Abstract
Mutations in isocitrate dehydrogenase 1 (IDH1) and isocitrate dehydrogenase 2 (IDH2) occur in most grade 2 and 3 gliomas, secondary glioblastomas, and a subset of acute myelogenous leukemias but have not been detected in other tumor types. The mutations occur at specific arginine residues and result in the acquisition of a novel enzymatic activity that converts 2-oxoglutarate to D-2-hydroxyglutarate. This study reports IDH1 and IDH2 genotyping results from a set of lymphomas, which included a large set of peripheral T-cell lymphomas. IDH2 mutations were identified in approximately 20% of angioimmunoblastic T-cell lymphomas (AITLs), but not in other peripheral T-cell lymphoma entities. These results were confirmed in an independent set of AITL patients, where the IDH2 mutation rate was approximately 45%. This is the second common genetic lesion identified in AITL after TET2 and extends the number of neoplastic diseases where IDH1 and IDH2 mutations may play a role.
Introduction
Heterozygous isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) mutations at specific active site arginine residues occur in most low-grade gliomas, secondary glioblastomas, and in some acute myeloid leukemias (AMLs).1-3 These mutations alter IDH enzymatic function, resulting in the conversion of 2-oxoglutarate to the rare metabolite D-2-hydroxyglutarate (D-2-HG), which accumulates to high levels in cells and tissues.4,5 D-2-HG may act as an oncometabolite, driving tumor progression by interfering with 2-oxoglutarate–dependent enzymes that affect hypoxia signaling (prolyl hydroxylases), histone methylation, and DNA methylation (TET2).6,7 Despite extensive genotyping, IDH1 and IDH2 mutations have not been identified in significant proportions of other neoplasms.3,8,9
Peripheral T-cell lymphomas (PTCLs) are non-Hodgkin lymphomas with a diverse presentation, histology, therapy response, and outcome.10 Because this group of diseases has not been comprehensively assessed for the presence of IDH mutations, patient samples from 3 independent groups were genotyped to determine whether these mutations are present.
Methods
Patients and clinical data
Lymphoma and leukemia DNA samples from frozen tissue were provided by a multicentric T-cell lymphoma consortium (Tenomic), and the University of Hong Kong. Diagnosis was confirmed by a panel of pathologists to ensure consistent classification of PTCL according to the WHO classification.
A complementary set of DNA from 22 angioimmunoblastic T-cell lymphomas (AITLs) was provided by the University of Nebraska Medical Center (UNMC) Lymphoma Tumor Bank.11 These AITL patients were identified by histopathologic assessment and confirmed by gene expression profiling.11
As there is no standard therapy for AITL, patients received heterogeneous treatment. AITL samples with greater than 50% estimated tumor cell content were prioritized (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All patients provided informed consent in accordance with the Declaration of Helsinki, and use of patient materials and information was approved by the institutional review boards of the University of Hong Kong, the UNMC, and the member institutions of the Tenomic Consortium.
IDH1 and IDH2 genotyping
For Tenomic and University of Hong Kong patients, IDH1 R132 and IDH2 R172 and R140 genotypes were determined at the Analytical Genetics Technology Center at the University Health Network (Toronto, ON) using a Sequenom MassARRAY platform (Sequenom) as previously described.5 Positive results were confirmed by Sanger sequencing of the mutated region. In the UNMC patients, IDH1 and IDH2 genotype was determined by Sanger sequencing of all exons and subsequently by Sequenom (Genomic Core facility, UNMC). Sequenom genotyping is more sensitive and specific than Sanger sequencing, with the ability to detect a mutation in 10% of the input DNA, which is an advantage for samples with stromal contamination.12
Statistical analysis
Fisher exact text and the Wilcoxon rank-sum test were used to test for differences between IDH2wt and IDH2 mutant patients. Estimates of overall and progression-free survival were calculated using the Kaplan-Meier method and compared using the log-rank test.
Results and discussion
Lymphoma samples from the Tenomic Consortium and University of Hong Kong were genotyped to determine whether mutations were present in IDH1 at R132 and in IDH2 at R172 and R140 (Table 1). This set of samples included a large group of PTCLs. Although other studies have suggested that IDH1/IDH2 mutations are not present in lymphomas and leukemias other than AML,3,9 PTCL has not been comprehensively studied.
No mutations were observed in lymphoma subtypes, including 46 B-cell lymphomas and 66 Hodgkin lymphomas, except for AITLs, where 16 of 79 (20%) samples from the Tenomic Consortium carried an IDH2 mutation. This is the second common mutation to be identified in AITL after TET213 and makes AITL the third disease where IDH1/IDH2 mutations have been identified in a significant proportion of patients. As has been observed in glioma and AML, all mutations were heterozygous. However, the spectrum of mutations observed in AITL was different. Unlike in glioma and AML, no IDH1 mutations were identified, and the IDH2 mutations were largely confined to alterations resulting in an R172 substitution (12 R172K, 2 R172G, 1 R172T, and 1 R140G).
To further validate these results, 22 AITL patients from UNMC were genotyped (Table 1). None of the patients carried IDH1 mutations, and in 10 of 22 cases (45%) IDH2 mutations were identified, confirming the results of the Tenomic data. Although this was an independent set of patients, and mutation detection was performed using Sanger sequencing of all exons, the mutation spectrum was consistent with that of the Tenomic patients, with the IDH2 mutations detected at R172 (4 R172K, 4 R172S, 1 R172T, and 1 R172G). There were no other mutations found in IDH1 or IDH2. The higher rate of mutation in this smaller set of patients may reflect differences in patient selection defined by a gene expression signature,11 which may identify a more homogeneous group of patients whoshare the IDH2 mutation at higher frequency.
AITL is 1 of the 3 most common PTCL subtypes, along with anaplastic large cell lymphoma and PTCL not otherwise specified.14 It normally presents as a systemic disease, with polyadenopathy and a variety of immunologic abnormalities, and carries a poor prognosis.15 Based on molecular marker expression (CD4, CD10, BCL6, PD1, and CXCL13) and microarray profiling, AITL is thought to arise from follicular T-helper cells normally present in germinal centers.11,16,17 The molecular pathogenesis and underlying genetic events driving AITL are largely unknown.
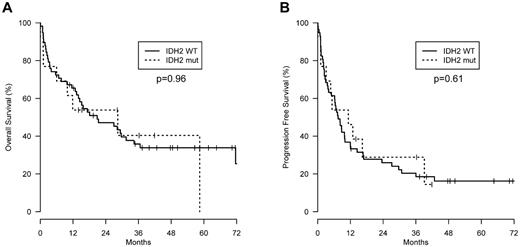
The clinical features of IDH2 wild-type and mutant AITL patients were assessed. Although all clinical parameters were not available for each patient, there were no significant differences between the 2 groups in the Tenomic patients, except that the Direct Coombs test was less frequently positive in IDH2 mutant patients (supplemental Table 1). These results were consistent with the smaller UNMC patient group (data not shown). On review, there were no pathologic differences between the groups. Furthermore, IDH2 status had no effect on progression-free or overall survival (Figure 1). This is at odds with the findings in glioma, where IDH1/IDH2 mutations predict for improved survival, but more consistent with AML, where there is no overall independent impact on outcome, although some studies show prognostic value when specific IDH mutations are combined with other prognostic markers.18 Prognostic impact may be difficult to detect in AITL, as patients are acquired from multiple centers and receive heterogeneous treatment.
Survival. (A) Overall survival and (B) progression-free survival of AITL patients with wild-type (n = 61) or mutant IDH2 (n = 16) from the Tenomic Consortium dataset. Wild-type IDH2 patients were not significantly different from IDH2 mutant patients for either parameter.
Survival. (A) Overall survival and (B) progression-free survival of AITL patients with wild-type (n = 61) or mutant IDH2 (n = 16) from the Tenomic Consortium dataset. Wild-type IDH2 patients were not significantly different from IDH2 mutant patients for either parameter.
Although the current results suggest that IDH2 may not provide prognostic information in AITL, a better understanding of the mechanisms underlying IDH1/IDH2-driven tumor progression may lead to new opportunities for AITL treatment. Future measurement of D-2-HG, the metabolite produced by these mutant enzymes, may provide a useful biomarker of disease progression and response to therapy. In addition, small-molecule inhibitors specific for the mutant IDH2 enzymes could represent important tools in the future management of AITL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Arda Shahinian for technical support and the Henri Mondor Hospital Biologic Resources platform.
This work was supported by the Canadian Institutes of Health Research, the Canadian Cancer Society, the Terry Fox Foundation, and the Leukemia & Lymphoma Society (T.W.M.); Institut National du Cancer, Program Hospitalier de Recherche Clinique, and Fondation pour la Recherche Médicale (P.G.); Institut Universitaire de France and the CITTIL program (P.B.); the Cancer Plan research Program (Belgium; L.d.L.); National Cancer Institute (grant 5U01/CA114778); National Institutes of Health (grant U01/CA84967); lymphoma SPORE (grant P50CA13641-02), and Eppley Core grant (W.-C.C. and J.I.).
National Institutes of Health
Authorship
Contribution: R.A.C., J.I., L.X., L.C.C., W.-C.C., P.B., P.G., and T.W.M. designed the research; R.A.C., J.I., C.K., L.d.L., M.P., F.L., L.X., and A.M. performed research and collected and analyzed the data; F.L., J.-P.J., and J.I. performed statistical analysis; and R.A.C. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tak W. Mak, Campbell Family Institute for Breast Cancer Research, 620 University Ave, Suite 706, Toronto, ON, Canada M5G 2C1; e-mail: tmak@uhnres.utoronto.ca.
References
Author notes
R.A.C., J.I., and F.L. contributed equally to this study.