them for degradation (for review, see Weiss and Dos Santos56 ). In addition, thalassemia and bortezomib induced the autophagosome marker LC3b additively, suggesting increased autophagic flux. Moreover, free α-globin is degraded by autophagy in thalassemic reticulocytes (Figure 6). Our findings are consistent with the concept that PQC pathways are interrelated and interactive. Proteasome inhibition induces both HSPs40 and macroautophagy57,58 by causing the accumulation of denatured proteins, which activate various cellular unfolded protein responses. These compensatory pathways are more likely to be induced in vivo in early-stage erythroid precursors during the relatively long time course of systemic bortezomib administration compared with brief ex vivo protease inhibitor treatment of isolated late-stage fetal liver erythroblasts or transcriptionally inert reticulocytes. In the future, it will be of interest to investigate how in vivo suppression of autophagy affects β-thalassemia. The use of lysosomal inhibitors such as chloroquine is complicated by the potential for autophagy-independent effects, such as interference with iron metabolism, which would impair erythropoiesis. Most likely, erythroid-specific suppression of autophagy pathways via genetic manipulation in mice will be the best initial approach to investigate whether β-thalassemia is sensitive to the loss of this pathway with or without proteasome inhibition.
The results of the present study indicate that β-thalassemic erythroid cells compensate for α-globin excess through multiple, functionally interconnected PQC pathways, and that decreased flux through a single pathway can be compensated for by enhanced activity of the others. Up to a certain threshold level, combined PQC pathways can detoxify most excess α-globin to minimize clinical phenotypes. Therefore, β-thalassemia, and potentially other hemoglobinopathies, can be studied using the same tools and models currently being applied to a broad range of protein-aggregation disorders. Given the high degree of clinical variability in β-thalassemic patients, some of which cannot be explained by β-globin gene mutations or compensatory γ-globin expression, it may be informative to examine genetic variation of PQC pathways in these patients. Moreover, improved understanding of how erythroblasts modulate PQC components in response to the accumulation of free α-globin may illustrate new pharmacologic and genetic means with which to up-regulate the activity of these pathways for treating β-thalassemia and other protein-aggregation disorders affecting nonerythroid tissues.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Don Baldwin at the University of Pennsylvania microarray core facility and Zhe Zhang at the Children's Hospital of Philadelphia Bioinformatics Core facility for assistance with design and analysis of the microarray experiments, and Gerd Blobel, David Nathan, and Vijay Sankaran for thoughtful comments on the manuscript.
This work was supported by the National Institutes of Health (grants DK061692, HL087427, and P30DK090969 to M.J.W.). E.K. was a trainee under a National Heart, Lung, and Blood Institute Medical Scientist Training Program grant (3T32GM007170-35S1). The DiGaetano family also provided generous support.
National Institutes of Health
Authorship
Contribution: E.K. designed and conducted the experiments, analyzed the data, and wrote the manuscript; C.S.T. designed and conducted the experiments and analyzed the data; J.D. conducted the experiments and analyzed the data; and M.J.W. designed the experiments, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mitchell J. Weiss, Division of Hematology, Children's Hospital of Philadelphia, 316B Abramson Research Center, Philadelphia, PA 19104; e-mail: weissmi@e-mail.chop.edu.