To the editor:
Forkhead box P3 (Foxp3) is a transcription factor critical for the differentiation of regulatory T cells (Tregs) and for the prevention of autoimmune disease. Although expression of Foxp3 was initially found to be restricted to CD4+CD25+ T cells,1 some research groups have reported Foxp3 expression in thymic,2 breast,3 lung and prostate epithelium,4 all of which have subsequently been shown by us and others to be unreproducible staining artifacts.5,6 Recently, Manrique et al (since retracted)7 described that Foxp3 is also expressed by a subtype of immunosuppressive macrophages. In light of our experiments on collagen-induced arthritis (CIA), where an important extramedullar expansion of various CD11b+ cells occurs,8 we set out to examine these Foxp3 expressing macrophages. Therefore, we used 2 reporter mice strains. First, the Foxp3GFP strain uses a Foxp3-GFP fusion protein knocked into the Foxp3 locus. Second, the Foxp3Cre strain coexpresses Foxp3 and YFP-Cre from the endogenous Foxp3 promoter. When crossed to the Cre-activated YFP reporter system (Foxp3Cre Rosa-YFP mice), this drives the expression of YFP in any cell lineage that has a history of Foxp3 expression, with essentially 100% efficiency.
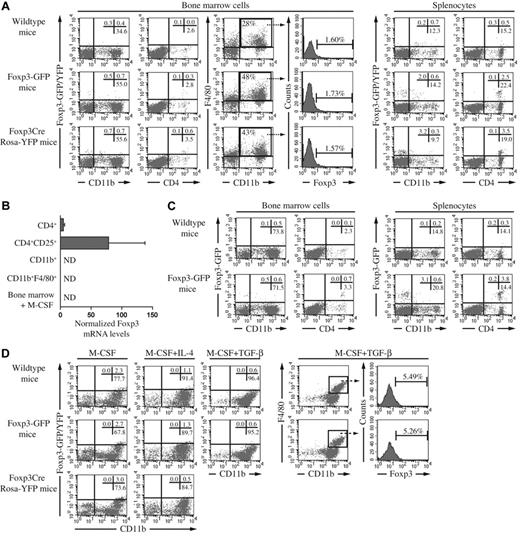
In a first set of experiments, bone marrow and spleen cells from naive Foxp3GFP, Foxp3Cre, Rosa-YFP, and wild-type mice were investigated for the expression of Foxp3 using flow cytometry (see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Our data revealed that the proportion of Foxp3+CD11b+ cells within the bone marrow did not exceed 0.7%, with background autofluorescence at 0.4% (Figure 1A), and no lineage history of Foxp3 expression could be detected when using Foxp3Cre Rosa-YFP mice. In splenocytes, the signal of GFP/YFP was restricted to CD4+ cells (Figure 1A). These findings were supported by detection of Foxp3 mRNA in CD4+ cells, but not in purified macrophages (Figure 1B). To increase the proportion and activation status of macrophages in vivo, CIA was induced in wild-type and Foxp3GFP mice. Five weeks postimmunization, bone marrow and spleen tissues of CIA mice showed increased proportions of CD11b+ cells (Figure 1C versus Figure 1A), but again these cells were found to be negative for Foxp3 expression. In contrast, CD4+ cells did express Foxp3-GFP (Figure 1C). In a third set of experiments, bone marrow cells were cultured for 3 days with M-CSF, a macrophage maturation factor, with M-CSF + IL-4, to enrich the number of alternatively activated macrophages,9 or with M-CSF + TGFβ, to provide the TGFβ signal suggested to induce Foxp3 expression in 50% of macrophages.7 Although the number of CD11b+ cells increased under the 3 conditions, the expression of Foxp3 in these cells was not different between transgenic and wild-type mice (Figure 1D). Moreover, when CD11b+F4/80+ cells were selected, no increased intensity could be observed in Foxp3-GFP animals (Figure 1D). Notably, as evident from the wild-type data, we observed higher background fluorescence in the macrophage cultures, indicative of the autofluorescence inherent to these cells.
Foxp3 expression in bone marrow cells and splenocytes of wild-type, Foxp3-GFP and Foxp3Cre Rosa-YFP mice in homeostatic and activating conditions. (A) Flow cytometric analysis was performed on bone marrow cells and splenocytes from wild-type, Foxp3GFP and Foxp3Cre RosaYFP mice (all on a C57Bl/6 background). Cells were stained with anti–CD11b-APC, anti–F4/80-PE or anti–CD4-PE. Percentages of single or double positive cells in the respective quadrants of the dot plots are indicated. Histograms show Foxp3 intensity in gated F4/80+CD11b+ cells. (B) Foxp3 mRNA levels were determined in macrophages and T cells by qRT-PCR. CD11b+ cell-enriched splenocytes were obtained by incubation with magnetic-activated cell sorting (MACS) CD11b beads and further purified after incubation with FITC-conjugated anti-CD11b antibodies and PE-conjugated anti-F4/80 antibodies and subsequent sorting by flow cytometry on a FACSVantage apparatus. CD4+ cells were obtained from lymph nodes and purified with the Mouse T cell CD4 Subset Column Kit. From this population, CD4+CD25+ Treg cells were purified by FACSVantage. The purity of the populations varied from 96% to 99%. Bone marrow cells were stimulated with 20 ng/mL M-CSF for 3 days. (C) Flow cytometric analysis was performed (as described in panel A) on bone marrow and splenocytes from wild-type and Foxp3GFP mice, 5 weeks post immunization with collagen type II in complete Freund's adjuvant containing heat-killed Mycobacterium tuberculosis. (D) Bone marrow cells of naive mice were stimulated for 3 days with M-CSF (20 ng/mL) or M-CSF and IL-4 (10 ng/mL). Bone marrow cells of immunized mice were stimulated for 3 days with M-CSF and TGF-β (1 ng/mL). Cells were stained as described in panel A. Percentages of single or double positive cells in the respective quadrants of the dot plots are indicated. Histograms show Foxp3 intensity in gated F4/80+CD11b+ cells stimulated with M-CSF and TGF-β. In this condition, Foxp3Cre Rosa-YFP mice were not included. In each of the panels, the data were obtained from a pool of 2 to 3 mice. ND indicates not detected.
Foxp3 expression in bone marrow cells and splenocytes of wild-type, Foxp3-GFP and Foxp3Cre Rosa-YFP mice in homeostatic and activating conditions. (A) Flow cytometric analysis was performed on bone marrow cells and splenocytes from wild-type, Foxp3GFP and Foxp3Cre RosaYFP mice (all on a C57Bl/6 background). Cells were stained with anti–CD11b-APC, anti–F4/80-PE or anti–CD4-PE. Percentages of single or double positive cells in the respective quadrants of the dot plots are indicated. Histograms show Foxp3 intensity in gated F4/80+CD11b+ cells. (B) Foxp3 mRNA levels were determined in macrophages and T cells by qRT-PCR. CD11b+ cell-enriched splenocytes were obtained by incubation with magnetic-activated cell sorting (MACS) CD11b beads and further purified after incubation with FITC-conjugated anti-CD11b antibodies and PE-conjugated anti-F4/80 antibodies and subsequent sorting by flow cytometry on a FACSVantage apparatus. CD4+ cells were obtained from lymph nodes and purified with the Mouse T cell CD4 Subset Column Kit. From this population, CD4+CD25+ Treg cells were purified by FACSVantage. The purity of the populations varied from 96% to 99%. Bone marrow cells were stimulated with 20 ng/mL M-CSF for 3 days. (C) Flow cytometric analysis was performed (as described in panel A) on bone marrow and splenocytes from wild-type and Foxp3GFP mice, 5 weeks post immunization with collagen type II in complete Freund's adjuvant containing heat-killed Mycobacterium tuberculosis. (D) Bone marrow cells of naive mice were stimulated for 3 days with M-CSF (20 ng/mL) or M-CSF and IL-4 (10 ng/mL). Bone marrow cells of immunized mice were stimulated for 3 days with M-CSF and TGF-β (1 ng/mL). Cells were stained as described in panel A. Percentages of single or double positive cells in the respective quadrants of the dot plots are indicated. Histograms show Foxp3 intensity in gated F4/80+CD11b+ cells stimulated with M-CSF and TGF-β. In this condition, Foxp3Cre Rosa-YFP mice were not included. In each of the panels, the data were obtained from a pool of 2 to 3 mice. ND indicates not detected.
In conclusion, we find no evidence for Foxp3 expression in macrophages, despite looking in vitro and in vivo under conditions of homeostasis and activation. Furthermore, using the fate-mapping system we find no evidence of transient historical expression in macrophages under any of these conditions. We would therefore postulate that the “Foxp3” expression reported by Manrique et al7 is an artifact because of the well-known property of macrophages as autofluorescent and “sticky” cells.10 We do not dispute the existence of an immunosuppressive population of macrophages, which because of activation status or marker expression may have a more “sticky” phenotype, but based on the experimental evidence presented here, we disagree with the conclusion that this population has genuine Foxp3 expression. In light of the repeated false positive reports of Foxp3 expression in nonT cell lineages, we would suggest the following criteria should be used in future reports of Foxp3 expression. First, a technically plausible explanation should exist for why previous surveys have not found Foxp3 expression, such as temporal restriction or investigation of an uninvestigated cell population. Second, protein expression should delineate a discrete population, present after using a negative gating strategy and absent in a Foxp3-deficient staining control. Third, sorted Foxp3+ cells should contain Foxp3 mRNA proportional to the protein expression, but should not contain CD3 mRNA. We believe that strict adherence to these 3 criteria would reduce the number of future false positive reports.
Authorship
The online version of this letter contains a data supplement.
Acknowledgments: The authors thank A. Rudensky for providing Foxp3GFP and Foxp3Cre mice.
Contribution: S.P., A.A., S.H-B., and E.S. performed the experiments; A.L. and P.M. designed the experiments; and S.P., A.A., A.L., and P.M. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Prof Dr Adrian Liston, VIB Autoimmune Genetics Laboratory; Katholieke Universiteit Leuven, Leuven, Belgium; e-mail: adrian.liston@vib.be.
References
Author notes
S.P. and A.A. contributed equally to this work.
A.L. and P.M. share equal contribution as senior author.