Abstract
Clinical-grade T cells are genetically modified ex vivo to express a chimeric antigen receptor (CAR) to redirect specificity to a tumor associated antigen (TAA) thereby conferring antitumor activity in vivo. T cells expressing a CD19-specific CAR recognize B-cell malignancies in multiple recipients independent of major histocompatibility complex (MHC) because the specificity domains are cloned from the variable chains of a CD19 monoclonal antibody. We now report a major step toward eliminating the need to generate patient-specific T cells by generating universal allogeneic TAA-specific T cells from one donor that might be administered to multiple recipients. This was achieved by genetically editing CD19-specific CAR+ T cells to eliminate expression of the endogenous αβ T-cell receptor (TCR) to prevent a graft-versus-host response without compromising CAR-dependent effector functions. Genetically modified T cells were generated using the Sleeping Beauty system to stably introduce the CD19-specific CAR with subsequent permanent deletion of α or β TCR chains with designer zinc finger nucleases. We show that these engineered T cells display the expected property of having redirected specificity for CD19 without responding to TCR stimulation. CAR+TCRneg T cells of this type may potentially have efficacy as an off-the-shelf therapy for investigational treatment of B-lineage malignancies.
Introduction
Allogeneic hematopoietic stem cell transplantation (HSCT) can cure some patients with high risk B-cell leukemia/lymphoma, but relapse remains a major cause of death. To improve the graft-versus-leukemia/lymphoma (GVL)–effect, donor-derived T cells can be genetically modified to express a tumor-specific chimeric antigen receptor (CAR) with specificity derived from the variable domains of a monoclonal antibody, thus focusing immunoreactivity toward the tumor in an major histocompatibility complex (MHC) nonrestricted manner.1 However, the endogenous αβ T-cell receptor (TCR) on infused allogeneic T cells may recognize major and minor histocompatibility antigens in the recipient leading to graft-versus-host-disease (GVHD). As a result, the majority of current clinical trials infuse autologous CAR+ T cells relying on immune tolerance to prevent TCR-mediated deleterious recognition of normal tissues after adoptive transfer.2 This approach has achieved initial clinical successes targeting CD19+ malignancies,3-7 but is limited by the time and expense to manufacture patient-specific T-cell products. Our goal is to generate off-the-shelf universal CAR+ T cells from allogeneic healthy donors, which can be administered to any patient without causing GVHD.
CD19 is constitutively expressed on most acute and chronic B-cell malignancies. Therefore, to target malignant B cells, we have adapted the Sleeping Beauty (SB) transposon/transposase system for human application to stably express a CD19-specific CAR (designated CD19RCD28).8-11 SB-modified CAR+ T cells can be numerically expanded to clinically sufficient numbers by the recursive addition of γ-irradiated artificial antigen presenting cells (aAPCs) that coexpress CD19 and desired T cell costimulatory molecules.12,13 These platforms have been adapted for human application as clinical trials based on the electroporation and propagation of CAR+ T cells have achieved institutional and federal regulatory approvals for the adoptive transfer of patient-derived and allogeneic CD19RCD28+ T cells after autologous and allogeneic HSCT (investigational new drug nos. 14193, 14577, 14739).2,8,10,11
To test the feasibility of using allogeneic CAR+ T cells we modified the culturing process for generating CAR+ T cells (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) to include the editing of the genome of CARneg and CAR+ T cells to irreversibly eliminate expression of the αβ TCR. To knockout the αβ TCR loci we used zinc finger nucleases (ZFNs),14 comprised of zinc finger protein DNA binding domains fused to the DNA cleavage domain from the Fok I endonuclease, targeting genomic sequences in the constant regions of the endogenous α or β subunits of the TCR. ZFNs mediate genome editing by catalyzing the formation of a DNA double strand break (DSB) in the genome. Targeting a DSB to a predetermined site within the coding sequence of a gene was previously shown to lead to permanent loss of functional target gene expression via repair by nonhomologous end joining, an error-prone cellular repair pathway that results in the insertion or deletion of nucleotides at the cleaved site.15,16
Here we demonstrate that ZFNs targeting either the α or β chains of endogenous TCRs in T cells resulted in the desired loss of TCR expression. As expected, these modified T cells did not respond to TCR stimulation, but maintained their CAR-mediated redirected specificity for CD19.
Methods
Human subjects
Peripheral blood mononuclear cells (PBMCs) were obtained from healthy adult volunteer donors who had provided informed consent from the Gulf Coast Regional Center (Houston, TX). Primary tumor cells were obtained after informed consent from patients at MD Anderson Cancer Center with chronic lymphocytic leukemia (CLL), mantle cell lymphoma (MCL), and diffuse large B-cell lymphoma. Clinical research was in accordance with the Declaration of Helsinki and approved by MD Anderson Cancer Center.
ZFNs targeting constant regions of α and β TCRs
ZFNs containing 5 or 6 fingers were assembled from an established archive of prevalidated 2-finger and 1-finger modules as described.17,18 The ZFN pairs designed to bind either a sequence within exon 1 of the TCR α constant region (TRAC: NG_001332.2; ZFNs designated as TRAC-ZFN-1 and TRAC-ZFN-2) or a consensus sequence common to exon 1 of both TCR β constant regions 1 and 2 (TRBC1 and TRBC2: NG_001333.2; ZFNs designated as TRBC-ZFN-1 andTRBC-ZFN-2), described in detail elsewhere.19 Genes encoding the ZFNs were assembled using polymerase chain reaction (PCR)–based methodology and cloned into a DNA expression plasmid (pVAX; Invitrogen). These plasmids were linearized with XhoI and the RiboMAX Large Scale RNA Production System-T7 (Promega) with ARCA cap analog (Ambion) was used to produce and cap mRNA. After in vitro transcription polyadenines were added using a poly A tailing kit (Ambion), the integrity and size of the mRNA species was validated on a denaturing 1% agarose gel with 3-(N-morpholino) propanesulphonic acid (MOPS) buffer and concentration was measured using a spectrophotometer (BioRad) at OD260. The mRNA was stored at −80°C in nuclease-free vials for single use.
Flow cytometry
The following monoclonal antibodies (mAbs) and reagents were used with indicated specificity and the appropriate isotype controls. From BD Bioscience: phycoerythrin (PE)–conjugated anti-CD3ϵ (clone SK7), PE–anti-CD19 (clone HIB-19), PE-Cy5 CD45RA (clone 5H9), PE-CD56 (clone B159), PE-CD62L (clone Dreg 56), PE-CD64 (clone 10.1), PE-CD86 (clone 2331), PE-CD137L (clone C65-485), fluorescein isothiocyanate (FITC)–conjugated anti-CD4 (clone RPA-T4), allophycocyanin (APC)–conjugated anti-CD4 (clone SK3), FITC–anti-CD8 (clone HIT8a), APC-conjugated anti-CD8 (clone SK1), PE–anti-TCRαβ (clone T10B9.1A-31), APC–anti-TCR γδ (clone B1), PE-mouse immunoglobulin (Ig) G2bκ, APC-mouse IgG1, and FITC-mouse IgG1. From Jackson ImmunoResearch Laboratories: PE-anti–mouse Fab (H+L), Alexa 488– or Alexa 647–conjugated CAR-specific antibody (clone 136-20-1) that recognizes an epitope within scFv region of CD19RCD28 was generated in our laboratory. TCR Vβ usage was analyzed by a panel of anti-Vβ mAb (IOTest Beta Mark; Beckman Coulter). We added propidium iodide (PI; Sigma-Aldrich) just before collecting cells on a flow-cytometer to exclude dead cells from analysis. Data were acquired on a fluorescence-activated cell sorter (FACS) Calibur (BD Biosciences) using CellQuest Version 3.3 (BD Bioscience) and analyzed by FCS Express Version 3.00 (De Novo Software) or FlowJo Version 7.6.1 (TreeStar).
Artificial antigen presenting cells
K562-derived aAPCs were previously modified by lentiviral transduction to constitutively coexpress CD19, CD64, CD86, CD137L, membrane-bound (MB) IL-15 and enhanced green fluorescent protein (EGFP; the latter encoded following the EMCV IRES element). A clone (#4) was obtained by limiting dilution and numerically expanded for use.20 For some experiments CD3-specific antibody (OKT3; eBioscience) was used to activate T cells by pulsing the mAb onto the CD64+ (FcR) clone #4 (supplemental Figure 2). Expression of desired transgenes and bound OKT3 was validated weekly by flow cytometry before use in coculture with T cells.
Propagation of primary T cells
Healthy donor derived PBMCs were isolated by density gradient separation using Ficoll-Paque Plus (GE Healthcare). T cells were numerically expanded in the presence of 50 IU/mL of recombinant human IL-2 (rhIL-2; added 3 times/week; Chiron) on γ-irradiated (100 Gy) aAPC (clone #4, 1:2 T cell: aAPC ratio) that had been preloaded with OKT3. T cells with aAPCs were cultured in complete medium (CM) defined as Hyclone-RPMI 1640 (Thermo Fisher Scientific) supplemented with 2mM l-glutamine (Glutamax-1: Invitrogen) and 10% heat-inactivated Hyclone-FBS (Thermo Fisher Scientific).
Generation and Propagation of CAR+ T cells
DNA supercoiled plasmids (15 μg of CD19RCD28/pSBSO and 5 μg of pKan-CMV-SB11)13 encoding the SB transposon (to stably express CD19RCD28) and the SB transposase (to transiently express SB11) were electrotransferred using an Amaxa Nucleofector II device (Lonza) at 2 × 107 PBMCs/cuvette as previously described (supplemental Figure 1).12 T cells expressing CD19RCD28 were preferentially propagated in CM by recursive addition every 7 or 14 days of clone #4 (not loaded with OKT3) at 1:2 T cell: aAPC (γ-irradiated to 100 Gy) ratio in the presence of rhIL-2 50 IU/mL, added 3 times a week.
Electrotransfer of messenger RNA species into primary or CAR+ T cells
Six days after stimulation of unmodified T cells with OKT3-loaded clone #4 or 2 to 4 days after the last stimulation of CD19RCD28+ T cells with clone #4, 5 × 106 T cells were mixed with 2.5 to 10.0 μg of each ZFN mRNA in 100 μL of human T-cell nucleofector solution (Lonza) and electroporated using the Nucleofector II device with program T-20. After electroporation, cells were immediately placed in 2 mL of prewarmed CM and cultured at 37°C, 5% CO2 for 4 to 6 hours and then 50 IU/mL of rhIL-2 was added with 2 mL of 20% FBS-RPMI. In some experiments to enhance ZFN-mediated enzymatic activity, after overnight culture, cells were transferred to 30°C, 5% CO2, and then cultured for 2 days then returned to 37°C 5% CO2.
Enrichment of CD3neg T cells
Cells washed with phosphate-buffered saline (PBS) supplemented with 2% FBS and 2mM ethylenediaminetetraacetic acid, were incubated for 10 minutes with CD3 microbeads (Milteneyi Biotec) at 4°C. After washing twice, cells were passed through a LD column (Milteneyi Biotec), and the flow-through fraction was collected for further use.
Surveyor nuclease assay
The levels of genomic disruption of TRAC, TRBC1, and TRBC2 in T cells were determined by surveyor nuclease assay (Transgenomics) using CEL I nuclease.21 The percentage target disruption was quantified by densitometry. The PCR primers used for the amplification of target locus were: TRAC forward 5′-GGGCAAAGAGGGAAATGAGA-3′; TRAC reverse 5′-CAATGGATAAGGCCGAGACC-3′; TRBC1 forward, 5′-CTGAACAAGGTGTTCCCACCC-3′; TRBC1 reverse 5′-GTGTGCGCTGGTTCCTTTCTT-3′; TRBC2 forward, 5′-CCTGGCCACAGGCTTCTACC-3′; TRBC2 reverse 5′-CCACCTTGTCCACTCTGGCTT-3′.
51Chromium release assay
Target cells were labeled with 0.1 mCi of 51Chromium (51Cr; PerkinElmer) for 2 hours. After washing thrice with ice-cold CM, labeled cells were diluted and plated at 103 cells/well in 100 μL CM in 96-well v-bottomed plates. T cells were added in 100 μL/well at indicated effector target ratios and the plate was spun (180g for 3 minutes without brake) to facilitate cell-to-cell contact. After 4 or 6 hours (when using primary tumor cells as targets) incubation at 37°C, 5% CO2, 50 μL of supernatants were counted on TopCount (PerkinElmer). All assays were performed in triplicate. The percentage specific lysis was calculated as follows: (experimental cpm − spontaneous cpm) / (maximum cpm − spontaneous cpm) × 100.
PKH-26 dilution assay
T cells were incubated with 2.0μM of the red-fluorescent lipophilic dye PKH-26 (Sigma-Aldrich) for 5 minutes at room temperature according to the manufacturer's instructions. Cells, 100% labeled with PKH-26, were stimulated with either OKT3 loaded aAPCs or CD19+ aAPCs in CM supplemented with 50 IU/mL rhIL-2 (added every other day). PKH-26–derived fluorescence was measured by flow cytometry 10 days after stimulation and CD19RCD28+ T cells were revealed using anti-CAR mAb clone 136-20-1.
Results
Disruption of the αβ TCR-CD3 complex on T cells using ZFNs
Two ZFN pairs targeting the constant regions of TCR α (TRAC-ZFN-1 and TRAC-ZFN-2) or TCR β (TRBC-ZFN-1 and TRBC-ZFN-2; Figure 1) were developed and tested in primary human T cells propagated ex vivo for 6 days on OKT3-loaded aAPCs (clone #4). Because transient expression of ZFNs is sufficient to mediate gene knockout, we used a “hit-and-run” delivery strategy to transiently express the ZFNs using electrotransfer of in vitro transcribed mRNA species coding for the ZFN pairs (Figure 2A). To measure TCR expression we used a mAb specific for CD3ϵ, which is only present on the cell surface when TCRαβ is expressed. Nine days after electrotransfer, flow cytometric analysis revealed that ZFN pairs targeting TRAC or TRBC eliminated CD3ϵ expression on primary T cells at levels reaching 19.4% and 5.2%, respectively. The efficiency of TCR knockout correlated with the amount of electrotransferred mRNA (Figure 2B top panel). Electro-transfer of mRNA to primary T cells was generally well-tolerated, though a slight reduction in cell viability was observed at higher doses. ZFN-mediated gene disruption has been reported to be more efficient when cells are transiently exposed to mild hypothermia.22 Thus, we cultured T cells for 2 days at 30°C after electrotransfer. ZFN-mediated disruption of CD3ϵ was up to 2.4-fold higher when electroporated T cells were cultured at 30°C versus 37°C. Using this approach, 37% and 15% of electroporated T cells lost expression of CD3ϵ using the ZFN pair targeting TRAC and TRBC, respectively (Figure 2B bottom panel) with no change in the levels of CD3 negative cells in the untransfected samples and without an appreciable decrease in viability (measured by Trypan blue).
ZFN pairs targeting sites within genomic loci of TCR-α and β constant region. Each exon is shown by a block. Black blocks represent coding regions. Gray columns represent noncoding regions. One ZFN pair was designed to bind exon 1 of the TCR α constant region (TRAC) and another ZFN pair binds a conserved sequence on exon 1 of the TCR β constant regions 1 (TRBC1) and 2 (TRBC2). Underlined nucleotide sequences represent the intended binding sequence of each ZFN.
ZFN pairs targeting sites within genomic loci of TCR-α and β constant region. Each exon is shown by a block. Black blocks represent coding regions. Gray columns represent noncoding regions. One ZFN pair was designed to bind exon 1 of the TCR α constant region (TRAC) and another ZFN pair binds a conserved sequence on exon 1 of the TCR β constant regions 1 (TRBC1) and 2 (TRBC2). Underlined nucleotide sequences represent the intended binding sequence of each ZFN.
Disruption of the TCR αβ-CD3 complex in primary T cells. (A) Schematic presentation of ZFN transfer. A pair of ZFN-encoding mRNA was electrotransferred 6 days after stimulation of CARneg T cells. T cells were then cultured with 50 IU/mL of IL-2 and incubated at 30°C or 37°C 5%CO2, as indicated. CD3 expression was analyzed by flow cytometry on day 7 to 9 after electroporation. (B) Down-regulation of CD3 after electrotransfer of mRNA encoding the TCR αβ targeted ZFNs. Day 9 after electrotransfer of the indicated doses of mRNA coding for TRAC or TRBC targeted ZFN pairs, TCR αβ-CD3 expression was analyzed by costaining for CD4, CD8, and CD3ϵ. Representative flow data at day 9 after ZFN electrotransfer is shown. Flow cytometry data are gated on cells excluding PI. Numbers in the lower right quadrant represent the percentage of CD3ϵ negative cells in T-cell populations. Top panels show CD3ϵ expression in T cells cultured at 37°C after ZFN transfer and bottom panels show CD3ϵ expression in T cells transiently cultured at 30°C from day 2 to 3 after ZFN transfer. (C) Surveyor nuclease assay to detect ZFN-mediated modification of TCR target sites in T cells. Arrows indicate the fragments produced by a surveyor nuclease digest of amplicons bearing a mismatch at the intended site of ZFN cleavage in the TRAC or TRBC1 locus, respectively. Lane headings indicate both the mRNA dose, specific ZFN pair delivered via electrotransfer, and temperature of incubation for the different samples. Numbers beneath each lane indicate the percentage of modified alleles in each sample.
Disruption of the TCR αβ-CD3 complex in primary T cells. (A) Schematic presentation of ZFN transfer. A pair of ZFN-encoding mRNA was electrotransferred 6 days after stimulation of CARneg T cells. T cells were then cultured with 50 IU/mL of IL-2 and incubated at 30°C or 37°C 5%CO2, as indicated. CD3 expression was analyzed by flow cytometry on day 7 to 9 after electroporation. (B) Down-regulation of CD3 after electrotransfer of mRNA encoding the TCR αβ targeted ZFNs. Day 9 after electrotransfer of the indicated doses of mRNA coding for TRAC or TRBC targeted ZFN pairs, TCR αβ-CD3 expression was analyzed by costaining for CD4, CD8, and CD3ϵ. Representative flow data at day 9 after ZFN electrotransfer is shown. Flow cytometry data are gated on cells excluding PI. Numbers in the lower right quadrant represent the percentage of CD3ϵ negative cells in T-cell populations. Top panels show CD3ϵ expression in T cells cultured at 37°C after ZFN transfer and bottom panels show CD3ϵ expression in T cells transiently cultured at 30°C from day 2 to 3 after ZFN transfer. (C) Surveyor nuclease assay to detect ZFN-mediated modification of TCR target sites in T cells. Arrows indicate the fragments produced by a surveyor nuclease digest of amplicons bearing a mismatch at the intended site of ZFN cleavage in the TRAC or TRBC1 locus, respectively. Lane headings indicate both the mRNA dose, specific ZFN pair delivered via electrotransfer, and temperature of incubation for the different samples. Numbers beneath each lane indicate the percentage of modified alleles in each sample.
To confirm that electroporated T cells had been genetically modified at the intended ZFN target sites (TCR α or β loci), a surveyor nuclease assay was performed using specific oligonucleotide primers flanking target sites within TRAC, TRBC1, or TRBC2. CEL I nuclease digestion products, representative of genetic changes induced by the ZFNs, were present only after electrotransfer of the TCR-specific ZFN pairs and the percentage disruption assessed by densitometry correlated with loss of cell surface CD3ϵ expression (Figure 2C). These experiments in primary T cells confirmed that ZFNs designed to target TRAC or TRBC lead to permanent disruption of αβTCR expression, as assessed by the CEL I–mediated surveyor nuclease assay and confirmed by flow cytometry analysis of CD3ϵ.
Enrichment of TCRneg T cells
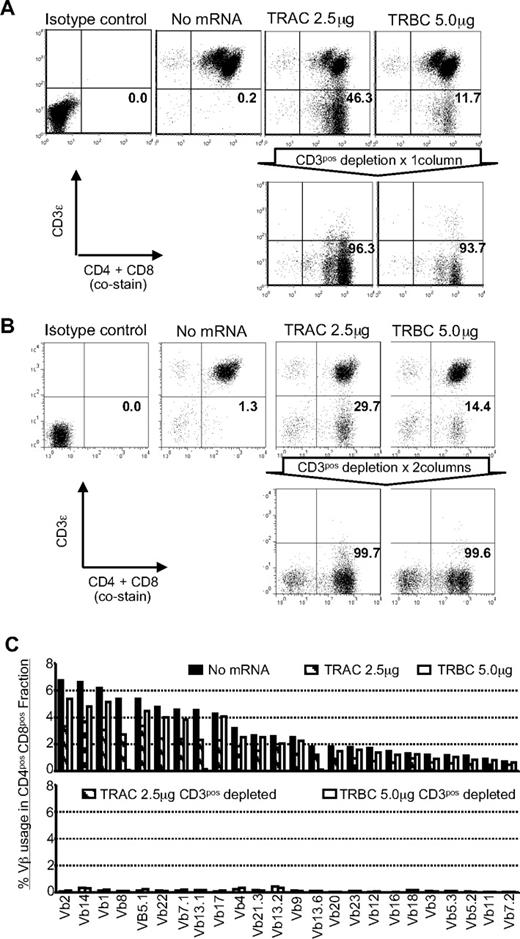
For future clinical applications, rapid and robust methods for isolating sources of a TCR-disrupted population will be needed. To address this issue, we enriched the TCR/CD3neg population by negative selection using paramagnetic beads and a depletion column. With a single depletion step, we enriched the CD3ϵneg population to more than 93% (Figure 3A). A CD3ϵneg population could not be enriched from control T cells that were not genetically edited with ZFNs. Back-to-back CD3-depletion resulted in > 99% enrichment without skewing the CD4+ or CD8+ T-cell subsets (Figure 3B). The depletion of CD3+ T cells will also deplete remaining γδTCR+ T cells. A flow cytometry analysis of TCR Vβ repertoire in enriched TCRneg T cells validated the elimination of TCRβ expression from the T-cell surface (Figure 3C). This degree of depletion is clinically appealing as the loss of TCR on donor-derived T cells will prevent GVHD in human leukocyte Ag (HLA)–disparate recipients.
TCRneg T cells can be enriched by depletion of CD3ϵ+ T cells. (A) CD3 expression before and after depletion using CD3-specific paramagnetic beads. Flow cytometry reveals expression of CD3ϵ in CD4+ and CD8+ T cells 15 days after stimulation by OKT3-loaded aAPC (9 days after ZFN transfection). Numbers in the bottom-right quadrant represent the percentage of CD3ϵneg T cells. Representative results using in vitro numerically expanded T cells. (B) CD3ϵneg T cells can be further enriched by additional round of depletion with CD3-specific paramagnetic beads. Flow cytometry revealing expression of CD3ϵ in CD4+ and CD8+ T cells after 2 rounds of depletion of CD3ϵpos T cells. Numbers in the lower right quadrant represents the percentage of CD3ϵ negative cells in CD4+ and CD8+ T-cell populations. (C) Vβ repertoire analysis in T cells modified with ZFNs. The Vβ usage clonogram was analyzed by a panel of TCR-specific mAbs, costained with CD4 and CD8. Percentage of specific Vβ+ T-cell fractions within CD4 and CD8 gating is shown. The nomenclatures of Vβ repertoire shown are based on Arden et al.50 Representative data from 3 independent experiments are shown.
TCRneg T cells can be enriched by depletion of CD3ϵ+ T cells. (A) CD3 expression before and after depletion using CD3-specific paramagnetic beads. Flow cytometry reveals expression of CD3ϵ in CD4+ and CD8+ T cells 15 days after stimulation by OKT3-loaded aAPC (9 days after ZFN transfection). Numbers in the bottom-right quadrant represent the percentage of CD3ϵneg T cells. Representative results using in vitro numerically expanded T cells. (B) CD3ϵneg T cells can be further enriched by additional round of depletion with CD3-specific paramagnetic beads. Flow cytometry revealing expression of CD3ϵ in CD4+ and CD8+ T cells after 2 rounds of depletion of CD3ϵpos T cells. Numbers in the lower right quadrant represents the percentage of CD3ϵ negative cells in CD4+ and CD8+ T-cell populations. (C) Vβ repertoire analysis in T cells modified with ZFNs. The Vβ usage clonogram was analyzed by a panel of TCR-specific mAbs, costained with CD4 and CD8. Percentage of specific Vβ+ T-cell fractions within CD4 and CD8 gating is shown. The nomenclatures of Vβ repertoire shown are based on Arden et al.50 Representative data from 3 independent experiments are shown.
Generation of TCRnegCAR+ T cells by ZFNs
To test the ability of ZFN pairs to knockout TCR αβ expression from allogeneic CD19RCD28+ T cells, we initially genetically modified PBMCs to stably express the CD19RCD28 CAR using the SB transposon/transposase system. The CD19RCD28+ T-cell population was specifically propagated by stimulating with γ-irradiated CD19+ aAPCs (clone #4) every 7 days (supplemental Figure 1B). After 4 rounds of stimulation, we observed more than 90% CAR expression in T cells similar to our previously published results.12 Within 2 to 4 days after the fifth stimulation with CD19+ aAPCs, when T cells were activated, we electroporated the cells with mRNA encoding the TRAC or TRBC ZFNs (Figure 4A). Flow cytometry analysis revealed that up to 30% and 26% of CD19RCD28+ T cells lost CD3ϵ expression after transfection of the TRAC or TRBC ZFNs, respectively (Figure 4B). The CD3ϵneg population was again readily enriched by paramagnetic beads, and the surveyor nuclease assay confirmed that the CD3ϵneg population contained a high percentage of modified alleles at the intended ZFN target sites within the TRAC and TRBC loci (Figure 4C). The frequency of TRBC1 and TRBC2 disruption at the DNA level was approximately 20%-25% and that of TRAC disruption was approximately 60%. These numbers fit with the observed frequencies of CD3ϵnegCD19RCD28+ T cells because in each cell only 1 of 4 TRBC alleles (2 TRBC1 and 2 TRBC2) is expressed. Similarly 1 of 2 TRAC alleles is expressed in each T cell. Therefore, disruption of the expressed allele is sufficient to achieve the CD3 negative phenotype.
Elimination of TCR αβ-CD3 complex from CD19-specific CAR+ T cells. (A) Schematic of electrotransfer of mRNA coding for ZFN pairs in CAR+ T cells. mRNA species encoding the indicated ZFN pairs were electrotransferred into CAR+ T cells 2 days after stimulation with CD19+ aAPC. After electroporation, cells were maintained with 50 IU/mL of IL-2 and incubated for 2 days at 30°C 5%CO2. CD3ϵ expression was analyzed 9 days after electroporation by flow cytometry. (B) Disruption of TCR αβ-CD3 complex expression after electrotransfer of mRNA encoding the TCR-specific ZFNs. Flow cytometry analysis of CD3ϵ expression in T cells 9 days after electrotransfer of mRNA species encoding the indicated ZFN pairs, gated on the PI-negative population. (C) Surveyor nuclease assay. Arrows indicate the fragments produced by a surveyor nuclease digest of amplicons bearing a mismatch at the intended site of ZFN cleavage in the TRAC or TRBC loci, respectively. Samples were analyzed 9 days after electroporation. The numbers at the bottom represent percentages of modified alleles in each sample.
Elimination of TCR αβ-CD3 complex from CD19-specific CAR+ T cells. (A) Schematic of electrotransfer of mRNA coding for ZFN pairs in CAR+ T cells. mRNA species encoding the indicated ZFN pairs were electrotransferred into CAR+ T cells 2 days after stimulation with CD19+ aAPC. After electroporation, cells were maintained with 50 IU/mL of IL-2 and incubated for 2 days at 30°C 5%CO2. CD3ϵ expression was analyzed 9 days after electroporation by flow cytometry. (B) Disruption of TCR αβ-CD3 complex expression after electrotransfer of mRNA encoding the TCR-specific ZFNs. Flow cytometry analysis of CD3ϵ expression in T cells 9 days after electrotransfer of mRNA species encoding the indicated ZFN pairs, gated on the PI-negative population. (C) Surveyor nuclease assay. Arrows indicate the fragments produced by a surveyor nuclease digest of amplicons bearing a mismatch at the intended site of ZFN cleavage in the TRAC or TRBC loci, respectively. Samples were analyzed 9 days after electroporation. The numbers at the bottom represent percentages of modified alleles in each sample.
TCRnegCAR+ T cells do not respond to TCR stimulation, but do maintain CD19 specificity
We anticipated that TCRnegCAR+ T cells could not respond to TCR stimulation. To test this, we measured the proliferative response of these cells after stimulation by cross-linking CD3 with OKT3 in comparison to activating CAR for sustained proliferation on docking with CD19. TCRnegCD19RCD28+ T cells proliferated in response to CD19, but not OKT3 (Figure 5A). Next, we assessed the ability of TCRnegCD19RCD28+ T cells to lyse CD19+ target cells in a standard 4-hour 51Cr release assay (Figure 5B). The capacity of TCRnegCAR+ T cells to specifically lyse CD19 target cells was similar to that observed for TCR+CD19RCD28+ T cells. Moreover, these TCRnegCAR+ T cells maintain cytotoxicity against CD19+ primary tumors (Figure 5C). Together, these data confirmed that the absence of a measurable TCR on TCRnegCD19RCD28+ T cells corresponds with abrogation of TCR activity, but does not impact the ability of the CAR to activate genetically modified T cells for proliferation and target cell killing.
Functional consequences of ZFN-mediated TCR knockout in CAR+ T cells. (A) Loss of responsiveness of TCRneg CAR+ T cells to TCR stimulation. Dilution of PKH26 was measured 10 days after stimulation with aAPC loaded with OKT3 (top panel) or expressing CD19 (bottom panel). Flow cytometry data were gated on CAR+ T cells. Parental: CAR+ T cells without modification; no mRNA: mock electroporated CAR+ T cells; TRAC CD3neg: CAR+ T cells electroporated with mRNA encoding ZFN pairs specific for TRAC, and depleted CD3pos population; TRBC CD3neg: CAR+ T cells electroporated with mRNA encoding ZFN pairs specific for TRBC, and depleted for CD3pos population. (B) Redirected specificity of TCRneg CAR+ T cells. Specific lysis by CAR+ T cells of an EL4 (mouse T-cell line) modified to express a truncated version of human CD19 (closed symbols) was measured by standard 4 hour 51Cr release assay. Specificity is shown by lack of lysis of CD19neg (parental) EL4 cells (open symbols). CAR+ T cells were modified by ZFNs (TRAC and TRBC) or unmodified CAR+ T cells (parental and no mRNA). The error bars represent SD. (C) Cytotoxicity by TCRneg CAR+ T cells against CD19+ primary B-cell tumors. Specific lysis by CAR+ T cells of B-cell malignances derived from patients was measured by 6 hour 51Cr release assay (effector: target ratio = 30:1). DLBCL: diffuse large B-cell lymphoma, CLL: chronic lymphocytic lymphoma, and MCL: mantle cell lymphoma. The error bars represent the standard deviation.
Functional consequences of ZFN-mediated TCR knockout in CAR+ T cells. (A) Loss of responsiveness of TCRneg CAR+ T cells to TCR stimulation. Dilution of PKH26 was measured 10 days after stimulation with aAPC loaded with OKT3 (top panel) or expressing CD19 (bottom panel). Flow cytometry data were gated on CAR+ T cells. Parental: CAR+ T cells without modification; no mRNA: mock electroporated CAR+ T cells; TRAC CD3neg: CAR+ T cells electroporated with mRNA encoding ZFN pairs specific for TRAC, and depleted CD3pos population; TRBC CD3neg: CAR+ T cells electroporated with mRNA encoding ZFN pairs specific for TRBC, and depleted for CD3pos population. (B) Redirected specificity of TCRneg CAR+ T cells. Specific lysis by CAR+ T cells of an EL4 (mouse T-cell line) modified to express a truncated version of human CD19 (closed symbols) was measured by standard 4 hour 51Cr release assay. Specificity is shown by lack of lysis of CD19neg (parental) EL4 cells (open symbols). CAR+ T cells were modified by ZFNs (TRAC and TRBC) or unmodified CAR+ T cells (parental and no mRNA). The error bars represent SD. (C) Cytotoxicity by TCRneg CAR+ T cells against CD19+ primary B-cell tumors. Specific lysis by CAR+ T cells of B-cell malignances derived from patients was measured by 6 hour 51Cr release assay (effector: target ratio = 30:1). DLBCL: diffuse large B-cell lymphoma, CLL: chronic lymphocytic lymphoma, and MCL: mantle cell lymphoma. The error bars represent the standard deviation.
TCRneg CD19RCD28+ T cells can be propagated on CD19 expressing aAPCs
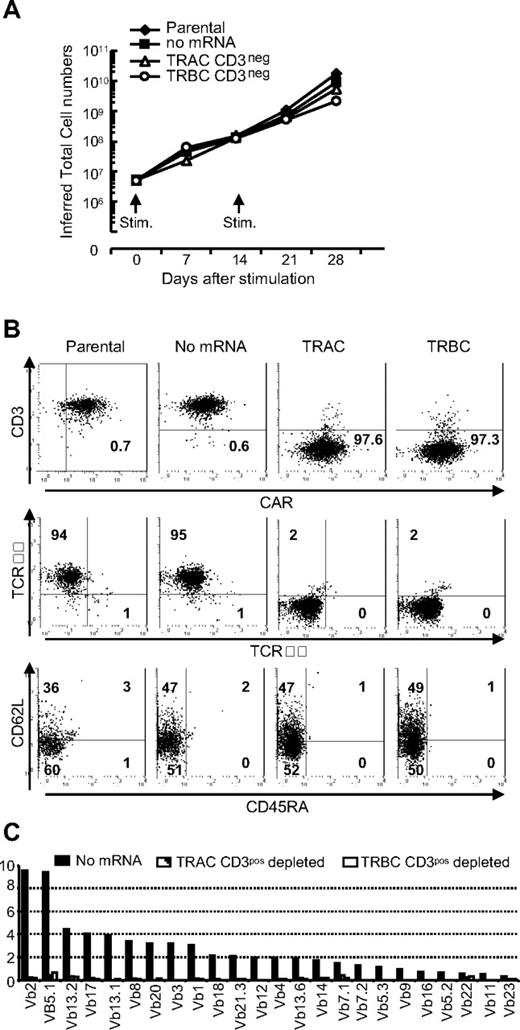
We validated that CD19RCD28+ T cells sustain their proliferative capacity to expand to the cell numbers required for clinical applications. Both the TCRnegCD19RCD28+ and parental TCR+CD19RCD28+ T cells exhibited similar growth kinetics in response to stimulation with the CD19+ aAPCs (Figure 6A). We did not observe any changes in CD3ϵ expression on TCRnegCD19RCD28+ T cells after aAPC-mediated propagation (Figure 6B top panel). As predicted, these T cells failed to express TCRαβ on their cell surface (Figure 6B middle panel; C). As expected, the ZFN-mediated disruption of αβTCR expression and depletion of CD3+ T cells led to loss of γδTCR+ T cells (Figure 6B middle panel). After propagation on aAPC, a subset of TCRnegCD19RCD28+ T cells exhibited memory phenotype based on expression of CD62L and absence of CD45RA23 (Figure 6B bottom panel), which may benefit persistence and thus the therapeutic potential of our approach to “off-the-shelf” adoptive T-cell therapy. These data confirm that TCRnegCAR+ T cells may be able to be propagated to achieve sufficient cell numbers from a single donor-derived modified T-cell pool for infusions into multiple recipients.
Propagation of TCRneg CAR+ T cells on CD19 expressing aAPC. (A) Sustained proliferation of TCRneg CAR+ T cells. CAR+ T cells with (TRAC and TRBC) or without (parental and no mRNA) TCR modification by ZFNs were stimulated with γ-irradiated CD19+ aAPCs every 2 weeks. Viable T cells were enumerated every 7 days and inferred total numbers were calculated. Representative data from 3 independent experiments are shown. (B) Analysis of TCRneg CAR+ T cells after propagation. Flow cytometry analysis of CD3ϵ expression (top panel), αβTCR and γδTCR expression (middle panel), and subset analysis for memory pool (bottom panel) of TCRnegCAR+ T cells after 28 days of propagation on aAPCs. Numbers are percent expression for the quadrant. (C) Vβ repertoire analysis in TCRneg CAR+ T cells after propagation on aAPCs. The Vβ usage clonogram was analyzed by a panel of TCR-specific mAbs, costained with CD4 and CD8. Percentage of identified Vβ+ T-cell fractions within CD4 and CD8 flow cytometry gates is shown. The nomenclatures of Vβ repertoire shown are based on Arden et al.50 Representative data from 3 independent experiments are shown.
Propagation of TCRneg CAR+ T cells on CD19 expressing aAPC. (A) Sustained proliferation of TCRneg CAR+ T cells. CAR+ T cells with (TRAC and TRBC) or without (parental and no mRNA) TCR modification by ZFNs were stimulated with γ-irradiated CD19+ aAPCs every 2 weeks. Viable T cells were enumerated every 7 days and inferred total numbers were calculated. Representative data from 3 independent experiments are shown. (B) Analysis of TCRneg CAR+ T cells after propagation. Flow cytometry analysis of CD3ϵ expression (top panel), αβTCR and γδTCR expression (middle panel), and subset analysis for memory pool (bottom panel) of TCRnegCAR+ T cells after 28 days of propagation on aAPCs. Numbers are percent expression for the quadrant. (C) Vβ repertoire analysis in TCRneg CAR+ T cells after propagation on aAPCs. The Vβ usage clonogram was analyzed by a panel of TCR-specific mAbs, costained with CD4 and CD8. Percentage of identified Vβ+ T-cell fractions within CD4 and CD8 flow cytometry gates is shown. The nomenclatures of Vβ repertoire shown are based on Arden et al.50 Representative data from 3 independent experiments are shown.
Discussion
We demonstrated that T cells and indeed CAR+ T cells can be genetically edited by ZFNs to eliminate expression of the endogenous αβ TCR. This has therapeutic implications where donor-derived T cells are infused to achieve an antitumor effect. Therapeutic success after allogeneic HSCT is defined as achieving a GVL-effect without causing clinically significant GVHD.24 Thus, separation of GVL and GVHD is a crucial issue after engraftment of allogeneic hematopoietic stem cells and strategies to accomplish this are based on infusing desired T-cell effector populations predicted to reduce unwanted allogeneic effects. This includes the adoptive transfer of donor-derived memory T cells using a narrowed TCR Vβ repertoire compared with naive T cells25,26 or in vitro depletion of T cells activated through allo-antigens.27-29 Adding to this approach, we previously demonstrated that CAR+ T cells expressing alloreactive TCRs can be rendered anergic to disparate HLA while maintaining specificity for CD19.13 This was achieved by blockade of costimulatory molecules on coculture of genetically modified T cells with stimulator cells expressing disparate HLA. An alternative to preselection includes conditional ablation of infused allogeneic CAR+ T cells if serious adverse events occur. This has been accomplished by genetic modification of allogeneic T cells to express “suicide genes,” such as thymidine kinase (TK),30 iCasp9,31 CD20,32 thymidylate kinase,33 and a modified Fas34 that can be triggered for conditional ablation via the administration of specific molecules (eg, ganciclovir to TK+ expressing cells).
We recognized that approaches to selectively deplete T cells expressing undesired αβTCRs may be incomplete and that complete knockout of the endogenous TCR might be advantageous to prevent GVHD. Therefore, we undertook a genetic approach using designer ZFNs to permanently disrupt the α and β constant region sequences in T cells thereby eliminating TCR expression. Because TCR αβ receptors need to form heterodimers to express a functional cell surface molecule, knocking out either TRAC or TRBC was sufficient to eliminate TCR αβ expression. This is supported by a recent publication showing that a mutation in the TRAC gene leads to the loss of TCR αβ expression.35
ZFNs have been demonstrated to disrupt target gene expression as a consequence of error-prone DNA DSB repair by nonhomologous end joining, which in most cases results in a frame shift mutation leading to a premature stop of translation.15 This technology is being evaluated in early stage clinical trials infusing HIV-resistant T cells generated by ZFN-mediated disruption of the CCR5 coreceptor for HIV-I.16,36 ZFNs target and thus disrupt gene expression at the genomic level, which is an advantage over techniques that involve transcriptional repression and require sustained expression of the inhibiting factor (eg, enforced expression of shRNA to mediate TCR down-regulation).37 That ZFNs can permanently disrupt gene expression after transient expression (without the inherent dangers of genomic integration) enabled our use of in vitro transcribed mRNA species in a “hit-and-run” manner for electrotransfer of ZFNs into T cells.
The human application of “universal” CAR+ T cells that have been genetically edited with ZFNs will depend on efficacy as well as safety. The genetically modified T cells specifically lysed primary targets and cell lines. Their therapeutic potential is also dependent on persistence after adoptive transfer. The second generation CAR chosen for this study activates T cells through chimeric CD28 and CD3-ζ. It remains to be determined in side-by-side clinical trials whether other CAR designs, such as signaling through CD137 and CD3-ζ, are superior. Safety depends on selective elimination of endogenous TCR and minimizing ZFN-mediated enzymatic activity at off-target sites. We evaluated the most probable “off-target” sites using CEL-I nuclease and did not detect cleavage. We did observe that the efficiency of enzymatic activity at TRAC or TRBC genomic loci is approximately 20%-40% after a single electrotransfer of mRNA species coding for ZFN pairs. However, continued cell-surface expression of TCRs from HLA-disparate candidate T-cell donors may cause GVHD after adoptive immunotherapy. Therefore, to prevent GVHD after infusion we used CD3-specific paramagnetic beads to deplete and redeplete T cells with residual expression of TCRs. This is a clinically appealing strategy as this approach can readily be undertaken in compliance with current good manufacturing practice for phase I/II trials. Our planed clinical trials will include release criteria of the manufactured T-cell product based on residual αβTCR expression and an assessment of the maximum number of genetically modified T cells that can be safely infused from an allogeneic donor into multiple recipients. In addition, the wellbeing of the patients can also be safeguarded by coexpressing CAR with a transgene capable of mediating conditional T-cell ablation.
Previous reports suggest that T-cell activation mediated through an endogenous TCR is required to obtain a fully functional CAR in a model system using Jurkat cell lines.38 In contrast, we observed that knocking out TCR αβ expression from CD19RCD28+ T cells did not appreciably alter the ability of these cells to specifically kill CD19+ targets or proliferate in response to CD19. One reason for this discrepancy other than the difference in host cells may be the use of a second generation CAR, which includes signaling not only through CD3ζ (signal 1) but also CD28 (signal 2; costimulation).39-41 A benefit to expressing a TCR with known specificity is that activation through the endogenous immunoreceptor can be used to propagate T cells to achieve an antitumor effect mediated by the CAR.42,43 It remains to be tested in humans whether coordinated costimulation achieved through multiple CAR signaling endodomains will be sufficient to sustain persistence in vivo or if triggering of T cells through TCRs is needed. However, the propagation of CD19RCD28+ T cells on aAPC modified to coexpress CD19 along with costimulatory molecules results in the significant expansion of CAR+ memory T-cell subsets predicted to have prolonged in vivo survival.44,45 Therefore, any loss of persistence of TCRnegCD19RCD28+ T cells may be offset by costimulatory properties of aAPCs and the encoded CD28 intracellular domain within the CAR.
Preparing antigen-specific T cells from a third-party donor is clinically appealing as these products can be generated, stored, and validated before use and infused to multiple patients immediately as needed.46 Indeed, third-party T cells have been successfully infused into patients with posttransplantation lymphoproliferative diseases.47,48 Although a majority of the viral antigen-specific TCRαβ chains demonstrate cross-reactivity to allo-HLA in vivo,49 clinically significant GVHD was not observed. This may be in part because of the ex vivo repetitive antigen stimulation resulting in the emergence of either an oligoclonal or monoclonal TCRαβ repertoire, which decreases the chance of T-cell alloreactivity. On the contrary, when we numerically expand CD19RCD28+ T cells through in vitro CD19 stimulation on γ-irradiated aAPC independent of TCR stimulation we did not observe skewing of the TCR Vβ usage as assessed by a panel of Vβ-specific antibodies.
In conclusion, we demonstrate that TCRnegCAR+ T cells can be generated using a genetic approach to remove (1) endogenous undesired TCR with ZFNs and (2) introduce a desired CAR with the SB system using a common electrotransfer platform. Our approach abolishes the danger of GVHD posed by adoptive transfer of large numbers of allogeneic T cells while maintaining desired effector functions mediated by CD19RCD28 CAR to target malignant B cells. This strategy provides an important step to developing a “universal” CAR+ T cell, which can be manufactured from one donor and administered on demand to multiple patients. Subsequent studies are focusing on preventing rejection of the infused allogeneic TCRnegCAR+ T cells by the recipient's immune system recognizing disparate HLA. This may be accomplished using genetic modifications including ZFN-mediated knockout of HLA and overexpression of conserved HLA homologues to inhibit natural killer cell activity.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Carl June at the University of Pennsylvania for assistance with K562-derived aAPC clone #4 and Dr Perry Hackett at the University of Minnesota for help with the SB system.
This work was supported by: Cancer Center Core Grant (CA16672); Department of Defense PR064229; RO1 (CA124782, CA120956, CA141303); R33 (CA116127); Burroughs Wellcome Fund; Cancer Prevention Research Institute of Texas; CLL Global Research Foundation; Gillson Longenbaugh Foundation; Harry T. Mangurian Jr Foundation, Institute of Personalized Cancer Therapy; Leukemia and Lymphoma Society; Lymphoma Research Foundation; Miller Foundation; Mr and Mrs Joe H. Scales; National Foundation for Cancer Research; Pediatric Cancer Research Foundation; Production Assistance for Cellular Therapies; Sister Institution Network Fund; Uehara Memorial Foundation; the William Lawrence and Blanche Hughes Children's Foundation; Italian Ministry of Health (GR07-5 BO and RO10/07-B-1); the Italian Ministry of Research and University (FIRB-IDEAS, linked to European Research Council [ERC] starting grants); Fondazione Cariplo and the Italian Association for Cancer Research; the EU (FP7: GA 222878, PERSIST and ERC advanced grant 249845 TARGETINGGENETHERAPY); and Italian Telethon (TELE11/12-12-D2).
National Institutes of Health
Authorship
Contribution: H.T. and A.R. designed and performed experiments analyzed data, and wrote the paper; P.-Q.L., Y.Z., L.Z., S.M., and J.C.M. performed the experiments; H.H. supported the experiments; P.K., B.R., D.A.L., and R.E.C. contributed discussion and edited the paper; E.J.R., P.D.G., and M.C.H. designed experiments, analyzed data, and edited the paper; C.B. and L.N. tested the TRAC and TRBC target ZFNs developed by Sangamo BioSciences, and edited the paper; and L.J.N.C. conceived the idea, coordinated the project, designed experiments, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Laurence J. N. Cooper, University of Texas MD Anderson Cancer Center, Pediatrics-Research, Unit 907, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: ljncooper@mdanderson.org.